Abstract
High mobility group (HMG) proteins of the HMGB family containing a highly conserved HMG box are chromatin-associated proteins that interact with DNA and nucleosomes and catalyze changes in DNA topology, thereby facilitating important DNA-dependent processes. The genome of Arabidopsis thaliana encodes 15 different HMG-box proteins that are further subdivided into four groups: HMGB-type proteins, ARID-HMG proteins, 3xHMG proteins that contain three HMG boxes and the structure-specific recognition protein 1 (SSRP1). Typically, HMGB proteins are localized exclusively to the nucleus, like Arabidopsis HMGB1 and B5. However, these Arabidopsis HMGB proteins showed a very high mobility within the nuclear compartment. Recent studies revealed that Arabidopsis HMGB2/3 and B4 proteins are predominantly nuclear but also exist in the cytoplasm, suggesting an as yet unknown cytoplasmic function of these chromosomal HMG proteins.
Key words: Arabidopsis, plant chromatin proteins, HMG proteins, localization, nuclear transport, partitioning
The dense packaging of genomic DNA in chromatin within the cell nucleus has crucial consequences for DNA-dependent processes, including gene transcription. Chromatin structure has to be very dynamic in order to be able to facilitate access to the DNA and high performance of the transcription machinery, for instance. HMG proteins are a heterogeneous class of chromatin-associated proteins. After the histones, HMG proteins are the second most abundant chromosomal proteins.1,2 They are thought to perform architectural functions and to serve as modulators of chromatin structure in the nucleus, thereby helping to facilitate important cellular processes, such as transcription, recombination and DNA repair.2–4
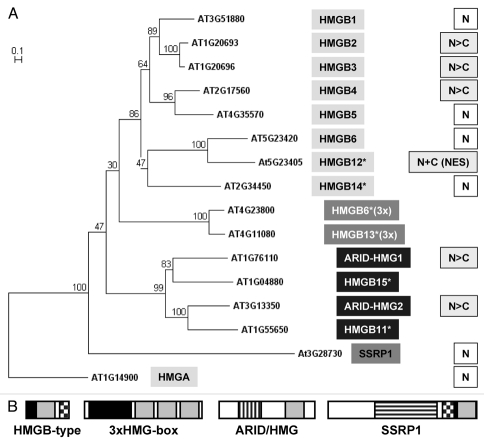
In plant genomes, genes encoding members of the HMGA and HMGB protein families were identified that are expressed throughout the entire plant at different levels.5 Members of the HMGN protein family that are found in vertebrate genomes are missing in plants.5 While the model plant Arabidopsis thaliana contains one HMGA protein, 15 different members of the HMG box family were identified that contain at least one HMG box, a characteristic DNA-binding domain that is highly conserved (Fig. 1). Like with other plant species, Arabidopsis HMG-box proteins can be subdivided into four different groups. HMGB-type proteins have a distinctive three-domain structure consisting of a basic N-terminal domain, a central HMG box and a C-terminal acidic domain. In contrast, 3xHMG-box proteins contain three consecutive HMG boxes and a basic N-terminus, and ARID-HMG proteins contain a HMG box and an additional A/T-rich interaction domain (ARID) that is also found in specific transcription factors.8,9 Finally, the unique structure-specific recognition protein 1 (SSRP1) also contains a HMG box and different additional domains10 (Fig. 1).
Figure 1.
(A)Phylogenetic tree of high mobility group (HMG) proteins of Arabidopsis thaliana. Protein alignments were performed with full-length protein sequences using ClustalW2 (www.ebi.ac.uk/Tools/clustalw2), and the tree was constructed with TreeCon using Poisson correction and neighbor joining, taking insertions and deletions into account.6 HMGA was used to root the tree. Distance bar is given top left and bootstrap values are indicated at the nodes. Protein names are given next to the Arabidopsis gene identifier in boxes. Protein names from the Plant Chromatin Database were used (indicated by asterisks) in cases where no published names were available. Subcellular localization of each protein is indicated on the far right side, if data were available. N, indicates complete nuclear localization; N>C, higher concentration of the protein in the nucleus relative to the concentration in the cytoplasm; N+C, relative protein concentration similar in both compartments; NES, nuclear export signal. (B) Architecture of the four groups of Arabidopsis HMG-box proteins (modified after ref. 7). HMGB-type proteins contain a single HMG box (light grey), an N-terminal basic region (black) and a C-terminal acidic region (checkered), 3xHMG-box proteins contain three consecutive HMG boxes and a basic N terminus, ARID/HMG proteins contain an ARID domain (hatched vertically) and a single HMG box, and SSRP1 contains an SSR domain (hatched horizontally) and a single HMG box separated by an acidic domain and a short basic region. Proteins are drawn to scale.
Consistent with their proposed function within the cell nucleus, Arabidopsis HMGA as well as HMGB1 and HMGB5 are exclusively nuclear proteins, as demonstrated by immuno-localization experiments and in vivo analyses using GFP fusion proteins.11 The latter two are typical members of the HMGB-type proteins. In contrast, recent studies of the localization of HMGB2/3 and HMGB4 revealed partitioning of these proteins between the cytoplasm and the nucleus.12 Although they are structurally related to the nuclear proteins HMGB1 and HMGB5, HMGB2/3 and HMGB4 only preferentially localized to the nucleus in a protoplast assay system and were also clearly detected in the cytoplasm. In transgenic plants expressing fusions of HMGB2 or HMGB4 with GFP, cytoplasmic localization was found to be increased in meristematic cells. Continuous traveling of HMGB2 and HMGB4 between the nucleus and the cytoplasm was also demonstrated in vivo in real time using fusions with photo-activatable GFP in combination with two-photon activation and onephoton fluorescence detection.12
The acidic C termini of HMGB2 and HMGB4 are mainly responsible for the cytoplasmic localization of the full-length proteins since their deletion resulted in complete nuclear accumulation of the partial proteins. The C terminus of HMGB5 could not restore HMGB2/B4 localization in domain swap experiments. On the other hand, the basic N terminus of HMGB2 or HMGB4 is necessary for the predominant nuclear localization of the full-length proteins since its deletion resulted in pronounced cytoplasmic steady state localization of the partial proteins.12 Replacement of the N terminus of HMGB2 or HMGB4 with the N terminus of HMGB1 that is necessary for exclusive nuclear accumulation of HMGB1 and acts as a strong nuclear targeting signal11 caused complete nuclear localization of the chimeric proteins.12 The molecular mechanisms for nucleo-cytoplasmic partitioning of HMGB2/3 and HMGB4 are unknown to date. Extensive basic regions are required for nuclear localization of these HMGB-type proteins, but no nuclear import pathway has been identified. Likewise, extensive acidic domains are required for cytoplasmic localization of HMGB2/3 and HMGB4 but a nuclear export pathway was not assigned.
To date, no cytoplasmic functions of plant HMGB-type proteins are identified. Other plant HMG-box proteins also show partitioning between the cytoplasm and the nucleus (Fig. 1). An example is the HMGB-type protein encoded by At5g23405 that shows an almost equal distribution of the protein between the two cellular compartments, while the related HMGB6 and the protein encoded by At2g34450 are exclusively nuclear, like HMGB1 and HMGB5.13,14 However, the protein encoded by At5g23405 is exceptional in the sense that it does not bind to DNA at all, despite its HMG box. In addition, the protein contains a nuclear export signal that is recognized by the nuclear export receptor exportin 1,6,15 explaining the relatively high cytoplasmic concentration of At5g23405.14 In addition, two members of another group of Arabidopsis HMG-box proteins, ARID-HMG1/2 primarily localize to the nucleus but, like HMGB2/3 and B4, are also clearly detectable in the cytoplasm.9 It should be noted here that human HMGB1 has a special function as a cytokine. It shuttles between the nucleus and the cytoplasm and is also targeted to the extracellular environment.17
The nucleo-cytoplasmic partitioning of HMGB2/3 and HMGB4 adds another unexpected feature to plant HMGB-type proteins, after the discovery that the Arabidopsis nuclear HMGB1 and HMGB5 proteins are highly mobile within the nucleus,11 similar to their mammalian counterparts.18,19 Thus, more than 30 years after their discovery and analysis in plants,20 the name “high mobility group proteins” gained an additional meaning.
References
- 1.Thomas JO, Travers AA. HMG1 and 2, and related “architectural” DNA-binding proteins. Trends Biochem Sci. 2001;26:167–174. doi: 10.1016/s0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- 2.Reeves R. Nuclear functions of the HMG proteins. Biochim Biophys Acta. 2010;1799:3–14. doi: 10.1016/j.bbagrm.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bustin M, Reeves R. High-mobility group chromosomal proteins: Architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 4.Agresti A, Bianchi ME. HMGB proteins and gene expression. Curr Opin Genet Dev. 2003;13:170–178. doi: 10.1016/s0959-437x(03)00023-6. [DOI] [PubMed] [Google Scholar]
- 5.Grasser KD, Launholt D, Grasser M. High mobility group proteins of the plant HMGB family: Dynamic chromatin modulators. Biochim Biophys Acta. 2007;1769:346–357. doi: 10.1016/j.bbaexp.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Van de Peer Y, De Wachter R. Construction of evolutionary distance trees with TREECON for Windows: Accounting for variation in nucleotide substitution rate among sites. Comput Appl Biosci. 1997;13:227–230. doi: 10.1093/bioinformatics/13.3.227. [DOI] [PubMed] [Google Scholar]
- 7.Štros M, Launholt D, Grasser KD. The HMG-box: A versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell Mol Life Sci. 2007;64:2590–2606. doi: 10.1007/s00018-007-7162-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 9.Hansen FT, Madsen CK, Nordland AM, Grasser M, Merkle T, Grasser KD. A novel family of plant DNA-binding proteins containing both HMG-box and AT-rich interaction domains. Biochemistry. 2008;47:13207–13214. doi: 10.1021/bi801772k. [DOI] [PubMed] [Google Scholar]
- 10.Röttgers K, Krohn NM, Lichota J, Stemmer C, Merkle T, Grasser KD. DNA interactions and nuclear localisation of the chromosomal HMG domain protein SSRP1 from maize. Plant J. 2000;23:395–406. doi: 10.1046/j.1365-313x.2000.00801.x. [DOI] [PubMed] [Google Scholar]
- 11.Launholt D, Merkle T, Houben A, Schulz A, Grasser KD. Arabidopsis chromatin-associated HMGA and HMGB use different nuclear targeting signals and display highly dynamic localization within the nucleus. Plant Cell. 2006;18:2904–2918. doi: 10.1105/tpc.106.047274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pedersen DS, Merkle T, Marktl B, Lildballe DL, Bergmann T, Tönsing K, et al. Nucleocytoplasmic distribution of the Arabidopsis chromatin-associated HMGB2/3 and HMGB4 proteins. Plant Physiol. 2010;154:1831–1841. doi: 10.1104/pp.110.163055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grasser KD, Grill S, Duroux M, Launholt D, Thomsen MS, Nielsen HK, et al. HMGB6 from Arabidopsis thaliana specifies a novel type of plant chromosomal HMGB protein. Biochemistry. 2004;43:1309–1314. doi: 10.1021/bi035931c. [DOI] [PubMed] [Google Scholar]
- 14.Grasser M, Lentz A, Lichota J, Merkle T, Grasser KD. The Arabidopsis genome encodes structurally and functionally diverse HMGB-type proteins. J Mol Biol. 2006;358:654–664. doi: 10.1016/j.jmb.2006.02.068. [DOI] [PubMed] [Google Scholar]
- 15.Haasen D, Köhler C, Neuhaus G, Merkle T. Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana. Plant J. 1999;20:695–706. doi: 10.1046/j.1365-313x.1999.00644.x. [DOI] [PubMed] [Google Scholar]
- 16.Merkle T. Nucleo-cytoplasmic transport of proteins and RNA in plants. Plant Cell Rep. 2011;30:153–176. doi: 10.1007/s00299-010-0928-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang H, Tracey KJ. Targeting HMGB1 in inflammation. Biochim Biophys Acta. 2010;1799:149–156. doi: 10.1016/j.bbagrm.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24:4321–4328. doi: 10.1128/MCB.24.10.4321-4328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phair RD, Scaffidi P, Elbi C, Vecerová J, Dey A, Ozato K, et al. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol Cell Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spiker S. High-mobility group chromosomal proteins of wheat. J Biol Chem. 1984;259:12007–12013. [PubMed] [Google Scholar]