Abstract
Circadian rhythms are a universal way for organisms, ranging from prokaryotes to humans, to maintain coordination with the daily changes of light and temperature. It is known that a functional circadian clock confers enhanced fitness. In both animals and plants, diverse physiological processes are affected by the clock and more than 10% of transcripts show a circadian rhythm. Recent advances in the field have revealed a link between circadian regulated gene expression and dynamic changes in chromatin. Jumonji C (JmjC) domain-containing proteins have been shown to be involved in chromatin remodeling, acting as histone demethylases. The recent discovery that a JmjC domain-containing protein functions as a novel clock component suggests that histone modification has evolved as an important mechanism at the core of the circadian machinery.
Key words: Arabidopsis, circadian clock, circadian rhythm, chromatin remodeling, Jumonji C domain, histone demethylase
The Circadian Clock
Biological rhythms with a period close to 24 h are called circadian rhythms. Circadian rhythms are autoregulatory, endogenous rhythms that allow organisms to anticipate rhythmic changes in the environment and accordingly adjust their cellular and physiological activities, thus providing them with an adaptive advantage.1–3 In all kingdoms of life, the circadian clock regulates a wide variety of physiological processes such as human sleep/wake cycles,4 fungal sporulation,5 plant growth and flowering time.6,7 Without a circadian system, organisms survive less well.2,3
Circadian systems can be divided conceptually into three parts: the input pathways that receive environmental cues (light and temperature) and entrain the oscillator; the central oscillator that generates rhythmicity; and the output pathways that create overt rhythmic processes. The central oscillator is the set of components that can maintain an endogenous rhythm of about 24 h even in the absence of external cues. In eukaryotic organisms, the central oscillator is, in principle, similar in different kinds of organisms and involves interaction of positive and negative elements that influence each other's expression or activity in interconnected feedback loops (Fig. 1). Crucial to the correct function of the oscillator is that the feedback loop takes approximately 24 h to complete and this is achieved by multiple levels of post-translational controls that are built into the system.
Figure 1.
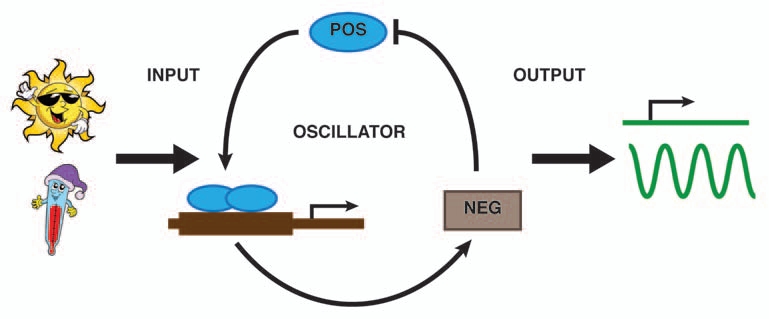
Simplified molecular model of a core circadian clock. Positive transcriptional elements (POS) rhythmically induce the expression of the negative elements (NEG), which in turn repress the POS.
The Molecular Mechanism of the Arabidopsis Clock
In Arabidopsis, three genes have been suggested as components of the central oscillator: CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY) and TIMING OF CAB EXPRESSION 1 (TOC1). CCA1 and LHY are closely related MYB-like transcription factors. They both have a circadian rhythm of expression peaking soon after dawn, and their overexpression leads to dramatically reduced levels of both transcripts and causes arrythmicity in gene expression, leaf movement and hypocotyl elongation.8,9 CCA1 and LHY reset the phase of the circadian clock after a transient increase in their expression,10 further supporting their roles as central oscillator components. However, the double mutant still retains robust rhythmicity with short period, suggesting that other MYB-related proteins of this multigene family may function redundantly.11,12 TOC1, also known as PSEUDORESPONSE REGULATOR 1 (PRR1), encodes a nuclear protein containing a receiver domain similar to that of response regulators from bacterial two-component signaling systems.13 Mutation of TOC1 causes period shortening and overexpression results arrhythmicity.14 Expression of TOC1 peaks at dusk, which is anti-phase with the expression of CCA1 and LHY.15 CCA1 and LHY repress TOC1 expression by direct binding to the EE (evening element) in its promoter,16,17 while TOC1 activates transcription of CCA1 and LHY through CHE (CCA1 HIKING EXPEDITION) and other unknown mechanisms.18
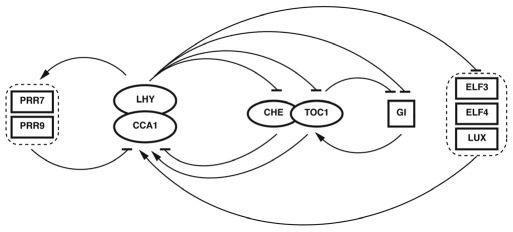
There are a number of other genes that have been shown to affect circadian cycling and three extra feedback loops have been proposed based on experimental observations and mathematical modeling (Fig. 2). Evening-phased clock components, EARLY FLOWERING 3 and 4 (ELF3 and ELF4) and a MYB transcription factor LUX ARRYTHMO (LUX; also known as PHYTOCLOCK1) contribute to the positive regulation of CCA1 and LHY rhythmic expression and are essential for maintaining robust rhythms in constant conditions.19–24 To close the second loop, CCA1 and LHY repress the expression of ELF3, ELF4 and LUX as they do TOC1. In the third loop, PRR7 and PRR9, which are two TOC1 homologs with morning expression, inhibit the expression of CCA1 and LHY through direct binding to their promoter.25 In turn, CCA1 and LHY have been shown to participate in the positive regulation of PRR7 and PRR9,26 suggesting the existence of this morning loop. The fourth loop involves the induction of TOC1 by an evening-expressed clock protein GIGANTEA (GI), which is known to be negatively regulated by TOC1, CCA1 and LHY.27 The presence of multiple interlocked loops is thought to make the clock more robust and less likely to be disturbed by the environmental noise. Despite the progress in linking interactions between the clock proteins,28 our understanding of the plant circadian system is far from complete. Identifying novel clock components and dissecting interactions among them will help us to elucidate the complex signal transduction network in the central oscillator.
Figure 2.
Four interlocked feedback loops at the core of the Arabidopsis oscillator. In the central loop, CCA1 and LHY act as negative elements to repress the expression of a positive element TOC1. TOC1 is involved in the positive regulation of CCA1 and LHY, partially through antagonizing CHE through protein interaction. Experimental observations and mathematical modeling have incorporated three interlocked loops on the top of the central loop. CCA1 and LHY repress the expression of ELF3, ELF4 and LUX, which in turn have positive effects on the expression of CCA1 and LHY through unknown mechanism. In the morning loop, PRR7 and PRR9 repress the expression of CCA1 and LHY, which in turn activate PRR7 and PRR9. In the evening loop, GI promotes TOC1 expression, which in turn represses GI. Arrows indicate transcriptional activation and horizontal bars indicate transcriptional repression.
Chromatin Remodeling and Circadian Rhythms
Circadian clock regulation of the transcriptome is a widespread phenomenon. More than 10% of transcripts oscillate in a circadian manner in both animals and plants.29,30 A recent study indicates that about 90% of Arabidopsis transcripts cycles in at least one condition when seedlings were exposed to different diurnal and circadian cycles.31 How does the circadian clock control the large number of oscillating transcripts? Recent advances in the field have linked the dynamic chromatin remodeling with the circadian clock.
Eukaryotic chromatin is composed of nucleosomes in which 146 bp of DNA wraps around an octamer of four histone proteins, H2A, H2B, H3 and H4. Histones, in particular their N-terminal tails, are subjected to various post-translational modifications, which play important roles in chromatin remodeling, DNA repair and transcriptional regulation.32,33 Histone modification, such as acetylation and methylation, can function as a molecular switch between a relaxed chromatin (transcriptionally permissive) and a compacted one (transcriptionally repressive).32,33 Circadian changes in histone modifications at the promoters of clock genes have been well documented. In Neurospora, an ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH (CSW) is required for clock function. CSW localizes to the promoter of the central clock gene FREQUENCY (frq) and regulates frq expression through controlling the accessibility of promoter DNA.34 In mammals, transcriptional regulation of the core clock genes is accompanied by rhythms in histone H3 acetylation and RNA polymerase II binding.35 In Arabidopsis, the expression of TOC1 is affected by clock-controlled cycles of histone acetylation, although the responsible enzymes are not known.17 Finally, transcription factor CLOCK protein, an essential component of the mammalian circadian system, has been shown to be a histone acetyltransferase,36 suggesting that histone acetylation is crucial for core clock mechanisms.
Although most studies have focused on histone acetylation in the circadian system, histone methylation has also been suggested to be important for clock function. Mammalian histone methyltransferase EZH2 (enhancer of zeste), a polycomb group enzyme that mediates the methylation of H3K27 at the promoter of the central clock gene Period is required for clock function.37 The WDR5 protein, a member of a histone methyltransferase complex, interacts with PERIOD1 (PER1) and mediates the rhythmic methylation of H3K4 and H3K9 at the promoter of PER1-regulated genes.38 In Arabidopsis, histone dimethylation at the CCA1, LHY, TOC1 and GI promoters positively correlates with their expression.39 Recent studies have shown that a Jumonji C (JmjC) domain-containing protein, which is generally known as a histone demethylase, functions in both the plant and human circadian systems, suggesting that histone methylation is an important regulatory mechanism in the eukaryote circadian clock.40,41
Jumonji C Domain-Containing Proteins
The methylation status of histones regulates chromatin structure and gene expression in eukaryotes. Histone methylation is a dynamic modification which can be associated with activation or repression of gene expression, depending on the methylated residue and the degrees of methylation.42 Jumonji C (JmjC) domain-containing proteins, a class of histone demethylases, directly reverse histone methylation through an oxidative reaction that requires Fe (II) and α-ketoglutarate as cofactors.43 The JmjC domain-containing proteins are evolutionarily conserved in species spanning from yeast to human. They are involved in a wide range of biological processes, such as embryonic stem cell self-renewal,44 animal posterior development,45 tumor suppression,46,47 X-linked mental retardation,48 neural stem cell differentiation,49 and metabolic gene expression and obesity resistance.50
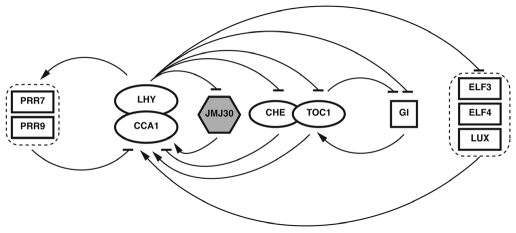
In Arabidopsis, there are twenty-one JmjC domain-containing proteins, which can be classified into five groups based on the sequence similarity in the JmjC domain.51 Although all 21 genes are actively expressed,52 only a few have been characterized and they have been shown to be involved in gametophyte development,53 cytosine methylation,54 brassinosteroid responses,55 RNA silencing56,57 and flowering time.58–60 Recent studies showed that the JmjC domain protein At JMJ30 (also known as JMJD5) regulates the pace of the circadian clock in both the Arabidopsis and the human circadian system.40,41 JMJ30 is the only gene of the 21 members that shows a robust circadian rhythm at the level of transcription. JMJ30 peaks at dusk and is co-regulated across developmental and circadian time with an evening-phased clock gene TOC1. Chromatin immunoprecipitation (ChIP) assays revealed that JMJ30 is a direct target of the morning-phased clock components, transcription factors CCA1 and LHY and JMJ30 expression is drastically reduced in seedlings overexpressing CCA1 or LHY, suggesting that CCA1 and LHY bind directly to the JMJ30 promoter to repress its expression.41 In turn, CCA1 and LHY have reduced expression in jmj30 loss-of-function mutants grown under high levels of red light, indicating that JMJ30 has a positive effect on CCA1 and LHY expression.40 Together, these findings suggest a potential negative feedback loop between CCA1/LHY and JMJ30 in the central oscillator (Fig. 3). Both loss- and gain-of-function mutants of JMJ30 shorten the free-running circadian period indicating that JMJ30 is a circadian clock component involved in controlling the pace of the clock. Interestingly, a similar period-shortening phenotype was observed in the mammalian cells deficient for the human ortholog of JMJ30, which has been shown to have histone demethylase activity.46 In addition, the JMJ30 human ortholog is able to rescue the short-period phenotype of Arabidopsis jmj30 loss-of-function mutants and vice versa, suggesting that JMJ30 has conserved function in both Arabidopsis and human circadian systems.40 These findings not only reveal the importance of histone methylation in the circadian clock mechanism, but also open up a gateway to explore the connection between chromatin remodeling and the circadian clock.
Figure 3.
Five interlocked feedback loops at the core of the Arabidopsis oscillator. One extra loop in which CCA1 and LHY repress the expression of JMJ30 and JMJ30 has positive effect on the expression of CCA1 and LHY has been incorporated on the top of four-loop model shown in Figure 2.
Conclusions and Future Perspectives
Circadian rhythms control key metabolic and physiological pathways in almost all organisms. Circadian function is largely based on the complex program of gene expression which involves dynamic changes in chromatin structure. In plants, chromatin remodeling plays important roles in various biological processes associated with photomorphogenesis, floral transition, hormone signaling and stress responses. The molecular nature of clock-controlled chromatin remodeling that responds to environmental stimuli has yet to be determined. Experimental description of the details of the molecular mechanism through which JMJ30 regulates the pace of the circadian clock will allow elucidation of the function of JMJ30 within the circadian clock. Further characterization of the functional and the evolutionary characteristics of JmjC domain-containing proteins in the circadian system will provide a new conceptual understanding of how histone methylation evolved in the core of the circadian machinery. Better understanding how different histone modifications interact to regulate the transcriptional state core clock genes will further our knowledge about how clocks respond to the daily and seasonal environmental changes and confer enhanced fitness onto the organism.
Acknowledgements
This work was supported by NIH grant GM23167 to E.M.T.
References
- 1.Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green RM, Tingay S, Wang ZY, Tobin EM. Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 2002;129:576–584. doi: 10.1104/pp.004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, et al. Plant circadian clocks increase photosynthesis, growth, survival and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 4.Moore RY. Circadian rhythms: basic neurobiology and clinical applications. Annu Rev Med. 1997;48:253–266. doi: 10.1146/annurev.med.48.1.253. [DOI] [PubMed] [Google Scholar]
- 5.Dunlap JC. Closely watched clocks: molecular analysis of circadian rhythms in Neurospora and Drosophila. Trends Genet. 1990;6:159–165. doi: 10.1016/0168-9525(90)90151-u. [DOI] [PubMed] [Google Scholar]
- 6.Imaizumi T, Kay SA. Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci. 2006;11:550–558. doi: 10.1016/j.tplants.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Dowson-Day MJ, Millar AJ. Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J. 1999;17:63–71. doi: 10.1046/j.1365-313x.1999.00353.x. [DOI] [PubMed] [Google Scholar]
- 8.Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, et al. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- 9.Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- 10.Knowles SM, Lu SX, Tobin EM. Testing Time: Can Ethanol-Induced Pulses of Proposed Oscillator Components Phase Shift Rhythms in Arabidopsis? J Biol Rhyth. 2008;23:463–471. doi: 10.1177/0748730408326749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Locke JC, Southern MM, Kozma-Bognar L, Hibberd V, Brown PE, Turner MS, et al. Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Syst Biol. 2005;1:13. doi: 10.1038/msb4100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu SX, Knowles SM, Andronis C, Ong MS, Tobin EM. CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 2009;150:834–843. doi: 10.1104/pp.108.133272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, et al. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- 14.Más P, Alabadi D, Yanovsky MJ, Oyama T, Kay SA. Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell. 2003;15:223–236. doi: 10.1105/tpc.006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Somers DE, Webb AA, Pearson M, Kay SA. The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development. 1998;125:485–494. doi: 10.1242/dev.125.3.485. [DOI] [PubMed] [Google Scholar]
- 16.Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 17.Perales M, Más P. A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell. 2007;19:2111–2123. doi: 10.1105/tpc.107.050807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323:1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA. 2005;102:10387–10392. doi: 10.1073/pnas.0503029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Onai K, Ishiura M. PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells. 2005;10:963–972. doi: 10.1111/j.1365-2443.2005.00892.x. [DOI] [PubMed] [Google Scholar]
- 21.Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognar L, Nagy F, et al. The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature. 2002;419:74–77. doi: 10.1038/nature00954. [DOI] [PubMed] [Google Scholar]
- 22.Kikis EA, Khanna R, Quail PH. ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J. 2005;44:300–313. doi: 10.1111/j.1365-313X.2005.02531.x. [DOI] [PubMed] [Google Scholar]
- 23.Covington MF, Panda S, Liu XL, Strayer CA, Wagner DR, Kay SA. ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell. 2001;13:1305–1315. doi: 10.1105/tpc.13.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR. ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell. 2001;13:1293–1304. doi: 10.1105/tpc.13.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara H. PSEUDO-RESPONSE REGULATORS 9, 7 and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell. 2010;22:594–605. doi: 10.1105/tpc.109.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farre EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol. 2005;15:47–54. doi: 10.1016/j.cub.2004.12.067. [DOI] [PubMed] [Google Scholar]
- 27.Locke JC, Southern MM, Kozma-Bognar L, Hibberd V, Brown PE, Turner MS, et al. Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Syst Biol. 2005;1:13. doi: 10.1038/msb4100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McClung CR. Plant circadian rhythms. Plant Cell. 2006;18:792–803. doi: 10.1105/tpc.106.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. Circadian programs of transcriptional activation, signaling and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12:551–557. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- 30.Edwards KD, Anderson PE, Hall A, Salathia NS, Locke JC, Lynn JR, et al. FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell. 2006;18:639–650. doi: 10.1105/tpc.105.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 2008;4:e14. doi: 10.1371/journal.pgen.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 33.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Belden WJ, Loros JJ, Dunlap JC. Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol Cell. 2007;25:587–600. doi: 10.1016/j.molcel.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 35.Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 36.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 37.Etchegaray JP, Yang X, DeBruyne JP, Peters AH, Weaver DR, Jenuwein T, et al. The polycomb group protein EZH2 is required for mammalian circadian clock function. J Biol Chem. 2006;281:21209–21215. doi: 10.1074/jbc.M603722200. [DOI] [PubMed] [Google Scholar]
- 38.Brown SA, Ripperger J, Kadener S, Fleury-Olela F, Vilbois F, Rosbash M, et al. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science. 2005;308:693–696. doi: 10.1126/science.1107373. [DOI] [PubMed] [Google Scholar]
- 39.Ni Z, Kim ED, Ha M, Lackey E, Liu J, Zhang Y, et al. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 2009;457:327–331. doi: 10.1038/nature07523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones MA, Covington MF, Ditacchio L, Vollmers C, Panda S, Harmer SL. Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc Natl Acad Sci USA. 2010;107:21623–21628. doi: 10.1073/pnas.1014204108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu SX, Knowles SM, Webb CJ, Celaya RB, Cha C, Siu JP, et al. The JmjC domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol. 2011;155:906–915. doi: 10.1104/pp.110.167015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 43.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 44.Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 46.Hsia DA, Tepper CG, Pochampalli MR, Hsia EY, Izumiya C, Huerta SB, et al. KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation. Proc Natl Acad Sci USA. 2010;107:9671–9676. doi: 10.1073/pnas.1000401107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki T, Minehata K, Akagi K, Jenkins NA, Copeland NG. Tumor suppressor gene identification using retroviral insertional mutagenesis in Blm-deficient mice. EMBO J. 2006;25:3422–3431. doi: 10.1038/sj.emboj.7601215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, et al. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 49.Jepsen K, Solum D, Zhou T, McEvilly RJ, Kim HJ, Glass CK, et al. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450:415–419. doi: 10.1038/nature06270. [DOI] [PubMed] [Google Scholar]
- 50.Tateishi K, Okada Y, Kallin EM, Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature. 2009;458:757–761. doi: 10.1038/nature07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hong EH, Jeong YM, Ryu JY, Amasino RM, Noh B, Noh YS. Temporal and spatial expression patterns of nine Arabidopsis genes encoding Jumonji C-domain proteins. Mol Cells. 2009;27:481–490. doi: 10.1007/s10059-009-0054-7. [DOI] [PubMed] [Google Scholar]
- 52.Lu F, Li G, Cui X, Liu C, Wang XJ, Cao X. Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J Integr Plant Biol. 2008;50:886–896. doi: 10.1111/j.1744-7909.2008.00692.x. [DOI] [PubMed] [Google Scholar]
- 53.Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, et al. Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development. 2005;132:603–614. doi: 10.1242/dev.01595. [DOI] [PubMed] [Google Scholar]
- 54.Saze H, Shiraishi A, Miura A, Kakutani T. Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science. 2008;319:462–465. doi: 10.1126/science.1150987. [DOI] [PubMed] [Google Scholar]
- 55.Yu X, Li L, Guo M, Chory J, Yin Y. Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc Natl Acad Sci USA. 2008;105:7618–7623. doi: 10.1073/pnas.0802254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Searle IR, Pontes O, Melnyk CW, Smith LM, Baulcombe DC. JMJ14, a JmjC domain protein, is required for RNA silencing and cell-to-cell movement of an RNA silencing signal in Arabidopsis. Genes Dev. 2010;24:986–991. doi: 10.1101/gad.579910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deleris A, Greenberg MV, Ausin I, Law RW, Moissiard G, Schubert D, et al. Involvement of a Jumonji-C domain-containing histone demethylase in DRM2-mediated maintenance of DNA methylation. EMBO Rep. 2010;11:950–955. doi: 10.1038/embor.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Noh B, Lee SH, Kim HJ, Yi G, Shin EA, Lee M, et al. Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell. 2004;16:2601–2613. doi: 10.1105/tpc.104.025353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang W, Jiang D, Jiang J, He Y. A plant-specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis. Plant J. 2010;62:663–673. doi: 10.1111/j.1365-313X.2010.04182.x. [DOI] [PubMed] [Google Scholar]
- 60.Lu F, Cui X, Zhang S, Liu C, Cao X. JMJ14 is an H3K4 demethylase regulating flowering time in Arabidopsis. Cell Res. 2010;20:387–390. doi: 10.1038/cr.2010.27. [DOI] [PubMed] [Google Scholar]