Abstract
During the last decade, it was established that the class III alcohol dehydrogenase (ADH3) enzyme, also known as glutathione-dependent formaldehyde dehydrogenase (FALDH; EC 1.2.1.1), catalyzes the NADH-dependent reduction of S-nitrosoglutathione (GSNO) and therefore was also designated as GSNO reductase. This finding has opened new aspects in the metabolism of nitric oxide (NO) and NO-derived molecules where GSNO is a key component. In this article, current knowledge of the involvement and potential function of this enzyme during plant development and under biotic/abiotic stress is briefly reviewed.
Key words: nitric oxide, nitrosative stress, S-nitrosoglutathione reductase
Introduction
The relevance of NO in the physiology of higher plants under optimal and stress conditions was established during the last decade.1–6 Nitric oxide has a family of NO-derived molecules designated as reactive nitrogen species (RNS) including radical and non-radical molecules;3 among them, the group of S-nitrosothiols (SNOs), which result from the interaction of NO with intracellular sulfhydryl-containing molecules, bears special interest because these molecules are generally more stable in solution than is NO, which has a very short in vivo half-life. Therefore, these SNOs can participate in the transport, storage and delivery of NO and consequently contribute to post-translational modifications involved in cell signaling and in stress processes.7,8 Among the different SNOs, S-nitrosoglutathione (GSNO) is formed by the S-nitrosylation reaction of NO with glutathione (GSH: the tripeptide γ-Glu-Cys-Gly) and appears to have a significant physiological relevance in plants, since GSNO is thought to function as a mobile reservoir of NO9,10 and therefore can affect the process of trans-nitrosation equilibrium between GSNO and S-nitrosylated proteins (Fig. 1). In this sense, very recently, it has been proposed a mechanism of GSNO formation mediated by cytochrome c.14
Figure 1.
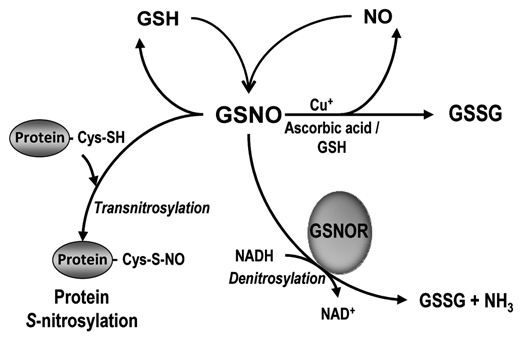
Schematic model of the metabolism of GSNO metabolism in plant cells and its regulation by GSNO reductase. Nitric oxide reacts with reduced glutathione (GSH) in the presence of O2 to form S-nitrosoglutathione (GSNO). This metabolite can be converted by the enzyme GSNO reductase (GSNOR) by a process of NADH-dependent denitrosylation into oxidized glutathione (GSSG) and NH3. Alternatively, GSNO in the presence of reductants, such as glutathione (GSH) and ascorbate, and Cu+, can be broken down to produce NO and GSSG.11–13 On the other hand, by a process of transnitrosylation the GSNO can transfer the NO to cysteine residues of proteins (protein S-nitrosylation). This is a post-translational modification that can modify the function of a broad spectrum of proteins.
Alcohol dehydrogenase 3 (ADH3) is a highly conserved and ubiquitous enzyme found in both prokaryotes and eukaryotes. This enzyme is involved in critical cellular pathways because it takes part in the detoxification of formaldehyde, catalyzing the following reaction: formaldehyde + GSH + NAD+ → S-formylglutathione + NADH + H+.15–17 However, the interest in this enzyme increased significantly when it was shown also to catalyze the NADH-dependent reduction of S-nitrosoglutathione (GSNO) to GSSG and NH3,18,19 which can be considered a reaction of denitrosylation (Fig. 1). Therefore, this activity can control the intracellular level of GSNO and consequently the effects of NO in cells. Thus, this GSNOR activity can change the transnitrosation equilibrium between GSNO and S-nitrosylated proteins and as a result participates in the cellular NO homeostasis. Additionally, because this reaction affects the equilibrium of GSH and NADH, the GSNOR could be indirectly involved in the cellular redox states.
GSNO Reductase in Plants
Prior to the new finding of the GSNOR activity by alcohol dehydrogenase 3 in plants, the glutathione-dependent formaldehyde dehydrogenase activity was studied as a mechanism of formaldehyde detoxification process.20 Thus, this protein was purified and/or characterized in several plant species including pea seeds,21,22 maize,23,24 rice25 and Arabidopsis thaliana plants.25,26
In higher plants, the first report on GSNO reductase activity involved Arabidopsis. Thus, the use of the heterologous expression of the corresponding putative cDNA (At5g43940) encoding a glutathione-dependent formaldehyde dehydrogenase showed that the recombinant protein reduces GSNO. This was confirmed by functional complementation of the hypersensitivity to GSNO of a yeast mutant with impaired GSNO metabolism.27 Thereafter, this activity began to attract the attention of many plant researchers because its capacity to metabolize GSNO opens new a perspective on the metabolism of NO in plants under physiological and environmental stress.
GSNOR during Plant Development
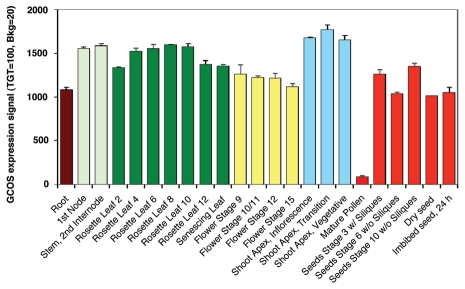
GSNOR activity appears to be necessary for normal development and fertility under optimal growth conditions.28 In Arabidopsis, the protein is encoded by one gene. Figure 2 shows the gene-expression analysis of GSNOR (At5g43940) in Arabidopsis using the Bio-Array Resource (BAR) for Plant Biology, indicating that this gene is significantly expressed in all organs with the exception of mature pollen. However, the analysis of GSNOR protein and activity in Arabidopsis by immunolocalization and histochemical methods showed that this protein is differentially expressed, being higher in the roots and leaves from the first stages of development.31 Moreover, in transgenic Arabidopsis plants, both overexpressing and knock-down GSNOR had a short-root phenotype that was correlated with a lowering of the intracellular GSH level and an alteration in its spatial distribution in the roots, suggesting that GSNOR might be involved in the regulation of redox state.31
Figure 2.
Gene expression of GSNO reductase (At5g43940) during Arabidopsis development. The data are from a Gene Expression Map of Arabidopsis Development and they are normalized by the Gene Chip Operating System (GCOS) method, with a target signal value of 100.29 Arabidopsis electronic Fluorescent Pictograph (eFP) browser at bar.utoronto.ca.30
At the subcellular level the analysis of the Arabidopsis protein sequence suggests that this protein is located in the cytosol. Nevertheless, some proteomic analyses of Arabidopsis peroxisomes have suggested that this protein could be located also in this organelle.32 However, additional experiments are needed to confirm this aspect.
More recently, it has been reported that this enzyme may also be involved in the regulation of plant cell death perhaps through a process of S-nitrosylation.33
Function of GSNOR under Biotic and Abiotic Stress Conditions
As mentioned above, the enzyme GSNOR can regulate the cellular level of SNOs and consequently GSNO content. In Arabidopsis thaliana, it has been reported that mutation of AtGSNOR1 modulates the level of cellular SNO formation and turnover, which appears to regulate multiple forms of plant disease resistance.34 Transgenic Arabidopsis plants with a decreased amount of GSNOR (using an antisense strategy) displayed more resistance against the pathogen Peronospora parasitica, apparently correlated with higher levels of intracellular SNOs.35,36 In this sense, in a sunflower (Helianthus annuus L.) cultivar resistant to the fungus Plasmopara halstedii, an inverse correlation was found in infected hypocotyls between GSNOR activity and GSNO distribution and content (Fig. 3). However, notably, GSNO accumulated in the cortex cells and, after interacting with the fungus, was redistributed in the epidermis, which is the site of infection by this pathogen. The authors suggest that this redistribution of GSNO before and after pathogen interaction could contribute to the resistance in this sunflower cultivar.37
Figure 3.
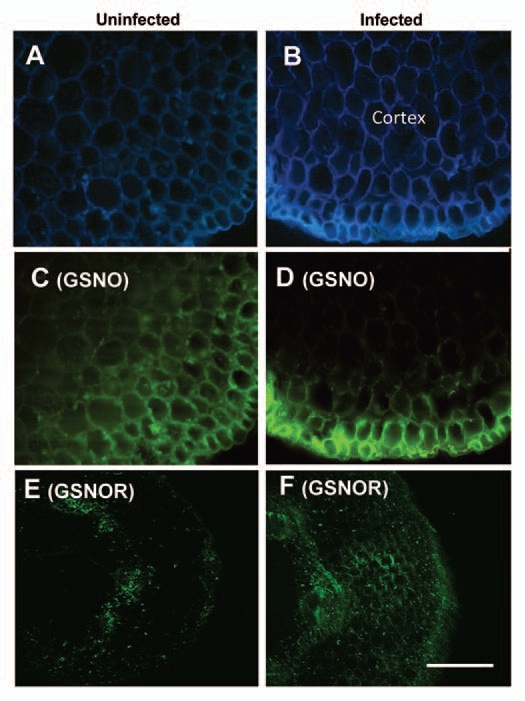
Confocal laser scanning microscope pictures illustrating the detection of GSNO and GSNOR in transverse sections of hypocotyls of the sunflower cultivar X55, which is resistant to infection by the pathogen Plasmopara halstedii. Autofluorescence appears as a blue color in (A and B). The bright green fluorescence corresponds to the detection of GSNO (C and D) and GSNOR (E and F) using specific antibodies with a dilution of 1:2,500 and 1:50, respectively. Bar = 200 µm.37 (Image available in color online at http://www.landesbioscience.com/journals/10/article/15161/.)
The modulation of GSNOR activity and expression has been also observed under different abiotic-stress conditions. In pea plants grown with 50 µM cadmium, which produces oxidative stress, the analysis of GSNOR activity and its transcript expression was found to be reduced 31%. This was accompanied by lower NO, GSH and GSNO contents.38 However, contrary behavior was noted in Arabidopsis seedlings grown in the presence of 0.5 mM arsenic. This metalloid caused a significant reduction in roots that was accompanied by oxidative stress, but the GSNOR activity significantly increased with a concomitant rise of NO content.39 Recently, in Arabidopsis a mutant sensitive to high temperatures was identified and consequently designated as HOT5. Its characterization revealed that this mutant was affected in the GSNOR, indicating that this enzyme was required for thermotolerance.28 Nevertheless, in wild-type Arabidopsis exposed to heat stress, the GSNOR protein expression was similar in both control and heat-stressed wild-type leaves. As a result, these authors hypothesized that elevated levels of GSNO increase heat sensitivity due to the perturbation of pathways sensitive to ROS/RNS.
The herbicide paraquat acts by inducing the production of superoxide and hydrogen peroxide. A screening of Arabidopsis using this herbicide allowed the identification of a knock-out mutant which showed resistance to paraquat, being designated par2.33 Its identification was surprising since it corresponded to the GSNOR gene, which was previously also described to be heat sensitive HOT5.28 The knock-out mutants (gsnor1/hot5/par2) showed greater NO content whereas overexpression of GSNOR1/HOT5/PAR2 lowered the NO level, indicating that the GSNOR activity is involved in the NO homeostasis.
In sunflower seedlings exposed to high temperature (38°C for 4 h), GSNOR activity, protein and gene expression have been found to be reduced in hypocotyls with the simultaneous accumulation of SNOs.40 The consequence was a rise in protein tyrosine nitration, which is considered a marker of nitrosative stress.41 Thus, it has been proposed that GSNOR regulates the level of SNOs, these being molecules that are a potential source of NO, necessary to mediate the process of tyrosine nitration.40 However, in pea seedlings exposed to the same stress conditions, the behavior was totally different because the GSNOR activity increased, as also did the SNO content.41 GSNOR activity has also been studied under low temperatures. In pea (Pisum sativum L.) seedlings subjected to 8°C for 48 h, the analysis of GSNOR activity in leaves showed a 67% increase compared to control plants. This was accompanied by the SNO content, which increased 5-fold.41 In pepper (Capsicum annuum L.) plants exposed to low temperature (8°C) for 24 h, the behavior was similar, since GSNO reductase activity rose 32%, although the NO content fell roughly 50%, with a concomitant rise in the GSH content.43
In Arabidopsis, the GSNOR gene has been shown to be regulated by wounding and salicylic acid, although the activity and SNO content was not analyzed.44 However, mechanically injured sunflower hypocotyls showed a downregulation of GSNOR activity, which boosted the SNO content.42 Previously, a similar situation was reported in tobacco leaves, in which after 2 h of mechanical damage both GSNOR mRNA and protein levels fell.44
Conclusions and Perspectives
S-nitrosoglutathione is a natural reservoir of NO, and therefore the mechanism of its regulation by GSNO reductase during plant development and under stress conditions will help to elucidate the complex metabolism of NO. Given the importance of S-nitrosoglutathione as a natural reservoir of NO, several key issues will need to be resolved: (1) A reliable method is needed to detect and quantify the GSNO in plant tissues to establish the direct relationship between the substrate and enzyme. (2) The correlation between NO content and the gene/protein expression of GSNO reductase needs to be elucidated in the different organs and tissues with special emphasis on its subcellular location. (3) The regulation mechanism of this enzyme, where processes such as S-nitrosylation and/or nitration of the own GSNO reductase must be involved, needs to be determined. Some of these aspects can contribute to the understating of NO metabolism in higher plants, since this molecule is involved in a wide range of processes such as cellular signaling and response mechanisms against adverse stress conditions.
Acknowledgements
This work was supported by ERDF-cofinanced grants from the Ministry of Science and Innovation (BIO2009-12003-C02-01, BIO2009-12003-C02-02 and ACI2009-0860).
References
- 1.Lamattina L, García-Mata C, Graziano M, Pagnussat G. Nitric oxide: The versatility of an extensive signal molecule. Annu Rev Plant Biol. 2003;54:109–136. doi: 10.1146/annurev.arplant.54.031902.134752. [DOI] [PubMed] [Google Scholar]
- 2.Corpas FJ, Barroso JB, Carreras A, Valderrama R, Palma JM, León AM, et al. Constitutive arginine-dependent nitric oxide synthase activity in different organs of pea seedlings during plant development. Planta. 2006;224:246–254. doi: 10.1007/s00425-005-0205-9. [DOI] [PubMed] [Google Scholar]
- 3.Corpas FJ, Barroso JB, Carreras A, Valderrama R, Palma JM, del Rio LA. Nitrosative stress in plants: A new approach to understand the role of NO in abiotic stress. In: Lamattina L, Polacco JC, editors. Nitric Oxide in Plant Growth, Development and Stress Physiology. New York: Springer; 2006. pp. 187–205. [Google Scholar]
- 4.Corpas FJ, Chaki M, Fernández-Ocaña A, Valderrama R, Palma JM, Carreras A, et al. Metabolism of reactive nitrogen species in pea plants under abiotic stress conditions. Plant Cell Physiol. 2008;49:1711–1722. doi: 10.1093/pcp/pcn144. [DOI] [PubMed] [Google Scholar]
- 5.Besson-Bard A, Pugin A, Wendehenne D. New insights into nitric oxide signaling in plants. Annu Rev Plant Biol. 2008;59:21–39. doi: 10.1146/annurev.arplant.59.032607.092830. [DOI] [PubMed] [Google Scholar]
- 6.Wilson ID, Neill SJ, Hancock JT. Nitric oxide synthesis and signaling in plants. Plant Cell Environ. 2008;31:622–631. doi: 10.1111/j.1365-3040.2007.01761.x. [DOI] [PubMed] [Google Scholar]
- 7.Foster MW, McMahon TJ, Stamler JS. S-nitrosylation in health and disease. Trends in Mol Med. 2003;9:160–168. doi: 10.1016/s1471-4914(03)00028-5. [DOI] [PubMed] [Google Scholar]
- 8.Benhar M, Forrester MT, Stamler JS. Nitrosative stress in the ER: A new role for S-nitrosylation in neurodegenerative diseases. ACS Chem Biol. 2006;1:355–358. doi: 10.1021/cb600244c. [DOI] [PubMed] [Google Scholar]
- 9.Durner J, Klessig DF. Nitric oxide as a signal in plants. Curr Opin Plant Biol. 1999;2:369–374. doi: 10.1016/s1369-5266(99)00007-2. [DOI] [PubMed] [Google Scholar]
- 10.Stamler JS, Lamas S, Fang FC. Nitrosylation the prototypic redoxbased signaling mechanism. Cell. 2001;106:675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- 11.Gorren ACF, Schrammel A, Schmidt K, Mayer B. Decomposition of S-nitrosoglutathione in the presence of copper ions and glutathione. Arch Biochem Biophys. 1996;330:219–228. doi: 10.1006/abbi.1996.0247. [DOI] [PubMed] [Google Scholar]
- 12.Holmes AJ, Williams DLH. Reaction of ascorbic acid with S-nitrosothiols: clear evidence for two distinct reaction pathways. J Chem Soc Perkin 1. 2000;2:1639–1644. [Google Scholar]
- 13.Smith JN, Dasgupta TP. Kinetics and mechanism of the decomposition of S-nitrosoglutathione by L-ascorbic acid and copper ions in aqueous solution to produce nitric oxide. Nitric Oxide. 2000;4:57–66. doi: 10.1006/niox.2000.0272. [DOI] [PubMed] [Google Scholar]
- 14.Basu S, Keszler A, Azarova NA, Nwanze N, Perlegas A, Shiva S, et al. A novel role for cytochrome c: Efficient catalysis of S-nitrosothiol formation. Free Radic Biol Med. 2010;48:255–263. doi: 10.1016/j.freeradbiomed.2009.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strittmatter P, Ball EG. Formaldehyde dehydrogenase, a glutathione dependent enzyme system. J Biol Chem. 1955;213:445–461. [PubMed] [Google Scholar]
- 16.Achkor H, Diaz M, Fernández MR, Biosca JA, Pares X, Martinez MC. Enhanced formaldehyde detoxification by overexpression of glutathione-dependent formaldehyde dehydrogenase from Arabidopsis. Plant Physiol. 2003;132:2248–2255. doi: 10.1104/pp.103.022277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staab CA, Hellgren M, Höög JO. Medium- and short-chain dehydrogenase/reductase gene and protein families: Dual functions of alcohol dehydrogenase 3: implications with focus on formaldehyde dehydrogenase and S-nitrosoglutathione reductase activities. Cell Mol Life Sci. 2008;65:3950–3960. doi: 10.1007/s00018-008-8592-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen DE, Belka GK, Du Bois GC. S-nitrosoglutathione is a substrate for rat alcohol dehydrogenase class III isoenzyme. Biochem J. 1998;331:659–668. doi: 10.1042/bj3310659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu L, Hausladen A, Zeng M, Que L, Heitman J, Stamler JS. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–494. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- 20.Giese M, Bauer-Doranth U, Langebartels C, Sandermann H., Jr Detoxification of Formaldehyde by the Spider Plant (Chlorophytum comosum L.) and by Soybean (Glycine max L.) cell-suspension cultures. Plant Physiol. 1994;104:1301–1309. doi: 10.1104/pp.104.4.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uotila L, Koivusalo M. Purification of formaldehyde and formate dehydrogenase from pea seeds by affinity chromatography and S-formylglutathione as the intermediate of formaldehyde metabolism. Arch Biochem Biophys. 1979;196:33–45. doi: 10.1016/0003-9861(79)90548-4. [DOI] [PubMed] [Google Scholar]
- 22.Shafqat J, El-Ahmad M, Danielsson O, Martínez MC, Persson B, Parés X, et al. Pea formaldehyde-active class III alcohol dehydrogenase: Common derivation of the plant and animal forms but not of the corresponding ethanol-active forms (classes I and P) Proc Natl Acad Sci USA. 1996;93:5595–5599. doi: 10.1073/pnas.93.11.5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fliegmann J, Sandermann H., Jr Maize glutathione-dependent formaldehyde dehydrogenase cDNA: A novel plant gene of detoxification. Plant Mol Biol. 1997;34:843–854. doi: 10.1023/a:1005872222490. [DOI] [PubMed] [Google Scholar]
- 24.Wippermann U, Fliegmann J, Bauw G, Langebartels C, Maier K, Sandermann H., Jr Maize glutathione-dependent formaldehyde dehydrogenase: Protein sequence and catalytic properties. Planta. 1999;208:12–18. doi: 10.1007/s004250050529. [DOI] [PubMed] [Google Scholar]
- 25.Dolferus R, Osterman JC, Peacok WJ, Denis ES. Cloning of the Arabidopsis and rice formaldehyde dehydrogenase genes: Implications of the origin of plant ADH enzymes. Genetics. 1997;146:1131–1141. doi: 10.1093/genetics/146.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martínez MC, Achkor H, Persson B, Fernández MR, Shafqat J, Farrés J, et al. Arabidopsis formaldehyde dehydrogenase. Molecular properties of plant class III alcohol dehydrogenase provide further insights into the origins, structure and function of plant class p and liver class I alcohol dehydrogenases. Eur J Biochem. 1996;241:849–857. doi: 10.1111/j.1432-1033.1996.00849.x. [DOI] [PubMed] [Google Scholar]
- 27.Sakamoto A, Ueda M, Morikawa H. Arabidopsis glutathione-dependent formaldehyde dehydrogenase is an S-nitrosoglutathione reductase. FEBS Letters. 2002;515:20–24. doi: 10.1016/s0014-5793(02)02414-6. [DOI] [PubMed] [Google Scholar]
- 28.Lee U, Wie C, Fernandez BO, Feelisch M, Vierling E. Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. Plant Cell. 2008;20:786–802. doi: 10.1105/tpc.107.052647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 30.Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2:718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Espunya MC, Diaz M, Moreno-Romero J, Martinez MC. Modification of intracellular levels of glutathione-dependent formaldehyde dehydrogenase alters glutathione homeostasis and root development. Plant Cell Environ. 2006;29:1002–1011. doi: 10.1111/j.1365-3040.2006.01497.x. [DOI] [PubMed] [Google Scholar]
- 32.Reumann S, Babujee L, Ma C, Wienkoop S, Siemsen T, Antonicelli GE, et al. Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways and defense mechanisms. Plant Cell. 2007;19:3170–3193. doi: 10.1105/tpc.107.050989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen R, Sun S, Wang C, Li Y, Liang Y, An F, et al. The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Res. 2009;19:1377–1387. doi: 10.1038/cr.2009.117. [DOI] [PubMed] [Google Scholar]
- 34.Feechan A, Kwon E, Yun BW, Wang Y, Pallas JA, Loake GJ. A central role for S-nitrosothiols in plant disease resistance. Proc Natl Acad Sci USA. 2005;102:8054–8059. doi: 10.1073/pnas.0501456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rustérucci C, Espunya MC, Díaz M, Chabannes M, Martínez MC. S-nitrosoglutathione reductase affords protection against pathogens in Arabidopsis, both locally and systemically. Plant Physiol. 2007;143:1282–1292. doi: 10.1104/pp.106.091686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong JK, Yun BW, Kang JG, Raja MU, Kwon E, Sorhagen K. Nitric oxide function and signaling in plant disease resistance. J Exp Bot. 2008;59:147–154. doi: 10.1093/jxb/erm244. [DOI] [PubMed] [Google Scholar]
- 37.Chaki M, Fernández-Ocaña AM, Valderrama R, Carreras A, Esteban FJ, Luque F, et al. Involvement of reactive nitrogen and oxygen species (RNS and ROS) in sunflower-mildew interaction. Plant Cell Physiol. 2009;50:265–279. doi: 10.1093/pcp/pcn196. [DOI] [PubMed] [Google Scholar]
- 38.Barroso JB, Corpas FJ, Carreras A, Rodríguez-Serrano M, Esteban FJ, Fernández-Ocaña A, et al. Localization of S-nitrosoglutathione and expression of S-nitrosoglutathione reductase in pea plants under cadmium stress. J Exp Bot. 2006;57:1785–1793. doi: 10.1093/jxb/erj175. [DOI] [PubMed] [Google Scholar]
- 39.Leterrier M, Airaki, Barroso JB, Palma JM, del Río LA, Corpas FJ. Arsenic imparirs the metabolismo f RNS and ROS in Arabidopsis plants. Salamanca: International Symposium on the Pathophysiology of Reactive Oxygen and Nitrogen Species. 2010. ISBN: 978-84-692-9284-6. [Google Scholar]
- 40.Chaki M. Function of reactive nitrogen species in sunflower (Helianthus annuus) in response to abiotic and biotic stresses. Ph.D. Thesis. University of Jaén, Spain: 2007. p. 241. [Google Scholar]
- 41.Corpas FJ, del Río LA, Barroso JB. Need of biomarkers of nitrosative stress in plants. Trends Plant Sci. 2007;12:436–438. doi: 10.1016/j.tplants.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 42.Chaki M, Valderrama R, Fernández-Ocaña AM, Carreras A, Gómez-Rodríguez MV, López-Jaramillo J, et al. Mechanical wounding induces a nitrosative stress by downregulation of GSNO reductase and a rise of S-nitrosothiols in sunflower (Helianthus annuus) seedlings. J Exp Bot. 2010;62:1803–1813. doi: 10.1093/jxb/erq358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Airaki M, Leterrier M, Mateos RM, Valderrama R, Chaki M, Barroso JB, del Río LA, Palma JM, Corpas FJ. Metabolism of reactive oxygen species and reactive nitrogen species in pepper (Capsicum annuum L.) plants under low temperature stress. Plant Cell Environ. 2011 doi: 10.1111/j.1365-3040.2011.02310.x. [DOI] [PubMed] [Google Scholar]
- 44.Díaz M, Achkor H, Titarenko E, Martínez MC. The gene encoding glutathione-dependent formaldehyde dehydrogenase/GSNO reductase is responsive to wounding, jasmonic acid and salicylic acid. FEBS Lett. 2003;543:136–139. doi: 10.1016/s0014-5793(03)00426-5. [DOI] [PubMed] [Google Scholar]