Abstract
The ER chaperone calreticulin plays vital roles in numerous cellular processes, including Ca2+-homeostasis, apoptosis and cell adhesion, in animal cells. Although calreticulin has been systematically characterized in animal cells, the focus has been on one of the isoforms. However, recent advances in the plant calreticulin field have revealed functional divergence of calreticulin isoforms. While two of the plant isoforms appear to work within a general ER chaperone framework, the third isoform is associated with folding of receptors for brassinosteroids and bacterial peptides. Hence, the discovery of functional specialization of plant calreticulins opens up new vistas for calreticulins also in the animal field.
Key words: chaperone, calreticulin, brassinosteroid, PAMP, EFR, calcium, protein folding
Calreticulins (CRTs) are ubiquitously expressed chaperones that fold newly synthesized proteins and regulate Ca2+ homeostasis in the Endoplasmic Reticulum (ER) of animal and plant cells.1 In addition, CRTs affect apoptosis, cell adhesion and regulation of gene expression in animal cells,2 reflecting the diverse processes that are affected by this protein family. While various functional aspects of the proteins have been long established in animals, it is only recently that comparable insights have been made in plants. Curiously, the results obtained in plants point towards functional specialization of the CRT homologs, something that has not been reported yet in animals.
The CRT family consists of two members in animals (CRT1 and 2; 3), and three members in higher plants (CRT1a, CRT1b and CRT3; 4). Based on expression of the two CRT genes in mouse it has been speculated that CRT1 is the main CRT isoform, whereas CRT2 may function in specialized tissue- and cell types.3 The function of CRTs in plants was for many years associated with protein folding5 and Ca2+ homeostasis,6 consistent with their animal homologs. In addition, modulation of CRT expression revealed that CRT functions are necessary for plant regeneration,7 and resistance to various environmental stresses,8,9 but the exact role for CRTs in these processes remain elusive. Interestingly, a recent report revealed that the third CRT isoform, CRT3, retains defective forms of the brassinosteroid receptor BRI1 in the ER in Arabidopsis.10 This paper also showed that neither the two other CRT isoforms, nor the membrane-spanning CRT homolog Calnexin (CNX), could retain the defective BRI1 suggesting functional specialization of the CRT3 isoform. Subsequently, several reports revealed that CRT3 also appears to mediate folding of the elf18 responsive EF-Tu receptor (EFR) associated with Pathogen-Associated Molecular Patterns (PAMPs) in Arabidopsis.11,12 In addition, CRT3 was unable to complement crt1a crt1b double mutant phenotypes, corroborating diverged functions of the Arabidopsis CRT3 isoform.13 Hence, it is clear that the CRT3 isoform perform different functions as compared to CRT1a and CRT1b in Arabidopsis.
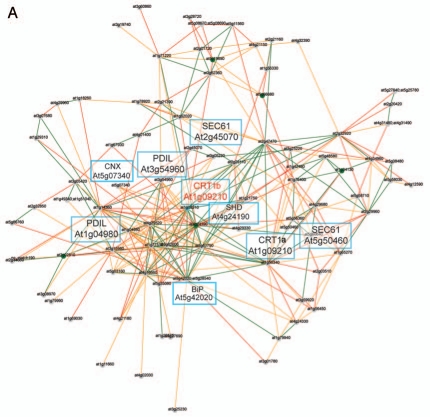
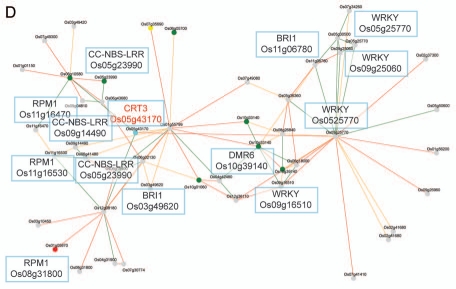
Transcriptionally coordinated genes tend to be functionally related.14,15 Indeed, while the Arabidopsis CRT1a and CRT1b genes appear co-expressed with many ER genes, which gene products are associated with protein folding and processing, the CRT3 gene is co-expressed with genes associated with pathogen responses and signal transduction (Fig. 1A,B).13 These data support a role for CRT1a and CRT1b in a more general ER chaperone framework and a more specialized role for CRT3 in Arabidopsis. To investigate whether a similar relationship also is evident in rice, we took advantage of the newly developed tool PlaNet that compare co-expressed subnetworks between plant species.16 From the comparative analysis, using CRT1b and CRT3 from Arabidopsis as bait genes, respectively, it is clear that the observed functional divergence for the Arabidopsis CRT1a/1b and CRT3 is also to be expected for the rice homologs (Figs. 1C,D). In other words, the rice CRT1 is co-expressed with many genes encoding ER chaperones and processive enzymes, while the rice CRT3 is co-expressed with many pathogen- and signal transduction-related genes (Figs. 1C,D). In particular, the rice CRT3 gene is co-expressed with a BRI1-related gene, perhaps pointing towards a function of CRT3 for folding of BRI1-related proteins also in rice.
Figure 1.
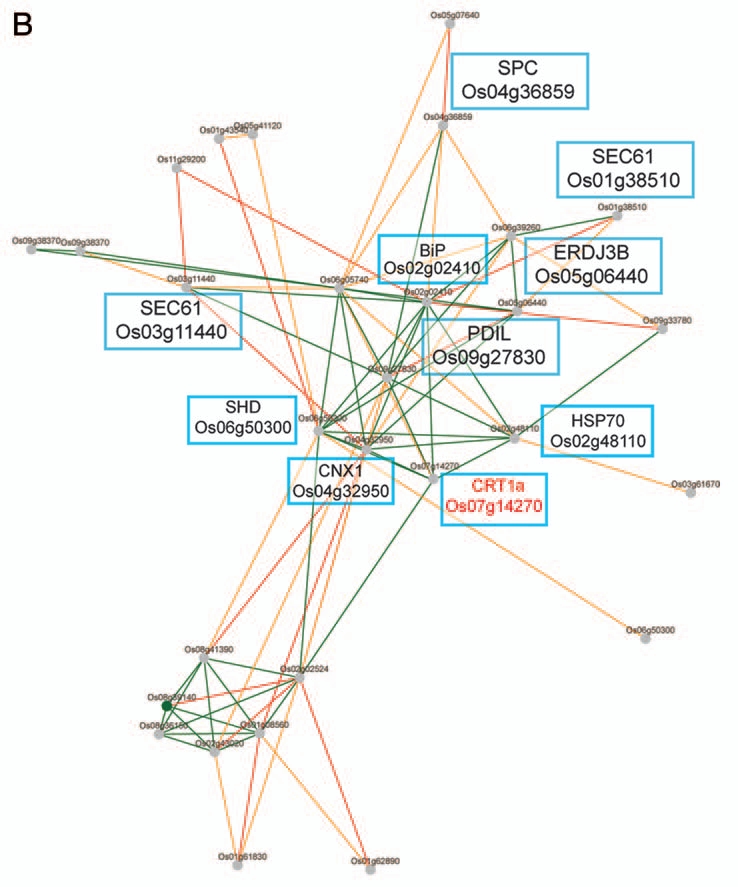
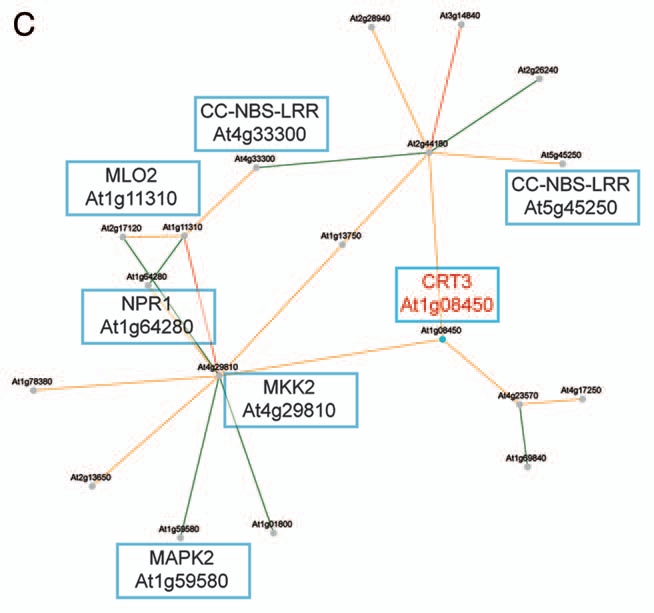
Co-expression networks of calreticulin isoforms in Arabidopsis and rice. (A) Co-expression network for Arabidopsis CRT1b. Nodes in the network represent genes, and edges represent significant co-expression between any two given genes. Different colors indicate different strength of co-expression. Genes of particular interest have been boxed and putative annotations indicated. The networks were generated via PlaNet derived tools.16 (B) Co-expression network for rice CRT1. Nodes in the network represent genes, and edges represent significant co-expression between any two given genes. Different colors indicate different strength of co-expression. Genes of particular interest have been boxed and putative annotations indicated. The networks were generated via PlaNet derived tools.16 (C) Co-expression network for Arabidopsis CRT3. Nodes in the network represent genes, and edges represent significant co-expression between any two given genes. Different colors indicate different strength of co-expression. Genes of particular interest have been boxed and putative annotations indicated. The networks were generated via PlaNet derived tools.16 (D) co-expression network for rice CRT3. Nodes in the network represent genes, and edges represent significant co-expression between any two given genes. Different colors indicate different strength of co-expression. Genes of particular interest have been boxed and putative annotations indicated. The networks were generated via PlaNet derived tools.16
The CRTs and CNXs classically hold three distinct domains; the N-, P- and C-domains.17 While the N- and C-domains simply refer to the domain-locations in the proteins, i.e., at the N-terminus and C-terminus, respectively; the P-domain was named due to two repeated proline-rich segments.18 The N-domain holds a typical signal sequence targeting the proteins to the ER, and the domain is likely to be involved in protein folding together with the P-domain.17 The C-domain contains an ER-retention signal (typically K/HDEL), and contributes to the main Ca2+-binding ability of the CRTs.13,17,19 The two proline-rich segments in the P-domain are normally repeated three times each, and are well conserved among animal and plant CRTs.4,17 These repeats are involved in interactions between CRTs and protein disulfide isomerases, such as the Erp57.20 These interactions may promote chaperone abilities in animal cells,21 and mutations in several amino acids in this domain affect the CRT-Erp57 interactions, for example mutations of Glu239, Asp241, Glu243 and Trp244 in rabbit CRT1.21 These amino acids correspond to Glu256, Asp258, Glu260 and Trp261 in the full-length mouse CRT1 sequence.
Mutations in crt3 result in reduced levels of the EFR, and consequently impaired EFR-triggered immune responses,11,12 presumably because CRT3 aids in the folding of EFR. Given that the P-domain plays a key role in CRTs ability to fold proteins,21 it is likely that sequence divergences in this region may explain the differences in observed phenotypes. To identify diverging sequence elements between the CRT isoforms, we aligned CRT sequences corresponding to the P-domain (spanning between amino acid 204 and 305 in mouse CRT1), and looked for amino acids that consistently were different between the CRT1a/1b and the CRT3 isoforms in four plant species, and compared these to the mouse CRT1 (Fig. 2). We subsequently used the resolved structure of the P-domain from mouse CRT1 and threaded the corresponding CRT3 sequence from Arabidopsis onto this structure (Fig. 2). While several amino acids differ between the two sequences, two differences may have functional implications. The first corresponds to a substitution of the positivity charged glutamic acid (Glu260) in mouse CRT1 to the non-polar leucine in Arabidopsis CRT3, located at the tip of the P-domain fold (A in Fig. 2). It is plausible that this substitution may influence a putative interaction between the CRT3 and protein disulfid isomerases, such as Erp57, that also are present in plants.22 Likewise, a negatively charged aspartic acid (Asp246) in mouse CRT1 is substituted to a positively charged lysine in Arabidopsis CRT3 (B in Fig. 2), which perhaps also may affect protein-disulfide isomerase interactions. It is important to note that these substitutions only occur in the plant CRT3 homologs and that the CRT1a/1b homologs have maintained the glutamic acid and aspartic acid in these positions (Fig. 2). Hence, it is plausible that these differences may account for diverging phenotypes in the crt1a/1b and crt3 mutants. In addition, several amino acids that are conserved among plant CRT isoforms have changed in mouse CRT1, for example the negatively charged glutamic acid (Glu257; number corresponding to mouse CRT1 sequence) in the plant CRTs corresponds to a methionine in the mouse CRT1 (C in Fig. 2). Such changes may signify diverging functions of CRTs across Kingdoms, which has been suggested through complementation experiments where plant CRTs only partially complemented adhesion phenotypes in CRT1 deficient mouse cells.13 Apart from the P-domain it is of course also plausible that differences in other domains may influence the functionality of the CRTs. For example, it appears likely that the highly charged C-domain in Arabidopsis CRT3 is involved in the retention of BRI1 in the ER.10
Figure 2.
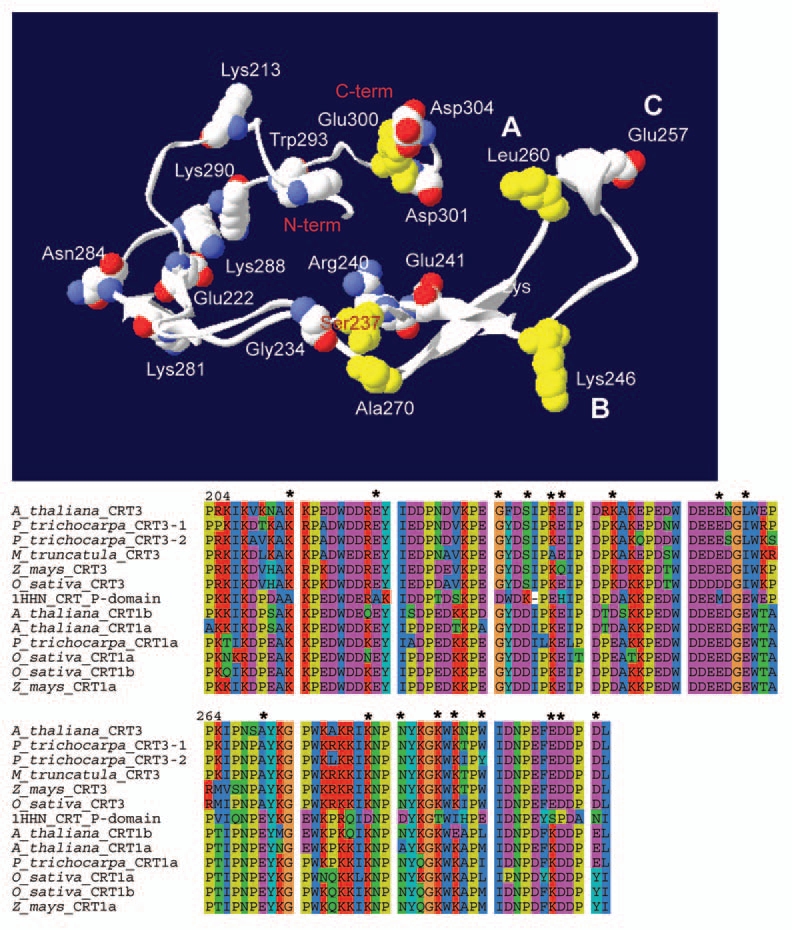
Amino acid differences between plant CRTs and mouse CRT1. Upper part displays the resolved structure of the mouse CRT1 P-domain part (aa 204 to 305) with the corresponding sequence for Arabidopsis CRT3 threaded onto it. The figure was generated using pyMol (DeLano Scientific, CA USA). β-sheets are indicated as flat arrows in the structure. Amino acids that differ from plant CRT3 isoforms (indicated in alignment in lower part), but are identical in plant CRT1a/1b and mouse CRT1 are indicated in yellow. Amino acids that differ in the mouse CRT1 (indicated in alignment in lower part), but are identical in plant CRT1a/1b and CRT3 are indicated in white/red/blue. These amino acids are marked as they appear in Arabidopsis CRT3. (A–C) in upper part indicates changed amino acids corresponding to the tip region of the P-domain. The lower part shows alignment of amino acids corresponding to the P-domain of CRTs from four plant species, and mouse CRT1 (1HHN CRT_P-domain). The amino acid numbers correspond to the numbers in mouse CRT1. Asterics indicate high-lighted amino acids in the upper part.
In brief, while both animal and plants contain two distinct isoform groups of CRTs, diverging functions of the members of these groups have only been shown in Arabidopsis. Based on conserved co-expression relationships we find it likely that these diverging functions also are evident in rice. Considering the importance of CRT1 in animal growth and development we hypothesize that also a detailed comparative characterization of the CRT1 and CRT2 in animal cells may reveal functional specialization.
Acknowledgements
S.P. and M.M. were funded through the Max-Planck Gesellschaft. M.S. and L.T. were supported by the Swedish Research Council Formas. We would like to thank Dr. Susanne Widell for critical reading of the manuscript.
References
- 1.Jia XY, He LH, Jing RL, Li RZ. Calreticulin: Conserved protein and diverse functions in plants. Physiol Plant. 2009;136:127–138. doi: 10.1111/j.1399-3054.2009.1223.x. [DOI] [PubMed] [Google Scholar]
- 2.Corbett EF, Michalak M. Calcium, a signaling molecule in the endoplasmic reticulum? Trends Biochem Sci. 2000;25:307–311. doi: 10.1016/s0968-0004(00)01588-7. [DOI] [PubMed] [Google Scholar]
- 3.Persson S, Rosenquist M, Sommarin M. Identification of a novel calreticulin isoform (Crt2) in human and mouse. Gene. 2002;4:151–158. doi: 10.1016/s0378-1119(02)00880-6. [DOI] [PubMed] [Google Scholar]
- 4.Persson S, Rosenquist M, Svensson K, Galvão R, Boss WF, Sommarin M. Phylogenetic analysis and expression studies reveal two distinct groups of calreticulin isoforms in higher plants. Plant Physiol. 2003;133:1385–1396. doi: 10.1104/pp.103.024943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denecke J, Vitale A. The use of protoplasts to study protein synthesis and transport by the plant endomembrane system. Methods Cell Biol. 1995;50:335–348. doi: 10.1016/s0091-679x(08)61041-9. [DOI] [PubMed] [Google Scholar]
- 6.Persso S, Wyatt SE, Love J, Thompson WF, Robertson D, Boss WF. The Ca2+ status of the endoplasmic reticulum is altered by induction of calreticulin expression in transgenic plants. Plant Physiol. 2001;126:1092–1104. doi: 10.1104/pp.126.3.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z, Komatsu S. Molecular cloning and characterization of calreticulin, a calcium-binding protein involved in the regeneration of rice cultured suspension cells. Eur J Biochem. 2000;267:737–745. doi: 10.1046/j.1432-1327.2000.01052.x. [DOI] [PubMed] [Google Scholar]
- 8.Jia XY, Xu CY, Jing RL, Li RZ, Mao XG, Wang JP, et al. Molecular cloning and characterization of wheat calreticulin (CRT) gene involved in drought-stressed responses. J Exp Bot. 2008;59:739–751. doi: 10.1093/jxb/erm369. [DOI] [PubMed] [Google Scholar]
- 9.Komatsu S, Yang G, Khan M, Onodera H, Toki S, Yamaguchi M. Overexpression of calcium-dependent protein kinase 13 and calreticulin interacting protein 1 confers cold tolerance on rice plants. Mol Genet Genomics. 2007;277:713–723. doi: 10.1007/s00438-007-0220-6. [DOI] [PubMed] [Google Scholar]
- 10.Jin H, Hong Z, Su W, Li J. A plant-specific calreticulin is a key retention factor for a defective brassinosteroid receptor in the endoplasmic reticulum. Proc Natl Acad Sci USA. 2009;106:13612–13617. doi: 10.1073/pnas.0906144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Zhao-Hui C, Batoux M, Nekrasov V, Roux M, Chinchilla D, et al. Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc Natl Acad Sci USA. 2009;106:15973–15978. doi: 10.1073/pnas.0905532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saijo Y, Tintor N, Lu X, Rauf P, Pajerowska-Mukhtar K, Häweker H, et al. Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 2009;28:3439–3449. doi: 10.1038/emboj.2009.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen A, Svensson K, Thelin L, Zhang W, Tintor N, Prins D, et al. Higher plant calreticulins have acquired specialized functions in Arabidopsis. PLoS One. 2010;5:11342. doi: 10.1371/journal.pone.0011342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 15.Persson S, Wei H, Milne J, Page GP, Somerville CR. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA. 2005;102:8633–8638. doi: 10.1073/pnas.0503392102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mutwil M, Klie S, Tohge T, Giorgi FM, Wilkins O, Campbell MM, et al. PlaNet: Combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell. 2011;23:895–910. doi: 10.1105/tpc.111.083667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michalak M, Corbett EF, Mesaeli N, Nakamura K, Opas M. Calreticulin: One protein, one gene, many functions. Biochem J. 1999;344:281–292. [PMC free article] [PubMed] [Google Scholar]
- 18.Michalak M, Milner RE, Burns K, Opas M. Calreticulin. Biochem J. 1992;285:681–692. doi: 10.1042/bj2850681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen A, Svensson K, Persson S, Jung J, Michalak M, Widell S, et al. Functional characterization of Arabidopsis calreticulin1a: A key alleviator of endoplasmic reticulum stress. Plant Cell Physiol. 2008;49:912–924. doi: 10.1093/pcp/pcn065. [DOI] [PubMed] [Google Scholar]
- 20.Frickel EM, Riek R, Jelesarov I, Helenius A, Wuthrich K, Ellgaard L. TROSY-NMR reveals interaction between ERp57 and the tip of the calreticulin P-domain. Proc Natl Acad Sci USA. 2002;99:1954–1959. doi: 10.1073/pnas.042699099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin V, Groenendyk J, Steiner SS, Guo L, Dabrowska M, Parker JM, et al. Identification by mutational analysis of amino acid residues essential in the chaperone function of calreticulin. J Biol Chem. 2006;281:2338–2346. doi: 10.1074/jbc.M508302200. [DOI] [PubMed] [Google Scholar]
- 22.Houston NL, Fan C, Xiang JQ, Schulze JM, Jung R, Boston RS. Phylogenetic analyses identify 10 classes of the protein disulfide isomerase family in plants, including single-domain protein disulfide isomerase-related proteins. Plant Physiol. 2005;137:762–778. doi: 10.1104/pp.104.056507. [DOI] [PMC free article] [PubMed] [Google Scholar]