Abstract
We previously reported that the SLEEPY1 (SLY1) homolog, F-box gene SNEEZY/SLEEPY2 (SNE/SLY2), can partly replace SLY1 in gibberellin (GA) hormone signaling through interaction with DELLAs RGA and GAI. To determine whether SNE normally functions in GA signaling, we characterized the phenotypes of two T-DNA alleles, sne-t2 and sne-t3. These mutations result in no apparent vegetative phenotypes, but do result in increased ABA sensitivity in seed germination. Double mutants sly1-t2 sne-t2 and sly1-t2 sne-t3 result in a significant decrease in plant fertility and final plant height compared to sly1-t2. The fact that sne mutations have an additive effect with sly1 suggests that SNE normally functions as a redundant positive regulator of GA signaling.
Key words: gibberellin signaling, GA, SLEEPY1, SNEEZY, DELLA, F-box protein
This paper describes genetic evidence that the SLEEPY1 (SLY1) homolog SNEEZY/SLEEPY2 (SNE/SLY2) functions redundantly with SLY1 to stimulate gibberellin signaling. GA responses such as seed germination, stem elongation and fertility are promoted by proteolysis of DELLA proteins, negative regulators of the GA signaling.1 In the classic GA signaling model, GA binding to the GA receptor GID1 increases GID1 affinity for DELLA protein. This GID1-GA binding to DELLA causes SLY1, the F-box subunit of an SCF E3 ubiquitin ligase complex, to recognize, bind and ubiquitinate DELLA proteins thereby targeting them for destruction by the 26S proteasome. Thus, loss of SLY1 function results in decreased GA responses, causing dwarfism, delayed flowering, infertility and seed dormancy. The sly1 mutants over-accumulate DELLA proteins due to failure to destroy them through the ubiquitin-proteasome pathway.
Overexpression of the SLY1 homolog, SNE, partially rescues the germination, dwarfism and infertility of the sly1-10 mutant.2–4 SNE overexpression in the sly1-10 background is associated with reduced accumulation of DELLA proteins RGA and GAI, but not of DELLA RGL2. Co-immunoprecipitation assays demonstrated that SNE directly binds RGA protein as well as the cullin subunit of the SCF E3 complex. These recently published data suggest that SNE forms a functional SCF E3 ubiquitin ligase complex that negatively regulates a subset of the DELLA proteins regulated by SLY1.4
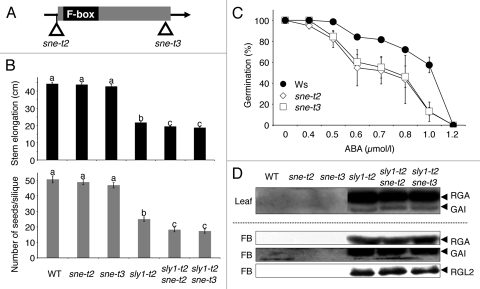
The finding that SNE overexpression rescues sly1-10 phenotypes through down-regulation of DELLA RGA and GAI suggests that SNE is normally a positive regulator of GA signaling. If this is true, then we expect sne mutations to cause phenotypes resulting from reduced GA response including reduced germination, stature and fertility. To examine this hypothesis, three sne T-DNA mutants were identified: sne-t1, sne-t2 and sne-t3. The sne-t1 allele is a SALK line containing a T-DNA insertion 183-bp upstream of the coding region.5 This line showed no apparent phenotype and was not further characterized. The sne-t2 allele is a Sussman T-DNA line4,6 containing a T-DNA insertion immediately before the ATG that is the SNE translational start codon (Fig. 1A). While this insertion does not disrupt the coding region, it likely disrupts SNE protein translation as the T-DNA contains multiple stop codons. The sne-t3 allele contains a T-DNA insertion within the SNE ORF before amino acid 146 of the 157 amino acid predicted protein (FLAG_461E03).7,8 This allele should result in loss of the last 11 SNE amino acids. We know that loss of the last 8 SLY1 amino acids in sly1-10 results in dwarfism, suggesting that loss of the last 11 SNE amino acids may also cause some loss of function in the small F-box protein. When the homozygous sne-t2 and sne-t3 lines were compared to wild-type Ws, no change was observed either in final plant height or fertility measured in seeds/silique (Fig. 1B). An ABA dose-response curve in seed germination detected a small but reproducible increase in ABA sensitivity during seed germination of sne-t2 and sne-t3 (Fig. 1C). The fact that the sne-t2 and sne-t3 mutants, like sly1-2 and sly1-10, show increased ABA sensitivity suggests that SNE and SLY1 may have similar functions in GA signaling during seed germination.2
Figure 1.
The phenotypes of sne-t2 and sne-t3 T-DNA mutants. (A) Schematic diagram of the sne-t2 T-DNA insertion at position −1 bp and of sne-t3 at position +435 bp with respect to the translation start site. (B) Final plant height (upper) and fertility (lower) of indicated genotypes. Letters indicate statistically different classes as determine by t-test. Bars represent standard error. (C) sne mutants show increase in ABA sensitivity. Seeds of wild-type Ws, sne-t2 and sne-t3 were after-ripened for 2 weeks then sown on MS-agar containing indicated concentrations of ABA as described by Steber et al.11 Germination was scored based on radical emergence after incubating 3 days at 4°C followed by 14 days at 22°C. (D) Mutations in SNE cause no significant effect on DELLA RGA, GAI and RGL2 protein accumulation. Total protein was extracted from leaves of 12-d-old seedlings (Top) or flower buds (FB, bottom) and detected as described in Ariizumi et al.4
One possible explanation for the lack of apparent GA-insensitive phenotypes in sne T-DNA insertion lines, is that SNE function is redundant with SLY1 in GA signaling.9 If so, we would expect sly1 sne double mutants to show stronger GA-insensitive phenotypes than the sly1 single mutation. Double mutants were constructed containing either the sne-t2 or sne-t3 mutation in the sly-t2 null background. The sly1-t2 allele was chosen because sly1-t2, sne-t2 and sne-t3 are all in the Ws ecotype. The sly1-t2 allele contains a T-DNA insertion within the F-box domain resulting in severe GA-insensitive phenotypes including failure to germinate, reduced stature and infertility.10 The sly1-t2 sne-t2 and sly1-t2 sne-t3 double mutants showed a small but significant decrease in final plant height and fertility (seeds/silique) compared to sly1-t2 (Fig. 1B). This increase in phenotype severity was not associated with an apparent increase in DELLA RGA, GAI or RGL2 protein accumulation (Fig. 1D). It could be that DELLA protein levels in sly1-t2 are so high that any slight increase due to sne mutations is undetectable. Our previous study of SNE overexpression lines showed that SNE has the ability to downregulate RGA and GAI protein accumulation. Figure 1 shows that the chromosomal SNE gene contributes to GA signaling presumably through ubiquitination of DELLA protein.
Taken together, the fact that sne mutants show only mild GA-insensitive phenotypes and that the natural SNE expression cannot compensate for lack of SLY1, indicate that SLY1 is the main E3 ubiquitin ligase stimulating GA signaling (this study, reviewed in ref. 4). We cannot rule out the possibility that stronger SNE alleles would show either stronger GA response phenotypes or phenotypes that are unrelated to GA signaling. Indeed, there is evidence to suggest that SNE may have unique functions. The sne-t3 (sne-1) allele results in a shortened root phenotype.8 That SNE is expressed in the endodermis and quiescent center of the root whereas SLY1 is expressed in the stele, suggests that SNE may function independently in the root.8 Moreover, SNE overexpression, but not SLY1 overexpression, results in decreased apical dominance and a prone growth habit suggesting that SNE may play a unique role in development.2,4 Our model is that in addition to regulating DELLA proteins RGA and GAI, SNE may also regulate a yet unidentified target involved in apical dominance (Fig. 2). Future research will need to elucidate the role of SNE in Arabidopsis growth and development.
Figure 2.
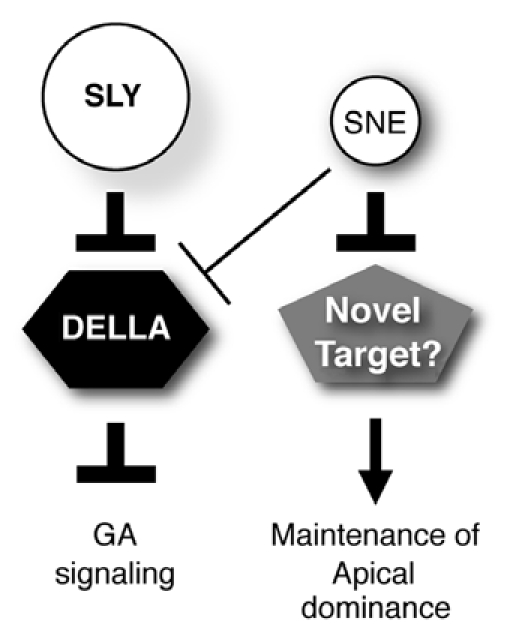
Model for SNE function in Arabidopsis. Both SLY1 and SNE act as positive regulators of GA responses via DELLA protein destruction. SNE may negatively regulate an unknown protein that maintains apical dominance.
Acknowledgements
We would like to thank R. Parveen for expert technical assistance. This work was supported by the Japan Society for the Promotion of Science (to T.A.) and the US Department of Agriculture—National Research Initiative (award no. 2002-01351 to C.M.S.).
References
- 1.Sun T. Gibberellin-GID1-DELLA: A pivotal regulatory module for plant growth and development. Plant Physiol. 2010;154:567–570. doi: 10.1104/pp.110.161554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strader LC, Ritchie S, Soule JD, McGinnis KM, Steber CM. Recessive-interfering mutations in the gibberellin signaling gene SLEEPY1 are rescued by overexpression of its homologue, SNEEZY. Proc Natl Acad Sci USA. 2004;101:12771–12776. doi: 10.1073/pnas.0404287101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu X, Richards DE, Fleck B, Xie D, Burton N, Harberd NP. The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell. 2004;16:1406–1418. doi: 10.1105/tpc.021386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ariizumi T, Lawrence PK, Steber CM. The role of two F-Box proteins, SLEEPY1 and SNEEZY, in Arabidopsis gibberellin signaling. Plant Physiol. 2011;155:765–775. doi: 10.1104/pp.110.166272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, et al. Genome-wide insertional mutagenesis. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 6.Sussman MR, Amasino RM, Young JC, Krysan PJ, Austin-Phillips S. The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol. 2000;124:1465–1467. doi: 10.1104/pp.124.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samson F, Brunaud V, Balzergue S, Dubreucq B, Lepiniec L, Pelletier G, et al. FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucl Acids Res. 2002;30:94–97. doi: 10.1093/nar/30.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui H, Benfey PN. Interplay between SCARECROW, GA and LIKE HETEROCHROMATIN PROTEIN 1 in ground tissue patterning in the Arabidopsis root. Plant J. 2009;58:1016–1027. doi: 10.1111/j.1365-313X.2009.03839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun TP, Steber CM. The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell. 2003;15:1120–1130. doi: 10.1105/tpc.010827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ariizumi T, Steber CM. Seed germination of GA-insensitive sleepy1 mutants does not require RGL2 protein disappearance in Arabidopsis. Plant Cell. 2007;19:791–804. doi: 10.1105/tpc.106.048009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steber CM, Cooney SE, McCourt P. Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics. 1998;149:509–521. doi: 10.1093/genetics/149.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]