Abstract
GRAS genes are a large family of streptophyte specific transcription factors that function in a diverse set of physiological and developmental processes. GRAS proteins of the HAIRY MERISTEM (HAM) sub-family are required for maintenance of shoot and root indeterminacy. The transcriptional targets of HAM proteins and the signaling inputs regulating HAM activity are completely unknown. Understanding the relationship of HAM proteins to other members of the GRAS family may inform hypotheses relating to cellular level HAM functions. Here I report a phylogenetic analysis of GRAS proteins employing the complete set of known and probable GRAS proteins from the sequenced genomes of the flowering plants Arabidopsis and Rice, the lycophyte Selaginella moellendorffii, and the bryophyte Physcomitrella patens. HAM proteins are most closely related to DELLA proteins, key components of gibberellin perception. However, GRAS proteins diversified into a minimum of 12 discreet monophyletic lineages, including the HAM and DELLA subfamilies, prior to divergence of the moss and flowering plant lineages. Substantial diversification of GRAS proteins at so early a point in land plant evolution suggests that relative relatedness among GRAS protein sub-families may not substantially reflect shared protein function.
Key words: GRAS, transcription factor, HAIRY MERISTEM, phylogenetics, Selaginella, Physcomitrella, land plant evolution
Improved GRAS Protein Phylogenies May Predict Cellular HAM Protein Functions
The Petunia HAIRY MERISTEM (HAM) protein, a member of the GRAS family of transcription factors, was first identified as a component of a novel signaling pathway in Petunia, promoting shoot indeterminacy.1 Petunia ham mutants exhibit a novel phenotype of cessation of organogenesis (meristem arrest) followed by or simultaneous with differentiation of the meristem as stem tissue. Three independent studies have recently extended analysis of HAM function from Petunia into Arabidopsis.2–4 My laboratory, in collaboration with Dr. Rossangela Sozzani, recently described the phenotypes of Arabidopsis ham1,2,3 and ham1,2,4 triple loss-of-function mutants, demonstrating that AtHAMs are required for maintenance of indeterminacy in the root as well as the shoot, and that aspects of ham loss-of-function phenotypes in both the shoot and the root are attributable to altered regulation of cell-division.2
Beyond a probable function as transcription factors,3,5 cellular-level functions of HAM proteins are entirely unknown. Direct transcriptional targets of HAM proteins, as well as proteins or other factors that directly interact with HAM proteins, remain to be identified. Hypotheses for testing cellular-level HAM functions may be informed by our understanding of the transcriptional targets and cellular interactions of related GRAS proteins that are understood to some detail.2 As the likelihood for shared function may correlate with the degree of relatedness, efforts to expand our understanding of HAM function provides impetus for understanding the phylogenetic relationship of HAM proteins with other subfamilies of GRAS proteins.
GRAS proteins contribute to the regulation of a diverse suite of physiological and developmental processes, including abiotic stress responses,6 phytochrome signal transduction,7 radial patterning,8,9 maintenance of organ indeterminacy,1,2 shoot meristem initiation,10,11 gibberellin signaling12 and nodulation.12 Genetic and biochemical characterization of GRAS protein function has been performed to date only in flowering plant species, but homologs of flowering plant GRAS genes have been identified in the lycophyte Selaginella, demonstrating that the GRAS family predates divergence of the lycophyte and euphyllophyte lineages.14 Given the impressive breadth of function displayed by GRAS proteins and their significance in regulating flowering plant development, questions of when and in what plant lineages specific classes of GRAS proteins arose, expanded and acquired novel functions, is of considerable interest within the broader contexts of elucidating GRAS function and reconstructing land plant evolution.
Major GRAS Protein Subfamilies Are Ancient
Earlier phylogenetic analyses of GRAS proteins, principally analyzing proteins from Arabidopsis and Rice, define eight major GRAS subfamilies largely congruent with the distinct functions of characterized GRAS proteins.15,16 However, the analysis of Tian and colleagues does not resolve relationships among the major GRAS protein subfamilies, while that of Bolle does not provide support values for inferred relationships. I therefore undertook a phylogenetic analysis of GRAS proteins, expanding upon earlier analyses with the incorporation of basal plant taxa GRAS protein sequences derived from the recently completed genome sequencing of the moss Physcomitrella patens and the lycophyte Selaginella moellendorffii, in order to gauge the extent of GRAS and HAM protein antiquity.
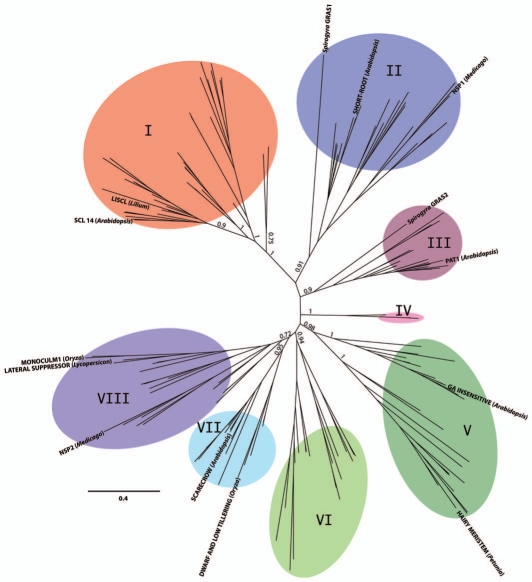
Bayesian analysis of 174 GRAS proteins, including the complete or near-complete set of GRAS-proteins from Arabidopsis thaliana, Oryza sativa, Selaginella moellendorffii and Physcomitrella patens, along with selected GRAS proteins from other taxa that have been subjected to genetic or biochemical analyses of function,2,10,11,13,17–19 and two GRAS proteins from the aquatic, filamentous Streptophyte Spirogyra generated a phylogenetic tree, from which eight major GRAS protein subfamilies, designated Clades I–VIII, were designated based upon the earlier analyses of Bolle and Tian and colleagues (Fig. 1, Sup. Fig. 1, Sup. Tab. 1, Sup. Methods). The phylogenetic tree in Figure 1 is largely congruent with the earlier analysis of Bolle.15 All of the major subfamilies of our analysis are well supported, with posterior probabilities of 0.9 or greater, the sole exception being Clade VIII with a posterior probability of 0.72. Though well-supported, Clade IV, consisting solely of three proteins, two from Rice and one from Selaginella, is perhaps artifactual, the branch lengths indicating relatively rapid rates of protein evolution, which may have generated Clade IV as a result of long-branch attraction.20
Figure 1.
Phylogeny of GRAS proteins. Unrooted tree of aligned protein sequences from 174 GRAS proteins, derived from Bayesian inference. Sequences include the inferred complete set of GRAS proteins from Arabidopsis thaliana (32 proteins), Oryza sativa (50 proteins), Selaginella moellendorffii (43 proteins) and Physcomitrella patens (39 proteins), along with characterized GRAS proteins from Lilium longiflorum, Petunia hybrida, Lycopersicon esculentum, Medicago truncatula and two GRAS proteins from Spirogyra.20,21 Eight monophyletic clades are delineated and numerically labeled in clockwise fashion, following the subfamily groupings established by Bolle.15 Support values for designated clades denote posterior probabilities. Genetically or biochemically characterized GRAS proteins are indicated for references 1, 6–13, and 17–19. Scale bar indicates substitutions per site.
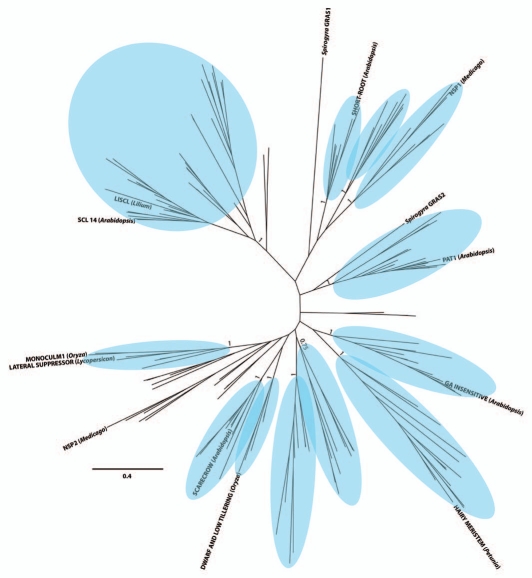
The tree contains 12 well-supported monophyletic clades that include proteins from Physcomitrella, Selaginella and flowering plants (Fig. 2), consistent with the common ancestor of the moss and vascular plant lineages possessing no fewer than 12 GRAS genes. The presence of 12 or more GRAS genes at such an early date, is consistent with either a rapid diversification in GRAS genes shortly following the transition of streptophytes to land or alternatively, with GRAS genes having undergone substantial diversification in aquatic streptophytes preceding the transition to land. Grouping of two GRAS proteins from the aquatic, filamentous streptophyte algae Spirogyra in well-supported internal monophyletic clades suggests that the origin of the GRAS gene family and at least a portion of GRAS gene diversification preceded the transition of streptophytes to terrestrial environments.
Figure 2.
GRAS protein phylogram shown in Figure 1 with 12 monophyletic clades containing Physcomitrella, Selaginella and flowering plant GRAS proteins delineated with blue ovals. Posterior probabilities of delineated clades are indicated.
Substantial expansion of GRAS genes has occurred within specific GRAS subfamilies in a manner specific both to the particular GRAS subfamily and to the particular taxa analyzed, suggesting that GRAS genes have been utilized extensively in the evolutionary history of mosses, lycophytes and euphyllophytes, but in highly lineage specific ways (Fig. 3). S. moellendorffii in particular appears to have relied heavily upon the GRAS family during its evolutionary history, as the S. moellendorffii genome contains more probable GRAS encoding genes relative to genome size than P. patens, Rice or Arabidopsis.
Figure 3.
Relative representation of Physcomitrella, Selaginella, Arabidopsis and Rice GRAS homologs within the major clades delineated in Figure 1. Clade IV is not included in this analysis.
Rooting the tree shown in Figure 1 would allow us to determine which clades and associated functions are more basal and which are more recently derived. Bolle's analysis rooted GRAS proteins with a metazoan STAT protein.15,23 This root is problematic, as the relationship between STAT and GRAS proteins, while plausible, remains uncertain and the phylogenetic distance between GRAS and STAT proteins may be too great for STAT proteins to function as meaningful outgroups to GRAS proteins. We incorporated Spirogyra GRAS proteins into our analysis with the intent of rooting the GRAS family tree. Our results suggest that a GRAS protein from a Streptophyte lineage more basal than Spirogyra will be required to root GRAS proteins. To date we have failed to detect probable GRAS homologs outside of the Streptophyte lineage in either the Chlamydomonas reinhardii or Ostreococcus genomes.
The very ancient diversification of GRAS proteins has implications for understanding the evolution of GRAS protein function, including possible cellular-level functions of HAM proteins. HAM proteins are most closely related to DELLA proteins, both classes residing in GRAS Clade V. DELLAs are transcriptional repressors of growth promoting proteins whose activity is negatively regulated by gibberellins via a ubiquitin-mediated degradation pathway.24 However, both HAM and DELLA proteins possess strongly supported homologs in S. moellendorffii and P. patens, indicating that divergence of the HAM and DELLA subfamilies from a common ancestral protein occurred prior to divergence of the Moss and vascular plant lineages. Supporting the nesting of S. moellendorffii and P. patens proteins within the HAM and DELLA subfamilies, S. moellendorffii and P. patens HAM homologs possess miR170/171 binding sites, a key synapomorphy of the HAM subfamily,2,20 while Selaginella and Physcomitrella DELLA homologs possess the DELLA and TVHYNPS domains, defining synapomorphies of DELLA subfamily proteins.24
The deep evolutionary divergence between closely related GRAS subfamilies, preceding the evolution of traits such as a sporophyte-dominant life-cycle, heterospory, leaves, roots and the gibberellin, ethylene, jasmonate and brassinosteroid signaling pathways, suggests the likelihood of substantial functional divergence both within and among GRAS protein subfamilies that is not reflected by the considerable sequence conservation between common GRAS protein domains.14,16,24,25 Consequently, shared ancestry is potentially a poor predictor of shared function for GRAS proteins and GRAS proteins may lack a unifying function beyond transcriptional regulation.
Acknowledgements
Charles Delwiche and Ruth Timme provided Spirogyra GRAS gene EST sequences.
References
- 1.Stuurman J, Jaeggi F, Kuhlemeier C. Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev. 2002;16:2213–2218. doi: 10.1101/gad.230702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engstrom EM, Andersen CM, Gumulak-Smith J, Hu J, Orlova E, Sozzani R, Bowman JL. Arabidopsis homologs of the Petunia HAIRY MERISTEM gene are required for maintenance of shoot and root indeterminacy. Plant Physiol. 2011;155:735–750. doi: 10.1104/pp.110.168757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L, Mai YX, Zheng YC, Luo Q, Yang HQ. MicroRNA171c-targeted SCL6-II, SCL6-III and SCL6-IV genes regulate shoot branching in Arabidopsis. Molecular Plant. 2010;3:794–806. doi: 10.1093/mp/ssq042. [DOI] [PubMed] [Google Scholar]
- 4.Schulze S, Schafer BN, Parizotto EA, Voinnet O, Theres K. LOST MERISTEMS genes regulate cell differentiation of central zone descendants in Arabidopsis shoot meristems. Plant J. 2010;64:668–678. doi: 10.1111/j.1365-313X.2010.04359.x. [DOI] [PubMed] [Google Scholar]
- 5.Lee MH, et al. Large-scale analysis of the GRAS gene family in Arabidopsis thaliana. Plant Mol Biol. 2008;67:659–670. doi: 10.1007/s11103-008-9345-1. [DOI] [PubMed] [Google Scholar]
- 6.Fode B, Siemsen T, Thurow C, Weigel R, Gatz C. The Arabidopsis GRAS Protein SCL14 Interacts with Class II TGA Transcription Factors and Is Essential for the Activation of Stress-Inducible Promoters. Plant Cell. 2008;20:3122–3135. doi: 10.1105/tpc.108.058974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolle C, Koncz C, Chua N. PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev. 2000;14:1269–1278. [PMC free article] [PubMed] [Google Scholar]
- 8.Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, et al. The SCARECROW Gene Regulates an Asymmetric Cell Division That Is Essential for Generating the Radial Organization of the Arabidopsis Root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- 9.Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, et al. The SHORT-ROOT Gene Controls Radial Patterning of the Arabidopsis Root through Radial Signaling. Cell. 2000;101:555–567. doi: 10.1016/s0092-8674(00)80865-x. [DOI] [PubMed] [Google Scholar]
- 10.Schumacher K, Schmitt T, Rossberg M, Schmitz G, Theres K. The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc Nat Acad Sci USA. 1999;96:290–295. doi: 10.1073/pnas.96.1.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong H, Jin Y, Liu W, Li F, Fang J, Yin Y, et al. DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 2009;58:803. doi: 10.1111/j.1365-313X.2009.03825.x. [DOI] [PubMed] [Google Scholar]
- 12.Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997;11:3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalo P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, et al. Nodulation Signaling in Legumes Requires NSP2, a Member of the GRAS Family of Transcriptional Regulators. Science. 2005;308:1786–1789. doi: 10.1126/science.1110951. [DOI] [PubMed] [Google Scholar]
- 14.Floyd SK, Bowman JL. The ancestral developmental tool kit of land plants. Int J Plant Sci. 2007;168:1–35. [Google Scholar]
- 15.Bolle C. The role of GRAS proteins in plant signal transduction and development. Planta. 2004;218:683. doi: 10.1007/s00425-004-1203-z. [DOI] [PubMed] [Google Scholar]
- 16.Tian C, Wan P, Sun S, Li J, Chen M. Genome-Wide Analysis of the GRAS Gene Family in Rice and Arabidopsis. Plant Mol Biol. 2004;54:519–532. doi: 10.1023/B:PLAN.0000038256.89809.57. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Qian Q, Fu Z, Wang Y, Xiong G, Zeng D, et al. Control of tillering in rice. Nature. 2003;422:618–621. doi: 10.1038/nature01518. [DOI] [PubMed] [Google Scholar]
- 18.Morohashi K, Minami M, Takase H, Hotta Y, Hiratsuka K. Isolation and Characterization of a Novel GRAS Gene That Regulates Meiosis-associated Gene Expression. J Biol Chem. 2003;278:20865–20873. doi: 10.1074/jbc.M301712200. [DOI] [PubMed] [Google Scholar]
- 19.Smit P, Radts J, Protyanko V, Debelle F, Gough C, Bisseling T, Geurts R. NSP1 of the GRAS protein family is essential for Rhizobial Nod Factor-induced transcription. Science. 2005;308:1789–1791. doi: 10.1126/science.1111025. [DOI] [PubMed] [Google Scholar]
- 20.Bergsten J. A review of long-branch attraction. Cladistics. 2005;21:163–193. doi: 10.1111/j.1096-0031.2005.00059.x. [DOI] [PubMed] [Google Scholar]
- 21.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of Plant MicroRNA Targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 22.Harberd NP, Belfield E, Yasumura Y. The Angiosperm Gibberellin-GID1-DELLA Growth Regulatory Mechanism: How an “Inhibitor of an Inhibitor” Enables Flexible Response to Fluctuating Environments. Plant Cell. 2009;21:1328–1339. doi: 10.1105/tpc.109.066969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richards DE, Peng J, Harberd NP. Plant GRAS and metazona STATs: one family? BioEssays. 2000;22:573–577. doi: 10.1002/(SICI)1521-1878(200006)22:6<573::AID-BIES10>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 24.Rensing, et al. The Physcomitrella Genome Reveals Evolutionary Insights into the Conquest of Land by Plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 25.Yasumara Y, Crumpton-Taylor M, Fuentes S, Harberd NP. Step-by-Step Acquisition of the Gibberellin-DELLA Growth-Regulatory Mechanism during Land-Plant Evolution. Curr Biol. 2007;17:1225–1230. doi: 10.1016/j.cub.2007.06.037. [DOI] [PubMed] [Google Scholar]