Abstract
Although neuroimaging is essential for prompt and proper management of traumatic brain injury (TBI), there is a regrettable and acute lack of robust methods for the visualization and assessment of TBI pathophysiology, especially for of the purpose of improving clinical outcome metrics. Until now, the application of automatic segmentation algorithms to TBI in a clinical setting has remained an elusive goal because existing methods have, for the most part, been insufficiently robust to faithfully capture TBI-related changes in brain anatomy. This article introduces and illustrates the combined use of multimodal TBI segmentation and time point comparison using 3D Slicer, a widely-used software environment whose TBI data processing solutions are openly available. For three representative TBI cases, semi-automatic tissue classification and 3D model generation are performed to perform intra-patient time point comparison of TBI using multimodal volumetrics and clinical atrophy measures. Identification and quantitative assessment of extra- and intra-cortical bleeding, lesions, edema, and diffuse axonal injury are demonstrated. The proposed tools allow cross-correlation of multimodal metrics from structural imaging (e.g., structural volume, atrophy measurements) with clinical outcome variables and other potential factors predictive of recovery. In addition, the workflows described are suitable for TBI clinical practice and patient monitoring, particularly for assessing damage extent and for the measurement of neuroanatomical change over time. With knowledge of general location, extent, and degree of change, such metrics can be associated with clinical measures and subsequently used to suggest viable treatment options.
Key words: magnetic resonance imaging, segmentation, TBI, visualization
Introduction
With an estimated 1.7 million people in the United States sustaining a traumatic brain injury (TBI) every year (Faul et al., 2010), the magnitude of this medical concern to the United States cannot be overstated (Chen and D'Esposito, 2010). Every year, TBI cases are associated with 1.2 million emergency room visits and over 50,000 deaths (Langlois et al., 2006). Interest in and public awareness of TBI-caused diffuse axonal injury (DAI) has surged with the publication of studies suggesting that DAI in professional and amateur athletes may have both acute and chronic effects on neurocognitive function (De Beaumont et al., 2009). Moreover, the large number of recent TBI cases in soldiers returning from military conflicts has created a significant clinical challenge for United States Veterans Administration hospitals (Taber et al., 2006) and has highlighted the critical need for improvement in TBI care and treatment.
Prompt and proper management of TBI sequelae can significantly alter their course, reduce mortality and morbidity, reduce lengths of hospital stay, and decrease health care costs (Watts et al., 2004). Neuroimaging of TBI is therefore vital for surgical planning, by providing important information for anatomical localization and surgical navigation, as well as for guiding decisions regarding the aggressiveness of TBI treatment (Chesnut, 1998). Moreover, because TBI often results in characteristic impairments dependent on the area of involvement (Warner et al., 2010), lesion analysis can assist in the localization of cognitive processes and in the attempt to obtain novel information on the relationship between brain anatomy and behavior (Bates et al., 2003). For all these reasons, reliable and precise methods of TBI assessment can play an essential role during both acute and chronic therapy for this condition.
Whereas neuroimaging is often critical for proper TBI clinical care, there is a regrettable and acute lack of robust methods for the exploration, visualization, and quantitative assessment of TBI-related anatomical insults and pathophysiology. Starting 2–3 days after the acute injury, MRI is generally considered to be superior to CT for the purpose of TBI assessment and analysis. Whereas CT is better at detecting the pathology of bones and certain bleeds, MRI can identify bleeds more easily and more accurately as blood composition gradually changes following TBI (Dubroff and Newberg, 2008). In this context, the application of automatic MRI segmentation algorithms to the clinical investigation of TBI cases remains an elusive goal because many existing methods are insufficiently robust to accurately capture TBI-related changes in brain anatomy. Therefore, because the state of the art in quantitative MRI analysis of TBI often involves labor-intensive manual tissue classification, the clinician's ability to generate robust and accurate three-dimensional (3D) models of neuropathology for TBI diagnosis and treatment remains extremely limited. Consequently, despite recent progress in the development of robust image analysis tools, it remains difficult to quantify TBI-related brain insults either uni- or multi-modally, particularly for the purpose of improving clinical outcome metrics.
To address the need for clinician-friendly TBI analysis tools, we here propose and illustrate the combined the use of multimodal, semi-automatic TBI analysis methods within 3D Slicer, a freely available software environment for the segmentation, registration, visualization, and quantification of MR images. In contrast with many other segmentation environments for TBI that have appeared in the literature (Ding et al., 2008), the 3D Slicer environment is both freely available as well as widely used by clinicians, scientists, and engineers. To showcase the ability to perform quantitative time point comparison and assessment of TBI in 3D Slicer, we present three cases of semi-automatic TBI volume segmentation and 3D brain model generation, while also highlighting the added clinical insight that this workflow can offer.
Methods
Research participants and MRI acquisition
Anonymized neuroimaging data from three representative TBI subjects were acquired at the Brain Injury Research Center of the UCLA Geffen School of Medicine. The study and data acquisition protocol were designed and performed in accordance with the Declaration of Helsinki and were approved by the UCLA Institutional Review Board. Signed informed consent was obtained from each patient or from their legally authorized representative before any procedure was performed. In Table 1, the age, sex, and types of MR scans acquired from each patient are specified, in addition to the number of days after injury when the acute and chronic scan sessions took place. In the case of the first two subjects, the sequences being used are representative of MR acquisition protocols that are appropriate for patients requiring critical care. In the case of patient 3, we illustrate the use of an extended MR scan protocol involving 12 distinct sequence types. This protocol requires significantly more acquisition time and can be envisioned as applicable for TBI patients in highly stable condition and/or who can withstand remaining motionless in the MR scanner for the extended amount of time required for the acquisition of images. Both protocols demonstrate the ability of our segmentation environment to accommodate, on the one hand, the use of a limited number of scan types and, on the other hand, the additional amount of clinically relevant information being supplied by a more enhanced protocol. Throughout the article, the set of 5 sequences that were used for the imaging of the first two patients is referred to as the “standard” protocol, whereas the set of 12 sequences used in the third patient is referred to as the “extended” protocol.
Table 1.
Case Study Subject Data Specifying the Age, Sex, Cause of Injury, Dates of Scanning Sessions as well as MR Modalities Acquired from Each TBI Study Case
| Patient | Age | Sex | Cause of injury | Day of acute scan | Day of chronic scan | T1 pre-contrast | T2 TSE | FLAIR | GRE T2 | SWI | Perfusion | T1 post-contrast | OEF EP SE | DTI | DWI 1 | DWI 2 | mIP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 45 | M | blunt trauma | 2 | 176 | √ | √ | √ | √ | √ | |||||||
| 2 | 31 | M | gun shot | 2 | 201 | √ | √ | √ | √ | √ | |||||||
| 3 | 33 | M | blunt trauma | 3 | 238 | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ |
Scanning session dates are specified as the number of days after acute injury when the acute and chronic MR volumes were acquired, respectively. Check marks indicate use of each MR scan type (see text for clarification).
TSE, turbo spin echo; FLAIR, fluid-attenuated inversion recovery; GRE, gradient-recalled echo; SWI, susceptibility-weighted imaging; OEF, Oxygen extraction factor; EP, echo-planar; SE, spin echo; DTI, diffusion tensor imaging; DWI, diffusion weighted imaging; mIP, maximum intensity projection.
MR volumes were acquired at 3 T from each subject using a Siemens Trio TIM scanner (Siemens AG, Erlangen, Germany). To assess the time evolution of TBI between the acute and the chronic stage, scanning sessions were held both several days (acute baseline) as well as several months (chronic follow-up) after the traumatic injury event. To eliminate the effect of different scanner parameters during each scanning session, every subject was scanned using the same scanner for both acute and chronic time points. The MP-RAGE sequence (Mugler and Brookeman, 1990) was used to acquire T1-weighted images. In addition, MR data were also acquired using fluid-attenuated inversion recovery (FLAIR) (De Coene et al., 1992), T2-weighted turbo spin echo (TSE), also known as fast spin echo (FSE), (Jones et al., 1992), gradient-recalled echo (GRE) T2-weighted images, as well as susceptibility-weighted imaging (SWI) (Sehgal et al., 2005). In the case of the enhanced acquisition protocol for Patient 3, seven additional scan sequences were also used, including diffusion weighted imaging (DWI), diffusion tensor imaging (DTI), and maximum intensity projection (mIP). A complete list of these sequences is provided for each patient in Table 1, whereas Table 2 specifies standard MR parameters associated with each sequence.
Table 2.
Standard MR Acquisition Parameters Associated with Each Sequence Type Specified in Table 1
| |
parameter |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sequence | TR [ms] | TE [ms] | TI [ms] | FA [deg] | ETL | Thickness [mm] | Phase FOV [%] | Sampling [%] | Acquisition type | Matrix |
| pre-contrast T1 | 1900 | 3.52 | 900 | 9 | 1 | 1 | 100 | 100 | 3D | 256×256 |
| T2 TSE | 3330 | 89 | 120 | 18 | 5 | 100 | 75 | 2D | 512×512 | |
| FLAIR | 8000 | 70 | 2375 | 130 | 16 | 3 | 75 | 75 | 2D | 384×512 |
| GRE T2 | 1500 | 7 | 20 | 1 | 3 | 75 | 80 | 2D | 384×512 | |
| SWI | 27 | 20 | 15 | 1 | 1.5 | 75 | 95 | 3D | 192×256 | |
| Perfusion | 2000 | 32 | 90 | 1 | 6 | 100 | 100 | 2D | 128×128 | |
| post-contrast T1 | 1900 | 3.52 | 900 | 9 | 1 | 1 | 100 | 100 | 3D | 256×256 |
| OEF EP SE | 10000 | 50 | 90 | 1 | 3 | 100 | 100 | 2D | 128×128 | |
| DTI | 8000 | 95 | 90 | 1 | 3 | 100 | 100 | 2D | 128×128 | |
| DWI 1 | 4000 | 80 | 90 | 1 | 6 | 100 | 100 | 2D | 130×130 | |
| DWI 2 | 4000 | 80 | 90 | 1 | 6 | 100 | 100 | 2D | 130×130 | |
| mIP | 27 | 20 | 15 | 1 | 12 | 75 | 95 | 3D | 256×192 | |
TR, repetition time; TE, echo time; TI, inversion time; FA, flip angle; ETL, echo train length; FOV, field of view; TSE, turbo spin echo; FLAIR, fluid-attenuated inversion recovery; GRE, gradient recalled echo; SWI, susceptibility-weighted imaging; OEF, oxygen extraction fraction; EP, echo-planar; SE, spin echo; DTI, diffusion tensor imaging; DWI, diffusion weighted imaging; mIP, maximum intensity projection.
Identification of pathology
TBI comprises a variety of cerebral lesion types, including hematoma, subarachnoid hemorrhage, contusion, and DAI. TBI edema, in particular, can further be characterized by its extent and distribution ranging from perifocal (regional) to diffuse (generalized) (Unterberg et al., 2004). In this study, non-hemorrhagic lesions on MRI scanning were coded as hyperintensities on FLAIR images. By using a long inversion time (TI), the FLAIR sequence achieves high contrast between lesions and healthy tissue as well as cerebrospinal fluid (CSF). In this type of neuroimaging, lesions appear as hyperintense regions in the white matter (WM), surrounded by normal tissue of lower, more uniform intensity (Itti et al., 2001). On the other hand, T2-weighted GRE imaging is excellent for the detection of acute hemorrhagic lesions, and SWI is much more sensitive than conventional T2-GRE in detecting hemorrhagic DAI. By comparison, the sensitivity of DWI in identifying DAI lesions is similar to that of FLAIR, and inferior to that of T2-GRE for detecting hemorrhage (Huisman, 2003). Although GRE images do reveal some micro-hemorrhages, this sequence is poor at resolving between gray matter (GM) and lesions, on the one hand and WM and lesions, on the other hand, at least partly because GM lesions have higher relaxation times than those of normal WM. For all these reasons, brain lesions adjacent to CSF were segmented from volumes acquired using FLAIR, and the quality of the segmentations obtained was confirmed using GRE imaging. TSE T2-weighted volumes were also used to confirm lesion characterization, and DWI volumes were examined, where available, to additionally distinguish vasogenic (increased diffusion) from cytotoxic (restricted diffusion) edema (Huisman et al., 2003; Warach et al., 1995). Non-hemorrhagic shearing lesions were defined as hyperintense lesions that were visible primarily on T2-weighted or FLAIR images. This method of identification was adopted because some shearing lesions often do not show substantial amounts of hemorrhage but are primarily visible as areas of hyperintensity on FLAIR, T2-weighted, and GRE images.
The appearance of hemorrhages in MR imaging is highly variable across sequences (Greenberg et al., 2009), as it depends upon a number of variables, both intrinsic (hemoglobin oxygenation, erythrocyte integrity) and extrinsic (e.g., scanner field strength, receiver bandwidth, T1 or T2 weighing). Because SWI takes into account MR signal phase information, regional magnetic field alterations caused by iron concentration gradients in the brain are easier to capture with this technique. As a result, SWI sequences are very useful in TBI imaging because susceptibility effects make micro-hemorrhages more obvious in SWI compared to other paradigms. Because SWI is therefore generally superior to GRE and T2-weighted imaging in detecting hemorrhagic lesions (Tong et al., 2003), volumes acquired using this former modality were used to identify micro-hemorrhages that were poorly detectable or undetectable using other sequences. Specifically, following the previous guidelines of Tong and associates (2003), hemorrhagic lesions were defined as hypointense foci that were not compatible with vascular, bone, or artifactual structures on conventional GRE images. Whenever foci etiology was doubtful, the lesions in question were not considered to be hemorrhagic.
TBI volume segmentation is greatly improved with the availability of MR volumes acquired using various sequence types that are suitable for the identification of pathology. As already pointed out, although FLAIR imaging is suitable for the identification of CSF-perfused injuries, its ability to detect hemorrhage is greatly superseded in GRE T2 imaging and especially in SWI, because the latter has improved abilities to detect micro-bleeds. In the present analysis, it was found that non-hemorrhagic lesions were best delineated from FLAIR, and that the precise identification of hemorrhagic lesions required the inclusion of GRE T2 and SWI image channels in the segmentation. In the absence of these last two sequence types, the volume of non-hemorrhagic pathology was found to inappropriately include that of hemorrhagic lesions, to the obvious detriment of segmentation quality. On the other hand, inclusion of both GRE T2 and SWI but not of FLAIR caused non-hemorrhagic edema to be misclassified as either healthy-looking GM or WM, which is reasonable to expect given SWI's excellent sensitivity to the presence of iron compounds, although not to that of CSF perfusion of affected tissues. Finally, for the cases examined here, exclusion of SWI resulted in the inability to identify micro-bleeds, which points to the important role of this sequence type in identifying the locations of small focal injuries.
Image Processing
Because TBI involves injury to both neuronal cell bodies and axonal processes, global atrophy of GM and WM are both common in this condition. Partly for this reason, reliable segmentation of WM and GM lesions is necessary for accurate quantitative analysis of TBI lesions. Additionally, accurate quantification of total lesion volume (lesion load) is essential for within-subject comparison of TBI volumes acquired at different time points. To address these clinical needs, we used the semi-automatic segmentation tools available in 3D Slicer software, including the Atlas Based Classification (ABC) and Expectation Maximization (EM) segmenters, which have been described elsewhere (Pohl et al., 2007; Prastawa and Gerig, 2008; Zoellei et al., 2007) and used widely to detect pathology in MR scans (Prastawa et al., 2009). Briefly, the ABC algorithm can perform multimodal registration of MR images (Maes et al., 1997), tissue classification of the brain (Van Leemput et al., 1999), as well as lesion segmentation based on outlier detection (Prastawa et al., 2003, 2004; Prastawa and Gerig, 2008). The EM Segmenter, on the other hand, is an algorithm guided by prior information represented algorithmically within a tree structure to estimate optimal segmentation via a conventional classifier. In this latter case, the tree mirrors the hierarchy of anatomical structures and the sub-trees correspond to limited segmentation problems, as described by Pohl and associates (2007).
As opposed to other specialized segmenters to which access is often restricted from outside users, both ABC and EM segmenters are freely available as segmentation modules in 3D Slicer. Both methods are automatic and their executions require minimal user supervision. In addition, the ABC segmenter possesses the ability to perform co-registration of an arbitrary number of MR volumes acquired using various sequences. This makes it highly suitable to the present multimodal TBI imaging paradigms, where as many as 12 distinct sequence types are used. Before segmentation, all image volumes were co-registered to the pre-contrast MP-RAGE T1-weighted volume acquired during the acute baseline scanning session. Intensity normalization within and between scans was performed within 3D Slicer. Bias field correction was applied using a fourth-order polynomial model. Because the anatomy of severe TBI often diverges from healthy anatomy to a very large extent, segmentation errors can frequently occur, especially in severe TBI cases. To address this shortcoming, manual review in 3D Slicer by an operator with experience in the recognition of neurotrauma was performed with the purpose of editing and correcting segmentation errors. The guidelines of Filippi and associates (1998) for editing and segmentation were followed. Nevertheless, when assessing the extent of segmentation error, it is fair to note that, although often considered to be the gold standard, even manual outlining resulted in both moderate (3–10%) intra-observer error as well as larger inter-operator variabilities (Raff et al., 2000). In conclusion, it is reasonable to expect the presence of subtle segmentation inaccuracies even in the context of our rigorous segmentation paradigm.
Quantitative metrics
Tissue-type segmentation was used to calculate the total volume of three selected structure types (ventricular system, non-hemorrhagic lesions, and hemorrhagic lesions). The volume change was computed as the ratio of the difference in volume between the follow-up and acute baseline time points, to the volume at the latter time point. In addition to these measures, we also report five measures of atrophy, namely the bifrontal index (Hahn and Schapiro, 1976), the bicaudate index (Earnest et al., 1979), Evan's index (Synek et al., 1976), the ventricular index (Gyldensted, 1977) and Huckman's index (Huckman et al., 1975).
Let the bifrontal horn distance be the maximum width of the anterior horns of the lateral ventricles (HLV). Then the bifrontal index is the ratio of the maximum HLV width to the inner skull diameter at HLV level. The bicaudate index is defined as ratio of the minimum width of the lateral ventricles (MLV) to the width of the inner skull at that level. Evan's index is the ratio of the HLV to the maximum inner skull diameter (MISD). Finally, the ventricular index is equal to MLV/HLV whereas Huckman's index is the sum of the MLV and HLV measures.
Results
Because neurotrauma often exhibits multiple, overlapping forms of focal and/or diffuse injury, insight from TBI imaging can be enhanced through the use of multimodal techniques. In what follows, the results of each segmentation and 3D model generation are reviewed and the procedures used to perform tissue classification are illustrated. For each patient, we provide six canonical views of ventricles and non-hemorrhagic and hemorrhagic lesions, thus allowing the reader to fully appreciate the 3D nature of TBI as well as the abilities of 3D Slicer to dynamically render and capture the complexities of severe brain pathology.
Case 1
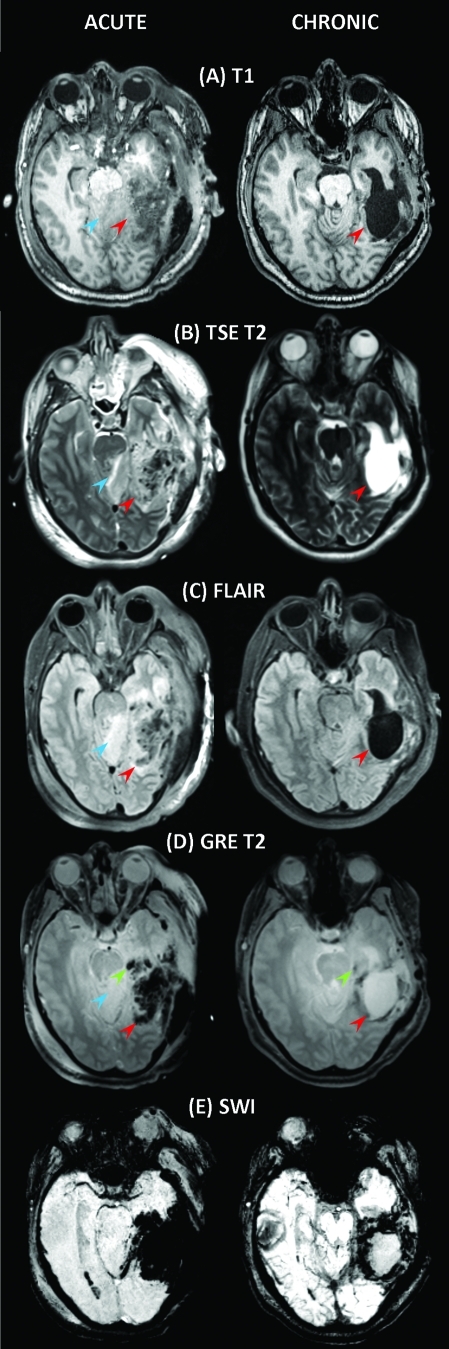
As Figure 1 reveals, Patient 1 exhibits a large deep-brain injury that is hyperintense in the FLAIR image (Fig. 1C) because of perfusion by CSF from the ventricular system. Contralaterally with respect to this injury, a smaller insult in the deep WM is also apparent in T1-weighted, FLAIR and GRE images (Fig. 1A, C, and D, respectively). The posterior portion of the left ventricle seems larger than the corresponding part of the right ventricle, possibly because of local WM inflammation. Additionally, FLAIR seems to reveal some DAI in the area surrounding the primary injury (yellow arrows), whereas GRE demonstrates the presence of hemorrhage both adjacent to and remote from the primary insult (Fig. 1 D, green arrows). Therefore, the inclusion of a GRE sequence is particularly useful in this case because DAI can be visualized indirectly through shear hemorrhages caused by blood vessel lesions (Scheid et al., 2003). Figure 1 suggests that the ability to characterize TBI is greatly enhanced by the use of SWI. As already quantified in detail by other authors (Tong et al., 2003, 2008) and illustrated in Figure 1E, this technique is very sensitive in the detection of extravascular blood products, which is illustrated by SWI's identification of significant hemorrhage surrounding the primary injury, as well as diffusely throughout the brain (Fig. 1E). A 3D animation of the segmentation for this patient is available in the Supplementary Material (see online supplementary material at http://www.liebertonline.com).
FIG. 1.
Sample MR images acquired from Patient 1 using the standard protocol. The first and second columns contain images acquired at acute baseline and chronic follow-up, respectively. Shown from top to bottom are T1-weighted MP-RAGE images (A), T2-weighted TSE (B), fluid-attenuated inversion recovery (FLAIR) (C), T2-weighted gradient-recalled echo (GRE) (D), and susceptibility-weighted imaging (SWI) (E). Arrows are color coded to represent the following: red, primary lesion(s); blue,smaller lesion(s); green, micro-hemorrhage(s); yellow, diffuse axonal injury (DAI). Note that only representative (i.e., not all) injuries of each type are indicated by arrows. Color image is available online at www.liebertonline.com/neu
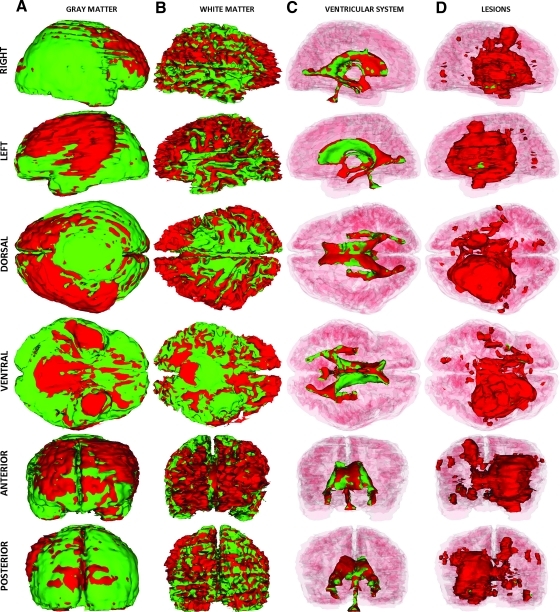
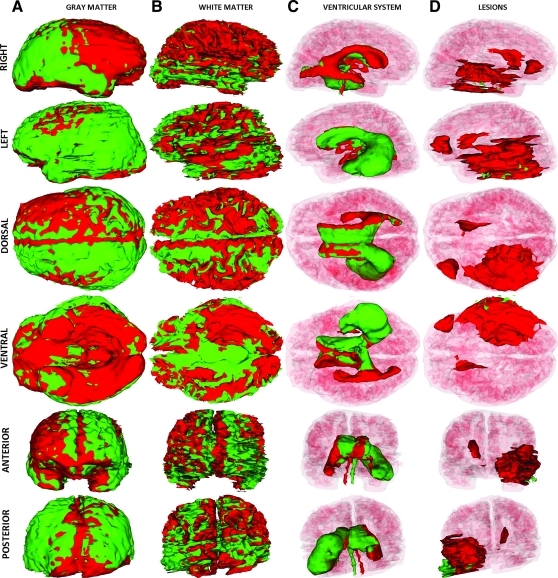
In Figure 2, the results of the TBI segmentation for Patient 1 are shown. Each row displays one of six canonical views (left, right, dorsal, ventral, anterior, and posterior) of the MR-segmented brain, with every column corresponding to one particular structure type (ventricles, non-hemorrhagic lesions, hemorrhagic lesions, and the full model). In each image, extracortical lesions, GM, and WM are transparent, whereas the ventricular system is additionally transparent in rows 2–5 to facilitate visualizing the relative position of each structure type with respect to other cortical landmarks. Figure 2A shows 3D views of the ventricular system, with arrows drawing attention to anatomical changes caused by pathology. As previously posited based on Figure 1, this volumetric analysis reflects the large extent of the primary injury, as well as the significant amount of bleeding present in the WM. The posterior part of the left ventricle is found to have been slightly enlarged as a consequence of the lesion compressing its anterior portion (Fig. 2A). By comparison, the posterior part of the right ventricle appears thinner, although its anterior portion is larger because of the primary insult being located contralaterally with respect to the latter (Fig. 2B and C). Figure 2B and C also illustrate the large volume of the primary injury, which covers a large spatial extent mainly in the left hemisphere and extends from the inferior to the superior extremity of the brain. In Figure 2D, standard views of the full 3D segmentation model are shown in which all structures are visible, with the exception of the WM and GM models, which are omitted for visual clarity. The 3D model of subdural edema is shown as transparent (green color) so as not to obstruct the view of the brain. The full model perspective has the advantage of allowing one to visualize and summarize all forms of TBI-related pathology as displayed in the other columns.
FIG. 2.
Three-dimensional models of TBI anatomy for Patient 1 (acute baseline time point), as generated in 3D Slicer. Each row displays one of six canonical views of the brain, whereas each column corresponds to a structure type (ventricles, non-hemorrhagic lesions, hemorrhagic lesions, and full model). In (A), ventricles are shown as opaque, whereas extracerebral injuries, gray matter (GM) and white matter (WM) volumes are shown as transparent to facilitate visualization of each structure in relationship to cortical landmarks. In (B–D), ventricles are also shown as transparent for similar reasons. Arrows indicate TBI-related pathology of interest (see text for elaboration). Color image is available online at www.liebertonline.com/neu
Figure 3 displays the segmentations of volumes acquired from Patient 1 during the chronic follow-up session. Although the ventricular system appears to exhibit improved bilateral symmetry (Fig. 3A) and the combined volume of bleeds as extracted from GRE and SWI imaging appears to be much smaller than in the acute scan (Fig. 3C), the latter cannot be stated regarding the volume of the scar caused by the primary injury (Fig. 3B). In fact, the volume of the lesion does not appear to have decreased appreciably at follow-up. This is confirmed in Figure 4, in which a visual time point analysis of the volumes associated with each structure is performed. For the purpose of the latter, all lesions in the acute baseline volume (whether hemorrhagic or non-hemorrhagic) are displayed jointly in red, whereas the lesions and bleeds at follow-up are shown in green. This avoids the highly problematic task of differentiating lesions in the follow-up model according to their provenience (i.e., from hemorrhagic or non-hemorrhagic regions in the acute baseline model), while still allowing one to perform a visual and quantitative comparison of the two time points.
FIG. 3.
As in Figure 2, for the chronic follow-up time point. Color image is available online at www.liebertonline.com/neu
FIG. 4.
Time point comparison of TBI in Patient 1 using 3D Slicer-generated volumes displayed in red (for the acute baseline time point), or green (for the chronic follow-up time point). As in Figures 2 and 3, each row displays one of six canonical views of the brain, with white matter (WM) and gray matter (GM) transparent in C and D to facilitate visualization of each structure in relationship to cortical landmarks. Color image is available online at www.liebertonline.com/neu
In Figure 4, results of the time point comparison for Patient 1 are displayed. The left view of the GM volume reveals important decrease in inflammation at the follow-up time point, particularly over dorsolateral frontal, prefrontal, parietal, and superior temporal cortex. Comparison of the acute and chronic WM volumes, on the other hand, suggests substantial decrease in inflammation over the entire frontal and prefrontal cortex, as well as more diffusely over the other cortical lobes. In this subject, comparison of the volumes associated with lesions portrays the acute baseline lesions as encasing those imaged at the chronic follow-up time point, which again suggests substantial decrease in injury extent. The longitudinal analysis in Figure 4 suggests that Patient 1 exhibits only a small longitudinal change in the volume of the ventricular system. This is confirmed by the quantitative analysis reported in Table 3, where it is seen that there is only a 1.3% increase in ventricular volume between the acute and chronic time points. By contrast, in agreement with Figure 4, both hemorrhagic and non-hemorrhagic lesions are found to decrease in volume substantially, that is, by over 76%. Among the metrics used for quantification of atrophy, Evan's index registers the largest percentage change (11.42%) from the acute to the chronic time point. (See Table 4 for numerical values of this and other measures in each subject.)
Table 3.
Longitudinal Analysis of Three Selected Volumetric Structures (Ventricular System, Non-Hemorrhagic Lesions, and Hemorrhagic Lesions) in Three TBI Patients
| |
|
Volume [cm3] |
|
|
|---|---|---|---|---|
| Structure | Patient | Acute | Chronic | Δ [%] |
| Brain | 1 | 1142.0 | 1009.3 | −11.6 |
| 2 | 1098.5 | 978.7 | −10.9 | |
| 3 | 1111.2 | 1018.1 | −8.4 | |
| Ventricular system | 1 | 29.7 | 30.1 | 1.3 |
| 2 | 45.8 | 95.2 | 107.6 | |
| 3 | 20.5 | 31.4 | 53.2 | |
| Non-Hemorrhagic lesions | 1 | 84.7 | 19.9 | −76.5 |
| 2 | 40.7 | 14.8 | −63.6 | |
| 3 | 54.7 | 0.9 | −82.7 | |
| Hemorrhagic lesions | 1 | 73.0 | 17.4 | −76.2 |
| 2 | 51.3 | <0.1 | −99.7 | |
| 3 | 22.5 | 0.3 | −88.1 | |
Values are reported for both acute and chronic segmented volumes, with the change Δ as a percentage being computed as (volume at the chronic time point—volume at the acute time point)×100/volume at the acute time point.
Table 4.
Indices of Atrophy Computed for Three Sample Subjects
| |
|
Time point |
|
|
|
|
|---|---|---|---|---|---|---|
| Metric | Patient | Acute | Chronic | Δ [%] | μ (Δ) [%] | σ (Δ) [%] |
| HLV [cm] | 1 | 2.323 | 2.541 | 9.63 | 6.63 | 4.97 |
| 2 | 2.965 | 2.985 | 0.89 | |||
| 3 | 3.077 | 3.364 | 9.35 | |||
| MLV [cm] | 1 | 0.541 | 0.554 | 2.41 | 19.32 | 18.67 |
| 2 | 0.845 | 0.983 | 16.19 | |||
| 3 | 0.765 | 1.067 | 39.36 | |||
| MISD [cm] | 1 | 14.546 | 14.313 | −1.60 | 0.44 | 2.68 |
| 2 | 14.068 | 13.960 | −0.55 | |||
| 3 | 13.209 | 13.669 | 3.48 | |||
| bifrontal index | 1 | 0.205 | 0.217 | 5.76 | 6.23 | 0.40 |
| 2 | 0.250 | 0.266 | 6.40 | |||
| 3 | 0.251 | 0.267 | 6.51 | |||
| bicaudate index | 1 | 0.047 | 0.046 | −3.72 | 25.44 | 28.47 |
| 2 | 0.068 | 0.086 | 26.89 | |||
| 3 | 0.059 | 0.091 | 53.17 | |||
| Evan's index | 1 | 0.159 | 0.177 | 11.42 | 6.18 | 5.00 |
| 2 | 0.211 | 0.214 | 1.46 | |||
| 3 | 0.233 | 0.246 | 5.67 | |||
| ventricular index | 1 | 0.233 | 0.218 | −6.58 | 12.01 | 17.23 |
| 2 | 0.286 | 0.329 | 15.16 | |||
| 3 | 0.249 | 0.317 | 27.44 | |||
| Huckman's index | 1 | 28.587 | 30.949 | 8.27 | 9.30 | 5.59 |
| 2 | 38.054 | 39.688 | 4.30 | |||
| 3 | 38.430 | 44.321 | 15.33 | |||
In addition to the numerical values of each parameter, also listed is the change Δ as a percentage, computed as (value at chronic time point – value at acute time point)×100/value at acute time point. The mean μ and standard deviation σ of the percentage change Δ computed over subjects are also listed.
HLV, maximum width of the anterior horns of the lateral ventricles; MLV, minimum width of the lateral ventricles; MISD, maximum inner skull diameter.
Case 2
Whereas Patient 1 illustrates the effects of a large WM lesion caused by a blunt, closed-head trauma, the imaging of Patient 2 illustrates the damage to the brain caused by a gunshot wound. In this latter case, all imaging modalities reveal extensive—both focal and diffuse—injuries in the left hemisphere, with particularly obvious damage to the temporal lobe, both laterally and medially (Fig. 5). In addition, all modalities reveal injuries to the cerebellum and brainstem, with FLAIR showing large portions of these structures being perfused by CSF (Fig. 5C). The medial aspect of the right temporal lobe as imaged using FLAIR and GRE T2 suggests the existence of an insult in this structure as well. In the acute scan slices displayed, the size and extent of the primary injury make the left ventricle essentially indiscernible. All modalities hint at the presence of extensive extracortical insults, whereas GRE T2-weighted and SWI images demonstrate the presence of significant hemorrhage (Fig. 5D–E). The images acquired during the chronic scan session illustrate significant left ventricular hypertrophy, presumably because of WM loss, whereas GM loss is also obvious, especially as seen on the lateral aspect of the left temporal lobe. A 3D animation of the segmentation for this patient is available in the Supplementary Material (see online supplementary material at http://www.liebertonline.com).
FIG. 5.
As in Figure 1, for Patient 2. Color image is available online at www.liebertonline.com/neu
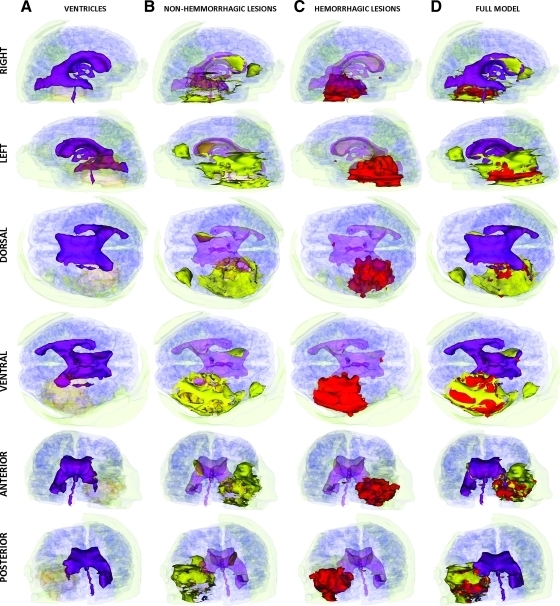
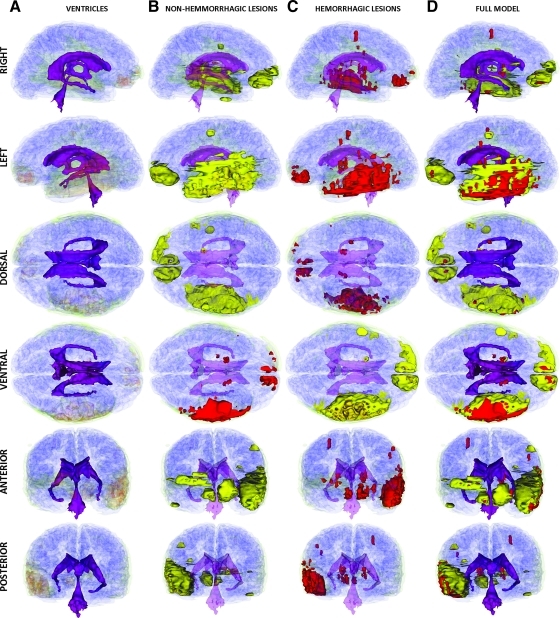
In Patient 2, the segmentations of the MR volumes acquired during the acute TBI scan (Fig. 6) reveal the presence of substantial loss of volume in the left ventricle as well as notable extracortical insults. Both hemorrhagic and non-hemorrhagic cerebral lesions are found to cover large portions of the temporal lobe, with some injuries also being present dorsofrontally in the left hemisphere as well as in the periventricular region of the anteromedial right hemisphere. The segmentation of the MR data set acquired at follow-up (Fig. 7) confirms that Patient 2 exhibits significant enlargement of the lateral ventricle ipsilateral to the primary insult, with large portions of the ventrolateral temporal lobe exhibiting low-density, CSF-perfused tissue (Fig. 7B).
FIG. 6.
As in Figure 2, for Patient 2 at the acute baseline time point. Color image is available online at www.liebertonline.com/neu
FIG. 7.
As in Figure 2, for Patient 2 at the chronic follow-up time point. Color image is available online at www.liebertonline.com/neu
Time point comparison of the GM volume in this subject hints to significant lateral shift of the longitudinal fissure (Fig. 8A, dorsal view), substantial decrease in frontal lobe GM volume as a consequence of decreased inflammation (Fig. 8A, right and left views), and large decrease in left temporal lobe lesion size (Fig. 8A, ventral view) amounting to a non-hemorrhagic lesion volume that is 63.6% smaller than that at the acute baseline time point (Table 3). Comparison of the ventricular system between acute baseline and chronic follow-up (Fig. 8C) confirms the significant increase in volume of the left ventricle and decrease in volume of the right ventricle. Whereas the former is probably the result of the replacement of injured WM by CSF, the latter is possibly caused at least in part, by the decrease in intracranial pressure between the two time points. As summarized in Table 3, the increase is over 50 cm3, which amounts to a 107% increase with respect to the ventricular volume at the acute time point. The obvious lateral shift in the position of the fourth ventricle (Fig. 8C, anterior and posterior views) appears to confirm the noteworthy finding that the midline has been shifted to the left during the chronic period. This impression is strengthened by the fact that lesion volume is smaller at follow-up compared to baseline (Fig. 8D). In Patient 2, the bicaudate and ventricular indices have the largest percentage changes at follow-up compared to baseline (26.89% and 15.15%, respectively).
FIG. 8.
As in Figure 4, for Patient 2. Color image is available online at www.liebertonline.com/neu
Case 3
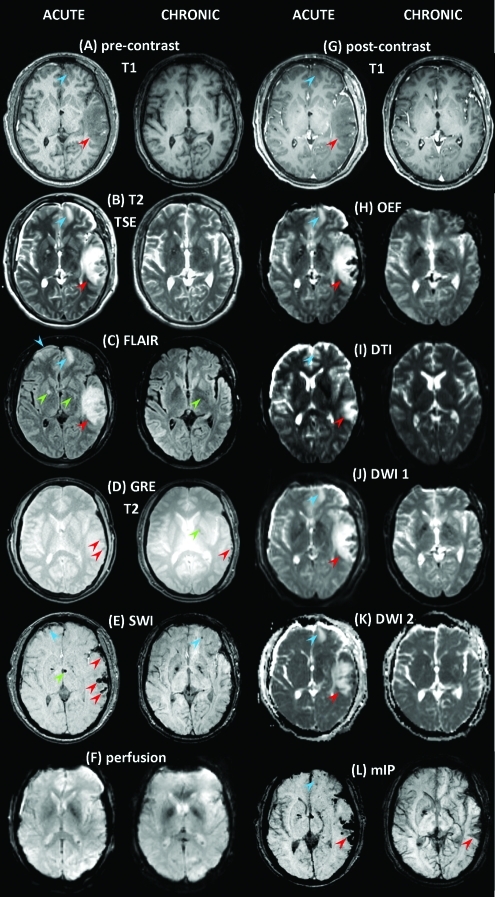
Shown in Figure 9 are MR images acquired from Patient 3. As in the previous patient, one can note an extensive primary TBI covering a significant portion of the left temporal lobe, including the inferior, middle, and superior temporal gyri and sulci. Smaller insults include lesions to the frontopolar region of both hemispheres, as well as bilateral subdural edema over substantial lateral portions of the frontal, parietal, temporal, and occipital lobes. Insults are visible as hypointensities in T1-weighted MP-RAGE images (Fig. 9A) and as hyperintensities in T2-weighted TSE images (Fig. 9B). FLAIR images (Fig. 9C) reveal hyperintense, CSF-perfused lesions located bilaterally in the frontal lobe, as well as a primary insult in the left temporal lobe. Focal and hyperintense periventricular lesions are also visible throughout the volume. Figure 9D presents T2-weighted GRE images, with hypointensities present in both the acute baseline and follow-up volumes. In the present case, this type of imaging reveals hypointense, CSF-perfused lesions (Fig. 9D) as well as some chronic injuries in proximity to the left ventricle. MR volumes available from the extended protocol (Fig. 9G–L) provide confirmation of these findings, with additional identification of micro-bleeds being made possible from the mIP angiography volume, which allows one to identify the existence of numerous additional micro-bleeds throughout the brain (Fig. 9L). Oxygen extraction factor (OEF) Fig. 9H), DTI images (Fig. 9I), and DWI-based apparent diffusion coefficient (ADC) maps (Fig. 9J and K) additionally confirm the existence and extent of lesions. Because ADC represents the algebraic sum of vasogenic (increased ADC) and cellular (decreased ADC) brain edema, the ADC maps (Fig. 9J and K) can be used to confirm the presence of edemic regions as identified using FLAIR, as well as T1 and T2 images. Whereas mIP (Fig. 9L) and DWI (Fig. 9J and K) are both useful for the identification of hemorrhage, Figure 9E demonstrates the improved ability of SWI to localize hemorrhages and micro-bleeds, some of which are not visible using the former two techniques. In addition, SWI is capable of identifying micro-bleeds present in the follow-up scans (Fig. 9E, left).
FIG. 9.
As in Figure 1, for Patient 3. Color image is available online at www.liebertonline.com/neu
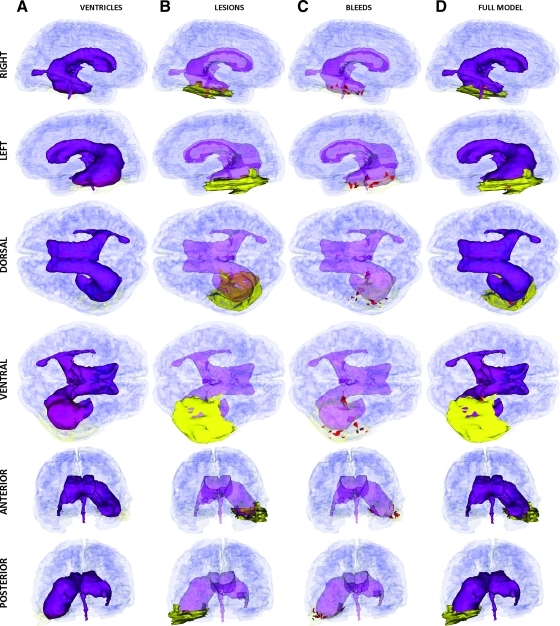
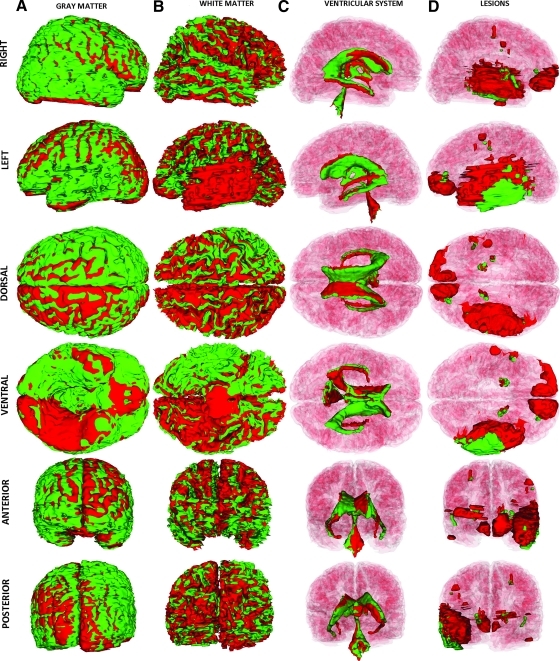
Inspection of the left and right views of the brain as reconstructed in Figure 10 reveals left–right asymmetry of the lateral ventricles, possibly partly caused by inflammation. The temporal horn of the left ventricle is positioned slightly above the horizontal plane of the right ventricle (left and right views), and its location is also seen to have shifted more medially (dorsal and ventral views). Figure 10B displays non-hemorrhagic cerebral lesions. The primary lesion occupies a significant volumetric extent within the temporal lobe of the left hemisphere, with smaller lesions in the frontopolar areas of both hemispheres as well as smaller, contrecoup lesions in the right temporal lobe. Figure 10C reveals that some portions of these lesions are hemorrhagic, with significant bleeding in the left temporal lobe and in the frontal lobes. A 3D animation of the segmentation for this patient is available in the Supplementary Material (see online supplementary material at http://www.liebertonline.com).
FIG. 10.
As in Figure 2, for Patient 3 at the acute baseline time point. Color image is available online at www.liebertonline.com/neu
Figure 11 showcases the results of segmenting the MR volumes acquired during the follow-up session. In the case of the ventricular system (Fig. 11A), as one might expect as a result of partial recovery, there is considerably more lateral symmetry than in the acute case. Approximately 8 months after acute injury, the lesion in the left temporal lobe is seen to have progressed into a structure (Fig. 11B) consisting of low-density WM and/or GM perfused by CSF. Diffuse bleeds are identified throughout the brain (Fig. 11C), mostly in areas that appear to hemorrhage in the acute baseline volume, although a few also appear in new locations (see arrows). Overall, the full model of the anatomy at follow-up (Fig. 11D) indicates noteworthy improvement after 8 months of recovery and treatment.
FIG. 11.
As in Figure 2, for Patient 3 at the chronic follow-up time point. Color image is available online at www.liebertonline.com/neu
Figure 12 shows the results of the time point comparison for the third patient. In the case of the GM volume, surface displacement is visible in the acute baseline case compared to the follow-up case. For example, the view of the left hemisphere reveals this to be true particularly in occipital, parietal, and dorsofrontal areas, where the segmented GM surface for the acute baseline model (red) lies above the corresponding surface for the follow-up case (green). For the right hemisphere, the dorsal view of the GM models has large portions of the acute baseline volume lying atop the follow-up volume, and vice versa for the left hemisphere. This obviates a clear rightward shift of the GM in the acute baseline case, presumably as a result of inflammation in the left temporal lobe. Confirmation of this impression is suggested by the ventral view of the brain, where it is seen that the temporal lobe and frontopolar regions are both larger in the acute baseline.
FIG. 12.
As in Figure 4, for Patient 3. Color image is available online at www.liebertonline.com/neu
The time point comparison of the WM volumes also indicates the presence of large differences between the two time points. The right and left views reveal a striped appearance of the two superposed models, with the crowns of gyri in the baseline volume consistently atop the corresponding gyral crowns in the follow-up volume, and vice versa for the troughs of gyri. This relative positioning of the two surfaces is consistent with the scenario of diffuse inflammation throughout the acute baseline WM volume, and possibly with the presence of DAI at the GM–WM boundary. In addition to these findings, the time point comparison of WM confirms the impressions formed from inspecting the GM volumes, where a general rightward shift of the brain had been found. This scenario is confirmed by exploring the time point comparison of the ventricular system where, in the acute baseline model, the left ventricle (ipsilateral to the primary insult) is positioned above the level corresponding to the follow-up model. This situation is reversed for the contralateral (right) hemisphere, suggesting a shearing transformation between the two models wherein the acute baseline brain mass is rotated counterclockwise about the anteroposterior axis as a result of the primary injury. This scenario is confirmed by the clear shift of the fourth ventricle in the acute baseline model (red) toward the left hemisphere, compared to the position of this ventricle in the follow-up model. Similar shifts are observed for the ventricular horns in the anterior and posterior views of Figure 12D.
As shown in Figure 12E, the time point comparison of Patient 3 reveals significant decreases in the total volume of injured brain regions, especially with regard to frontal lobe lesions as well as micro-bleeds located diffusely throughout the brain. As outlined in Table 3, the former are found to have a combined volume that is significantly smaller at chronic follow-up (0.9 cm3) than at acute baseline (54.7 cm3). A similar impression emerges for hemorrhagic lesions (0.3 cm3 at follow-up, 22.5 cm3 at baseline).
Discussion
Significance
Neuroimaging has become important for TBI surgical planning as well as for the provision of important prognostic indicators that may help the aggressiveness of injury management (Lee and Newberg, 2005). Brain contusions are relatively common in TBI, occurring in a large fraction of patients with blunt trauma, frequently as coup or contrecoup injuries. Similarly, subdural hematomas occur in 10–20% of patients with head trauma and are associated with high mortality (50–85%) (Gutman et al., 1992). Therefore, intracranial hemorrhage detection of the type performed here can be important in the process of identifying the mechanism of injury as well as the potential clinical outcome. Detection of micro-hemorrhages is dependent upon a large number of MR sequence parameters, including spatial resolution, slice thickness and TE, as outlined by Greenberg and associates (2009). Because the SWI sequence as used here takes into account MR signal phase information, regional magnetic field alterations caused by iron concentration gradients in the brain are easier to capture with this technique. As a result, the use of SWI is highly beneficial because susceptibility effects make micro-hemorrhages more obvious in SWI images compared to other paradigms. The 3D Slicer environment presented here has the ability to take into account simultaneously a variety of sequence types. This can allow clinicians and researchers to identify and quantify DAI, and this capability of the environment is notable because DAI contributes to disability in approximately 40% of closed head injuries (Buki and Povlishock, 2006). Moreover, the presence of hemorrhage in DAI lesions may portend a poor prognosis compared to their absence (Paterakis et al., 2000; Pierallini, 2000). Therefore, the ability to differentiate between hemorrhagic and non-hemorrhagic edema using 3D Slicer may be of critical use to clinicians in their attempts to evaluate the extent of each type of edema, and to formulate appropriate forms of treatment or critical care. In addition, because the number of traumatic micro-bleeds detected chronically in T2-weighted GRE and SWI images is significantly correlated with Glasgow Coma Scale (GCS) and Extended Glasgow Outcome Scale (GOS-E) scores, respectively (Itti et al., 2001; Tong et al., 2008), the capabilities of this imaging and visualization environment are well suited for the analysis of TBI data in view of exploring the relationship between information provided by neuroimaging techniques and results from neuropsychological testing.
Various quantitative measures that can be extracted from MR images using segmentation methods have been correlated with clinical outcome measures. For example, total lesion volume in TBI as extracted from FLAIR images has been found to correlate significantly with clinical scores (Pierallini, 2000). Because the signal that arises from CSF is suppressed in FLAIR imaging, more precise estimation of parenchymal damage is possible using this type of sequence. Similarly, a study of 69 TBI patients found a stepwise, dose–response relationship between parenchymal volume loss and TBI severity (Levine et al., 2008) and concluded that patterns of parenchymal volumetric changes can differentiate among levels of TBI severity, even in mild TBI. Using the 3D Slicer environment and the type of analysis described in this article to perform TBI quantification can be useful in such studies to compute volumetric measures as well as to obtain results comparable to those of manual analysis methods.
Comparison with previous work
The significance and novelty of the environment presented here can be emphasized through comparison with other segmentation techniques that have been applied for TBI analysis from MR volumes. The FreeSurfer package (Dale et al., 1999; Fischl et al., 2002, 2004), particularly, has achieved widespread popularity for automatic brain segmentation in both health and disease. An excellent study by Biglerand associates (2010) investigated the comparability of TBI volume segmentations as determined by 1) operator-controlled image quantification using the ANALYZE® package (Robb et al., 1989) and 2) automated image analysis using FreeSurfer. These authors found that both methods could detect atrophic changes but differed in the magnitude of the atrophic effect. Nevertheless, in the case of automatic segmentation, inaccuracies were found involving skull stripping, frank exclusion of brain parenchyma, WM/GM boundary identification, and inadvertent removal of brain sections. These inaccuracies required Bigler and associates to perform manual editing by 1) adding control points to aid FreeSurfer in identifying WM, 2) fixing the skull strip by removing remaining dura, and 3) manually adding back brain sections that had been inappropriately removed during automatic segmentation. The problematic characteristics of FreeSurfer TBI segmentations are confirmed by the work of Strangman and associates (2010), who reported crossover of pial and WM surfaces, inadequate skull stripping, major surface topology defects, and failures in subcortical segmentation. In contradistinction to manual editing required by Freesurfer as documented by these authors, our use of ABC within 3D Slicer did not involve operator-guided post-processing, because the latter algorithm incorporates methodologies that had been expressly conceived for robust detection of pathology in MR scans (Prastawa et al., 2009) as well as for lesion segmentation based on outlier detection (Prastawa et al., 2004). Therefore, although highly effective and widely popular for automatic segmentation of MR volumes in health and certain types of pathologies such as Alzheimer's disease (Lehmann et al., 2010), FreeSurfer incorporates neither pathology detection methods nor outlier detection algorithms, which conceivably makes ABC's methodological sophistication preferable to that of FreeSurfer in the case of TBI segmentation, especially where severe anatomical differences from normal structure are encountered, as in the present study. Finally, whereas FreeSurfer offers extensive flexibility for the task of automatic tissue classification of healthy GM/WM, our present method additionally allows differentiation among various pathology classes (e.g., hemorrhagic vs. non-hemorrhagic lesions), which is currently outside the scope of FreeSurfer capabilities.
Of essence in evaluating segmentation results and methods is the task of validation. When assessing the reliability of pathology segmentation, validation can often be difficult because of frequent lack of reliable ground truth, and manual tracings by trained experts have traditionally been considered to be appropriate from this standpoint. In the case of the methodologies used in the present study, a thorough investigation of their reliability has been undertaken by Prastawa and associates (2009), who conducted a thorough and extensive validation study using a system that combines physical and statistical modeling to generate synthetic multimodal 3D brain MRI with tumors and edema, along with the underlying anatomical ground truth. In that study, important effects of pathology on MRI were investigated, such as contrast enhancement, local healthy tissue distortion, infiltration of edema, and multimodal MRI contrast of healthy tissue and pathology. This sophisticated validation method was used to generate synthetic ground truth and to create MR volumes with edema that exhibit segmentation challenges that are comparable to real pathology as captured by MRI. To assess the reliability of ABC, both automatic and manual segmentation of five subjects exhibiting pathology was undertaken by Prastawa and associates (2003), with manual segmentation chosen as the gold standard. The volume of overlap between segmentations and the distances between the surfaces of the segmented structures were computed in addition to intra-rater variability metrics for each case. Based on this analysis, the distance between the surface generated using our present method and that generated using manual segmentation was found to be in the range of 0.5 to 1.4 mm (μ=0.92 cm σ=0.4 mm). In conclusion, although these validation results are not based on the data set analyzed in the present study, it is reasonable to expect that the error estimates obtained by Prastawa and associates are illustrative of those that one might expect from ABC segmentations of pathology.
Present segmentation results are consistent with the previous finding that approximately half of DAI lesions are located either in the deep WM or at the corticomedullary junction (GM–WM interface) of the frontal and temporal lobes (Adams et al., 1982). In this respect, our case studies illustrate findings that are quite typical of TBI pathology and therefore very well suited for demonstrating the segmentation and analysis capabilities of the 3D Slicer environment. The large lesions that are both hyperintense in FLAIR images and superficial with respect to the cortical surface of this patient are also typical of TBI pathology, as they involve the frontal and temporal lobes (Pierallini, 2000). One improvement of the present approach over previous studies consists of the superior multimodal ability to identify, distinguish, and characterize hemorrhagic versus non-hemorrhagic lesions. In this respect, sensitivity and specificity may be comparable to those of investigations that found improved detection of hemorrhage using SWI as compared to other studies that did not benefit from the availability of this technique (Tong et al., 2003, 2008).
Numerous studies have identified MRI-extracted correlates between TBI pathology and neuropsychological outcomes in the areas of information-processing speed, learning, and memory, as well as executive function. In particular, GM volume has been strongly linked to chronic damage to WM tracts (Warner et al., 2010). Similarly, changes in WM and GM volume were found to correlate well with functional TBI outcome as measured using the Functional Status Examination (FSE), and FLAIR-measured acute axonal lesions are strongly predictive of post-traumatic cerebral atrophy (Ding et al., 2008), which can also be quantified using the metrics previously described (e.g., bifrontal index, bicaudate index). In addition, global brain volume loss correlates well with admission GCS, coma duration, and amnesia (Ding et al., 2008), and total lesion volume as measured 2–3 months after injury using FLAIR correlates significantly with the 1-year Glasgow Outcome Score (GOS) (Pierallini, 2000). These analyses are reproducible and can be streamlined using the 3D Slicer environment, particularly as a consequence of the latter's ability to extract brain volumetrics straightforwardly and to perform time point comparisons. For example, cortical brain volumes are suggestive of learning, memory, and processing speed performance (Warner et al., 2010); similarly, total brain volume loss has been associated with loss of consciousness duration (MacKenzie et al., 2002). Therefore, the 3D Slicer environment will be of particular interest to researchers who quantify TBI using measures of these cognitive functions.
It has been suggested that lesion volume in acute DAI can be used to stratify injury severity when selecting patients for TBI clinical trials (Ding et al., 2008). Therefore, the ability of the 3D Slicer environment to offer useful information on DAI pathological change might be useful for DAI-directed therapies. A study involving 37 TBI patients found a mean decrease in brain volume of −1.43% between approximately 79 and 409 days post-TBI, with greater decline in brain volume being associated with longer duration of post-injury coma (Trivedi et al., 2007); the findings of our volumetric analysis are comparable to these results as well as to those of Sidaros and associates (2009), who found a %BVC ranging between −0.6% and −9.4% (mean of −4.0%) in a population of 24 TBI patients. For our three cases, the %BVC was found to have a mean of −10.3% and a standard deviation of 1.89% (Table 3). Although in our study the sample size is very small, these percentage changes in brain volume appear to be in line with previous %BVC ranges, especially given the extreme TBI severity being investigated in this article. In conclusion, because cerebral atrophy is known to reach significance 8–12 months after the traumatic event (Blatter et al., 1997), we expect that the cases described in this article may be reasonably representative of the typical atrophy patterns observed in TBI populations.
Conclusion
As demonstrated here, 3D Slicer offers improvement over existing analysis tools for the purpose of TBI analysis. Specifically, through the use of this freely available environment, multimodal registration and bias field correction of a large number of MR volumes acquired using different sequences can be accomplished. Joint visualization of multiple MRI sequences acquired at different time points and co-registered to standard coordinates can improve the ability to classify various lesion types as encoded in various channels and to track changes over time. Furthermore, user-guided model extraction and quantification can allow one to utilize multi-modal information for improved benefit. In addition to this, semiautomatic use of ABC as well as of other segmenters available through 3D Slicer allows one to perform robust joint visualization of co-registered MRI volumes, tissue classification, and 3D model generation. The calculation of anatomic measures (such as volume and area) associated with structures of interest is clinically relevant and methodologically straightforward.
Finally, an important application of our methodological paradigm involves the cross-correlation of volume and atrophy measures (as extracted from our models) with clinical measures of outcome or with the results of neuropsychological testing. This type of investigation may reveal in detail the relationship between quantitative MR segmentation results, on the one hand, and other measures of patient functioning and rehabilitation, on the other hand. Accurate injury classification using our methodologies may also allow clinicians to formulate treatment strategies based on the detailed structural profile and classification of injuries as provided via neuroimaging. In acute trauma, such a detailed profile may impact the short-term outcome of the subject by providing significant information for monitoring and stabilization, and such timely interventions may in turn translate into positive effects upon long-term outcome and rehabilitation.
Supplementary Material
Acknowledgments
We acknowledge the assistance of David McArthur, Maria Etcheparre, Silvain Gouttard, Steve Pieper, Stephen Aylward, Sonja Pujol, and the staff of the Laboratory of Neuro Imaging at UCLA. 3D Slicer is a multi-platform, free, and open source software package for visualization and medical image computing available from www.slicer.org. This work was supported by the National Alliance for Medical Image Computing (NA-MIC; www.na-mic.org), under National Institutes of Health Roadmap Initiative grant 2U54EB005149 to R. K. and sub-award to J. D. V. H., and by the NINDS, grant P01NS058489 to P. M. V.
Author Disclosure Statement
No competing financial interests exist.
References
- Adams J.H. Graham D.I. Murray L.S. Scott G. Diffuse axonal injury due to nonmissile head injury in humans: an analysis of 45 cases. Ann. Neurol. 1982;12:557–563. doi: 10.1002/ana.410120610. [DOI] [PubMed] [Google Scholar]
- Bates E. Wilson S.M. Saygin A.P. Dick F. Sereno M.I. Knight R.T. Dronkers N.F. Voxel-based lesion-symptom mapping. Nat. Neurosci. 2003;6:448–450. doi: 10.1038/nn1050. [DOI] [PubMed] [Google Scholar]
- Bigler E.D. Abildskov T.J. Wilde E.A. McCauley S.R. Li X. Merkley T.L. Fearing M.A. Newsome M.R. Scheibel R.S. Hunter J.V. Chu Z. Levin H.S. Diffuse damage in pediatric traumatic brain injury: a comparison of automated versus operator-controlled quantification methods. NeuroImage. 2010;50:1017–1026. doi: 10.1016/j.neuroimage.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Blatter D.D. Bigler E.D. Gale S.D. Johnson S.C. Anderson C.V. Burnett B.M. Ryser D. Macnamara S.E. Bailey B.J. MR-based brain and cerebrospinal fluid measurement after traumatic brain injury: correlation with neuropsychological outcome. AJNR Am. J. Neuroradiol. 1997;18:1–10. [PMC free article] [PubMed] [Google Scholar]
- Buki A. Povlishock J.T. All roads lead to disconnection?––Traumatic axonal injury revisited. Acta Neurochir. (Wien). 2006;148:181–193. doi: 10.1007/s00701-005-0674-4. discussion 193–184. [DOI] [PubMed] [Google Scholar]
- Chen A.J. D'Esposito M. Traumatic brain injury: from bench to bedside [corrected] to society. Neuron. 2010;66:11–14. doi: 10.1016/j.neuron.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Chesnut R.M. Implications of the guidelines for the management of severe head injury for the practicing neurosurgeon. Surg. Neurol. 1998;50:187–193. doi: 10.1016/s0090-3019(98)00075-5. [DOI] [PubMed] [Google Scholar]
- Dale A.M. Fischl B. Sereno M.I. Cortical surface-based analysis – I. Segmentation and surface reconstruction. NeuroImage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- De Beaumont L. Theoret H. Mongeon D. Messier J. Leclerc S. Tremblay S. Ellemberg D. Lassonde M. Brain function decline in healthy retired athletes who sustained their last sports concussion in early adulthood. Brain. 2009;132:695–708. doi: 10.1093/brain/awn347. [DOI] [PubMed] [Google Scholar]
- De Coene B. Hajnal J. Gatehouse P. Longmore D. White S. Oatridge A. Pennock J. Young I. Bydder G. MR of the brain using fluid-attenuated inversion recovery (FLAIR) pulse sequences. AJNR Am. J. Neuroradiol. 1992;13:1555–1564. [PMC free article] [PubMed] [Google Scholar]
- Ding K. Marquez de la Plata C. Wang J.Y. Mumphrey M. Moore C. Harper C. Madden C.J. McColl R. Whittemore A. Devous M.D. Diaz–Arrastia R. Cerebral atrophy after traumatic white matter injury: correlation with acute neuroimaging and outcome. J. Neurotrauma. 2008;25:1433–1440. doi: 10.1089/neu.2008.0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubroff J.G. Newberg A. Neuroimaging of traumatic brain injury. Semin. Neurol. 2008;28:548–557. doi: 10.1055/s-0028-1083698. [DOI] [PubMed] [Google Scholar]
- Earnest M.P. Heaton R.K. Wilkinson W.E. Manke W.F. Cortical atrophy, ventricular enlargement and intellectual impairment in the aged. Neurology. 1979;29:1138–1143. doi: 10.1212/wnl.29.8.1138. [DOI] [PubMed] [Google Scholar]
- Faul M. Xu L. Wald M.M. Coronado V.G. Traumatic Brain Injury in the United States: Emergency Department Visits, Hospitalizations and Deaths 2002–2006. Atlanta, GA: Centers for Disease Control and Prevention, National Center for Injury Prevention and Control; 2010. [Google Scholar]
- Filippi M. Gawne–Cain ML. Gasterini C. Effect of training and different measurement strategies on the reproducibility of brain MRI lesion load measurements in multiple sclerosis. Neurology. 1998;50:238–244. doi: 10.1212/wnl.50.1.238. [DOI] [PubMed] [Google Scholar]
- Fischl B. Salat D.H. Busa E. Albert M. Dieterich M. Haselgrove C. van der Kouwe A. Killiany R. Kennedy D. Klaveness S. Montillo A. Makris N. Rosen B. Dale A.M. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Fischl B. van der Kouwe A. Destrieux C. Halgren E. Segonne F. Salat D.H. Busa E. Seidman L.J. Goldstein J. Kennedy D. Caviness V. Makris N. Rosen B. Dale A.M. Automatically parcellating the human cerebral cortex. Cereb. Cortex. 2004;14:11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- Greenberg S.M. Vernooij M.W. Cordonnier C. Viswanathan A. Al-Shahi Salman R. Warach S. Launer L.J. Van Buchem M.A. Breteler M.M. Cerebral microbleeds: a guide to detection and interpretation. Lancet Neurol. 2009;8:165–174. doi: 10.1016/S1474-4422(09)70013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman M.B. Moulton R.J. Sullivan I. Hotz G. Tucker W.S. Muller P.J. Risk factors predicting operable intracranial hematomas in head injury. J. Neurosurg. 1992;77:9–14. doi: 10.3171/jns.1992.77.1.0009. [DOI] [PubMed] [Google Scholar]
- Gyldensted C. Measurements of the normal ventricular system and hemispheric sulci of 100 adults with computed tomography. Neuroradiology. 1977;14:183–192. doi: 10.1007/BF00496982. [DOI] [PubMed] [Google Scholar]
- Hahn F.J. Schapiro R.L. The excessively small ventricle on computed axial tomography of the brain. Neuroradiology. 1976;12:137–139. doi: 10.1007/BF00341858. [DOI] [PubMed] [Google Scholar]
- Huckman M.S. Fox J. Topel J. Validity of criteria for evaluation of cerebral atrophy by computed tomography. Radiology. 1975;116:85–92. doi: 10.1148/116.1.85. [DOI] [PubMed] [Google Scholar]
- Huisman T.A. Diffusion-weighted imaging: basic concepts and application in cerebral stroke and head trauma. Eur. Radiol. 2003;13:2283–2297. doi: 10.1007/s00330-003-1843-6. [DOI] [PubMed] [Google Scholar]
- Huisman T.A. Sorensen A.G. Hergan K. Gonzalez R.G. Schaefer P.W. Diffusion-weighted imaging for the evaluation of diffuse axonal injury in closed head injury. J. Comput. Assist. Tomogr. 2003;27:5–11. doi: 10.1097/00004728-200301000-00002. [DOI] [PubMed] [Google Scholar]
- Itti L. Chang L. Ernst T. Segmentation of progressive multifocal leukoencephalopathy lesions in fluid-attenuated inversion recovery magnetic resonance imaging. J. Neuroimaging. 2001;11:412–417. doi: 10.1111/j.1552-6569.2001.tb00071.x. [DOI] [PubMed] [Google Scholar]
- Jones K.M. Mulkern R.V. Schwartz R.B. Oshio K. Barnes P.D. Jolesz F.A. Fast spin-echo MR imaging of the brain and spine: current concepts. AJR Am. J. Roentgenol. 1992;158:1313–1320. doi: 10.2214/ajr.158.6.1590133. [DOI] [PubMed] [Google Scholar]
- Langlois J.A. Rutland–Brown W. Thomas K. Traumatic brain injury in the United States: emergency department visits, hospitalizations, and deaths. Atlanta, GA: Centers for Disease Control and Prevention, National Center for Injury Prevention and Control; 2006. [Google Scholar]
- Lee B. Newberg A. Neuroimaging in traumatic brain imaging. NeuroRx. 2005;2:372–383. doi: 10.1602/neurorx.2.2.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M. Douiri A. Kim L.G. Modat M. Chan D. Ourselin S. Barnes J. Fox N.C. Atrophy patterns in Alzheimer's disease and semantic dementia: a comparison of FreeSurfer and manual volumetric measurements. NeuroImage. 2010;49:2264–2274. doi: 10.1016/j.neuroimage.2009.10.056. [DOI] [PubMed] [Google Scholar]
- Levine B. Kovacevic N. Nica E.I. Cheung G. Gao F. Schwartz M.L. Black S.E. The Toronto traumatic brain injury study: injury severity and quantified MRI. Neurology. 2008;70:771–778. doi: 10.1212/01.wnl.0000304108.32283.aa. [DOI] [PubMed] [Google Scholar]
- MacKenzie J.D. Siddiqi F. Babb J.S. Bagley L.J. Mannon L.J. Sinson G.P. Grossman R.I. Brain atrophy in mild or moderate traumatic brain injury: a longitudinal quantitative analysis. AJNR Am. J. Neuroradiol. 2002;23:1509–1515. [PMC free article] [PubMed] [Google Scholar]
- Maes F. Collignon A. Vandermeulen D. Marchal G. Suetens P. Multimodality image registration by maximization of mutual information. IEEE Trans. Med. Imaging. 1997;16:187–198. doi: 10.1109/42.563664. [DOI] [PubMed] [Google Scholar]
- Mugler J.P., 3rd Brookeman J.R. Three-dimensional magnetization-prepared rapid gradient-echo imaging (3D MP RAGE) Magn. Reson. Med. 1990;15:152–157. doi: 10.1002/mrm.1910150117. [DOI] [PubMed] [Google Scholar]
- Paterakis K. Karantanas A.H. Komnos A. Volikas Z. Outcome of patients with diffuse axonal injury: the significance and prognostic value of MRI in the acute phase. J. Trauma. 2000;49:1071–1075. doi: 10.1097/00005373-200012000-00016. [DOI] [PubMed] [Google Scholar]
- Pierallini A. Pantano P. Fantozzi L. M. Bonamini M. Vichi R. Zylberman R. Pisarri F. Colonnese C. Bozzao L. Correlation between MRI findings and long-term outcome in patients with severe brain trauma. Neuroradiology. 2000;42:860–867. doi: 10.1007/s002340000447. [DOI] [PubMed] [Google Scholar]
- Pohl K.M. Bouix S. Nakamura M. Rohlfing T. McCarley R.W. Kikinis R. Grimson W.E. Shenton M.E. Wells W.M. A hierarchical algorithm for MR brain image parcellation. IEEE Trans. Med. Imaging. 2007;26:1201–1212. doi: 10.1109/TMI.2007.901433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prastawa M. Bullitt E. Gerig G. Simulation of brain tumors in MR images for evaluation of segmentation efficacy. Med. Image Anal. 2009;13:297–311. doi: 10.1016/j.media.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prastawa M. Bullitt E. Ho S. Gerig G. A brain tumor segmentation framework based on outlier detection. Med. Image Anal. 2004;8:275–283. doi: 10.1016/j.media.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Prastawa M. Bullitt E. Moon N. Van Leemput K. Gerig G. Automatic brain tumor segmentation by subject specific modification of atlas priors. Acad. Radiol. 2003;10:1341–1348. doi: 10.1016/s1076-6332(03)00506-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prastawa M. Gerig G. Brain lesion segmentation through physical model estimation. Lect. Notes Comput. Sci. 2008;5358:562–571. [Google Scholar]
- Robb R.A. Hanson D.P. Karwoski R.A. Larson A.G. Workman E.L. Stacy M.C. Analyze: a comprehensive, operator-interactive software package for multidimensional medical image display and analysis. Comput. Med. Imaging Graph. 1989;13:433–454. doi: 10.1016/0895-6111(89)90285-1. [DOI] [PubMed] [Google Scholar]
- Scheid R. Preul C. Gruber O. Wiggins C. von Cramon D.Y. Diffuse axonal injury associated with chronic traumatic brain injury: evidence from T2*-weighted gradient-echo imaging at 3 T. AJNR Am. J. Neuroradiol. 2003;24:1049–1056. [PMC free article] [PubMed] [Google Scholar]
- Sehgal V. Delproposto Z. Haacke E.M. Tong K.A. Wycliffe N. Kido D.K. Xu Y. Neelavalli J. Haddar D. Reichenbach J.R. Clinical applications of neuroimaging with susceptibility-weighted imaging. J. Magn. Reson. Imaging. 2005;22:439–450. doi: 10.1002/jmri.20404. [DOI] [PubMed] [Google Scholar]
- Sidaros A. Skimminge A. Liptrot M.G. Sidaros K. Engberg A.W. Herning M. Paulson O.B. Jernigan T.L. Rostrup E. Long-term global and regional brain volume changes following severe traumatic brain injury: a longitudinal study with clinical correlates. Neuroimage. 2009;44:1–8. doi: 10.1016/j.neuroimage.2008.08.030. [DOI] [PubMed] [Google Scholar]
- Strangman G.E. O'Neil–Pirozzi T.M. Supelana C. Goldstein R. Katz D.I. Glenn M.B. Regional brain morphometry predicts memory rehabilitation outcome after traumatic brain injury. Front. Hum. Neurosci. 2010;4:182. doi: 10.3389/fnhum.2010.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synek V. Reuben J.R. Du Boulay G.H. Comparing Evans' index and computerized axial tomography in assessing relationship of ventricular size to brain size. Neurology. 1976;26:231–233. doi: 10.1212/wnl.26.3.231. [DOI] [PubMed] [Google Scholar]
- Taber K.H. Warden D.L. Hurley R.A. Blast-related traumatic brain injury: what is known? J. Neuropsychiatry Clin. Neurosci. 2006;18:141–145. doi: 10.1176/jnp.2006.18.2.141. [DOI] [PubMed] [Google Scholar]
- Tong K.A. Ashwal S. Holshouser B.A. Shutter L.A. Herigault G. Haacke E.M. Kido D.K. Hemorrhagic shearing lesions in children and adolescents with posttraumatic diffuse axonal injury: improved detection and initial results. Radiology. 2003;227:332–339. doi: 10.1148/radiol.2272020176. [DOI] [PubMed] [Google Scholar]
- Tong K.A. Ashwal S. Obenaus A. Nickerson J.P. Kido D. Haacke E.M. Susceptibility-weighted MR imaging: a review of clinical applications in children. AJNR Am. J. Neuroradiol. 2008;29:9–17. doi: 10.3174/ajnr.A0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi M.A. Ward M.A. Hess T.M. Gale S.D. Dempsey R.J. Rowley H.A. Johnson S.C. Longitudinal changes in global brain volume between 79 and 409 days after traumatic brain injury: relationship with duration of coma. J. Neurotrauma. 2007;24:766–771. doi: 10.1089/neu.2006.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterberg A.W. Stover J. Kress B. Kiening K.L. Edema and brain trauma. Neuroscience. 2004;129:1021–1029. doi: 10.1016/j.neuroscience.2004.06.046. [DOI] [PubMed] [Google Scholar]
- Van Leemput K. Maes F. Vandermeulen D. Suetens P. Automated model-based tissue classification of MR images of the brain. IEEE Trans. Med. Imaging. 1999;18:897–908. doi: 10.1109/42.811270. [DOI] [PubMed] [Google Scholar]
- Warach S. Gaa J. Siewert B. Wielopolski P. Edelman R.R. Acute human stroke studied by whole brain echo planar diffusion-weighted magnetic resonance imaging. Ann. Neurol. 1995;37:231–241. doi: 10.1002/ana.410370214. [DOI] [PubMed] [Google Scholar]
- Warner M.A. Marquez de la Plata C. Spence J. Wang J.Y. Harper C. Moore C. Devous M. Diaz–Arrastia R. Assessing spatial relationships between axonal integrity, regional brain volumes, and neuropsychological outcomes after traumatic axonal injury. J. Neurotrauma. 2010;27:2121–2130. doi: 10.1089/neu.2010.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts D.D. Hanfling D. Waller M.A. Gilmore C. Fakhry S.M. Trask A.L. An evaluation of the use of guidelines in prehospital management of brain injury. Prehosp. Emerg. Care. 2004;8:254–261. doi: 10.1016/j.prehos.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Zoellei L. Shenton M. Wells W. Pohl K. The impact of atlas formation methods on atlas-guided brain segmentation. In: Ayache N., editor; Ourselin S., editor; Maeder A., editor. Lecture Notes in Computer Science; International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI); 2007. pp. 39–46. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.