Abstract
Endospore formation is a characteristic shared by some Bacilli and Clostridia that involves the creation of two cell types, the forespore and the mother cell. Hundreds of protein-encoding genes have been shown to be transcribed in a cell-specific fashion during this developmental process in Bacillus subtilis. We have used a phylogenetic profiling procedure to identify clusters of B. subtilis coding and non-coding sequences that co-occur in other endospore formers. One such cluster shows a strong bias for sporulation-related genes (42% among 156 genes) and is enriched in potential non-coding RNAs. We have studied one RNA candidate, encoded in the ylbG-ylbH interval. In vivo analysis using a transcriptional fusion to the Escherichia coli lacZ gene demonstrates that this region of the chromosome contains a gene, csfG, encoding a 147-nucleotide RNA that is transcribed only during sporulation, specifically in the forespore. csfG is present in many endospore formers, mostly Bacilli and some Clostridia, whereas it is absent from bacteria that do not produce endospores. All CsfG RNAs contain a strongly conserved, pyrimidine-rich, central motif that overlaps a potential stem-loop structure. The remarkable conservation of this sequence in widely divergent bacteria suggests that it plays a conserved physiological role, presumably by interacting with an unidentified target in the forespore, where it contributes to the acquisition of the spore properties.
Key words: small RNA, sporulation, germination, forespore, Bacilli, Clostridia
Introduction
Endospore formation is a primitive developmental process observed in some Bacilli and Clostridia when subjected to nutrient starvation. These bacteria belong to the phylum of Firmicutes that also contains many bacteria unable to sporulate, among which members of the Listeria, Staphylococcus, Streptococcus and Lactobacillus orders. The morphological process of sporulation appears to be extremely similar in all endospore formers, starting with a polar division that creates two cells of different sizes and different fates, the smaller forespore and the larger mother cell. Engulfment of the forespore by the mother cell is followed by the synthesis of peptidoglycan and protein layers surrounding the forespore and contributing to its resistance properties after its release in the environment by lysis of the mother cell.1 Sporulation has been extensively studied in Bacillus subtilis because of the amenability of this bacterium to classical and reverse genetics. Transcriptome experiments have identified several hundreds of protein-encoding genes that are expressed at some stage of sporulation in one or the other cell.2–4 It is now well established that the developmental process is initiated by accumulation of the Spo0A∼P regulatory protein and the σHtransition sigma factor, whereas a cascade of four sigma factors, σF followed by σG in the forespore, and σE followed by σK in the mother cell, is responsible for the temporal and spatial activation of sporulation-specific genes after septation.5 Analysis of the complete genomes of more than 50 endospore formers, including many Clostridia,6 indicates the conserved presence of the major regulatory players identified in B. subtilis, in accordance with an early suggestion that all modern endospore formers derive from a single sporulating ancestor.7
Until recently, only proteins were considered to be involved in the control of sporulation. However, with the identification of many small non-coding RNAs (sRNAs) in a wide variety of bacteria, where they modulate gene expression of multiple regulons, including complex processes such as biofilm formation,8 morphological development9 or host-pathogen interaction,10 it becomes essential to investigate the presence and the role of sRNAs during sporulation. This question has been addressed by Silvaggi et al. who combined microarray transcriptional profiling of intergenic regions in B. subtilis with a computational screen for potential RNA secondary structures conserved in other endospore formers.11 Importantly for what follows, Silvaggi and co-workers eliminated the candidates for which a role as a cis-regulatory RNA element had been previously reported. They identified one sRNA that accumulates early during sporulation before septation and for which no ortholog was indicated, a second sRNA synthesized under the control of σK in the mother cell and also present in Bacillus anthracis, and finally a complex pattern of sRNAs synthesized from an intergenic region under the control of σGin the forespore and σK in the mother cell and conserved in Bacillus halodurans. Recently, an additional sRNA, identified only in B. subtilis, was reported to be expressed under the control of σE in the mother cell.12 Disrupting the intergenic regions encoding these various sRNAs did not lead to any obvious sporulation defect.
Another study13 focused exclusively on B. subtilis intergenic regions larger than 500 bp and did not detect any sporulation related sRNA. A comprehensive, high-resolution tiling array analysis has been recently reported in reference 14 and, although performed on cells growing exponentially in rich or minimal medium, it provides useful hints about genome regions potentially transcribed only during sporulation because of their conspicuous silence in both growing conditions in the thorough published data set. Here we have built upon our previously published phylogenetic study of B. subtilis intergenic and coding regions15 to identify potential sporulation-related sRNAs. We report the characterization of one such sRNA, present exclusively in endospore formers, expressed in the forespore and containing a highly-conserved pyrimidine motif.
Results
A cluster enriched in sporulation genes.
We reasoned that sporulation-related sRNAs would be encoded by genes co-occuring with protein-encoding genes known to be involved in sporulation and present in several endospore formers. We used the NAPP (Nucleic Acids Phylogenetic Profiling) procedure to identify clusters of B. subtilis coding and non-coding sequences that co-occur in other species.15 Briefly, NAPP involves collecting all conserved noncoding elements (CNEs) and coding sequences (CDS) in a reference species and seeking homologous sequences in other available species. CNEs and CDSs are then clustered based on the similarity of their occurrence profiles. When using B. subtilis as the reference species, CNEs corresponding to actual sRNA genes or riboswitches were significantly over-represented in two clusters.15
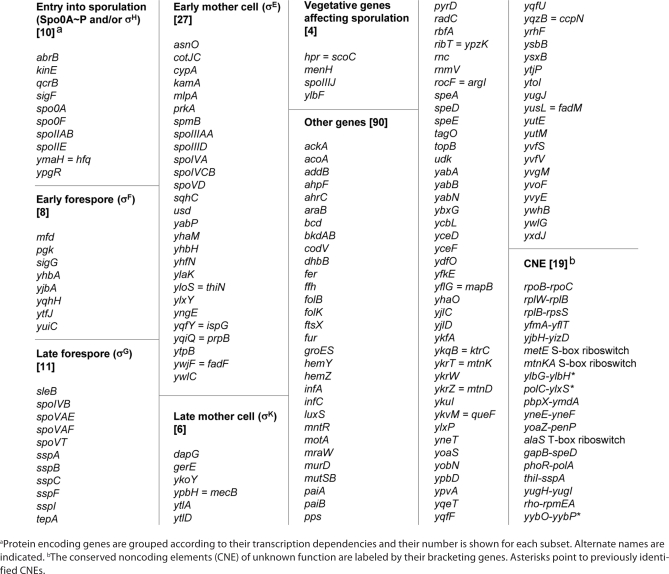
Here we further analyzed phylogenetic and functional biases in these two RNA-rich clusters. We have identified a subcluster of genes co-occuring in many endospore formers, mainly Bacilli and some Clostridia, and highly enriched in sporulation-related genes. Out of 156 protein-encoding genes, 66 (i.e., 42%) are essential for sporulation (bona fide spo genes) or known to be transcribed at some stage of this developmental process (Table 1). The cluster also contains 19 CNEs, among which three riboswitches (for alaS, metE and the mtnKA operon), two genetic intervals previously reported as potentially expressing a non-coding RNA (next to ylbH and to yybP),16 and one region already identified as producing several sRNAs during sporulation (the polC-ylxS interval).11 The latter result confirms the existence of a specific functional bias among the 175 members of the cluster and suggests that sporulation related sRNAs might be encoded by some of the other CNEs.
Table 1.
A B.subtilis cluster enriched in sporulation genes
 |
A forespore-specific sRNA.
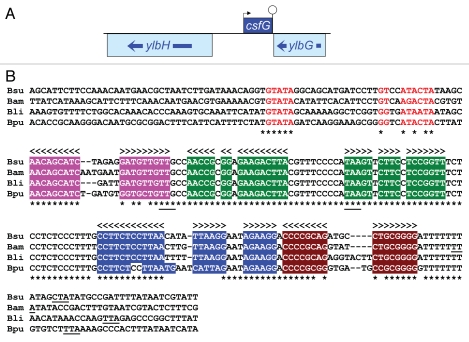
Our attention was drawn to the CNE corresponding to the ylbG-ylbH interval (Fig. 1A). This CNE overlaps an annotated cis-acting RNA named “ylbH leader” (RFAM entry RF00516),16 and was therefore excluded from the analysis of Silvaggi et al. However, we observed that this CNE is most likely expressed from the reverse strand as a trans-acting RNA, since it is followed on this strand by a run of Ts that could be part of a strong rho-independent transcription terminator. Moreover, aligning this region with the orthologous regions from three closely related Bacilli reveals an impressive sequence conservation that starts immediately downstream of a putative σF-controlled promoter and stops immediately after the putative transcription terminator (Fig. 1B). A consensus, fully-conserved -35 recognition motif (GTATA) is followed at the optimal 15 bp distance by a sequence closely related to the consensus −10 recognition motif, Gg-A-AHTR, where H is A or C or T and R is A or G.4 Some σF-controlled promoters are also recognized by σG because of an overlap of sequence recognition by the two sigma factors. This is apparently the case here since the conserved putative promoter motifs fit well with the consensus for σG-dependent promoters, tGHATA (17–18 bp) MAWAMTA, where M is A or C, and W is A or T.4
Figure 1.
An sRNA gene in the ylbH-ylbG interval. (A) The protein coding sequences of the B. subtilis ylbG and ylbH genes are shown as boxes (drawn to scale) with arrows showing their transcription orientation. The location of the putative csfG gene on the opposite strand is indicated by a box bracketed by its transcription initiation (arrow) and termination (lollipop) signals. (B) The sequence immediately adjacent to ylbG is shown for B. subtilis, Bacillus amyloliquefaciens, Bacillus licheniformis and Bacillus pumilus. Asterisks point to nucleotides identical in the four sequences. Arrow-heads indicate potential base pairing and the four corresponding stem-loop structures are highlighted by a specific colored background. Nucleotides that constitute a σF-controlled promoter are shown in red. For clarity the overlapping σG-dependent promoter is not shown. The start and stop codons of a conserved short open reading frame are underlined under the alignment. The stop codon of ylbG (located downstream and in convergent orientation) is underlined under each sequence.
If this promoter is actually functional it would lead to the synthesis of a 147-nt RNA starting with an A residue at position 1569347 in the revised B. subtilis genome sequence (NCBI accession number NC_000964) and ending with a run of seven Us at position 15692001, in the opposite orientation to the adjacent ylbG and ylbH genes (Fig. 1A). To confirm the existence of this RNA we carried out a northern blot analysis on total RNA extracted from cells harvested at early stages of sporulation. In our growth conditions polar septation takes place one hour after the entry into stationary phase. As shown in Figure 2 an RNA of the expected size is present 30 minutes after septation (lane 1.5), but absent 30 minutes earlier (lane 0.5). This small RNA accumulates during the next hour (lane 2.5) but is not synthesized in the absence of σF. The probe hybridizes with a less abundant, larger RNA (ca. 400 nt), that follows the same temporal pattern. Since no promoter sequence can be identified at the corresponding position upstream, on the strand complementary to ylbH, it is likely that this larger RNA results from readthrough transcription and therefore ends around the beginning of the ylbG gene. In any case it is a very minor species as compared to the smaller RNA.
Figure 2.
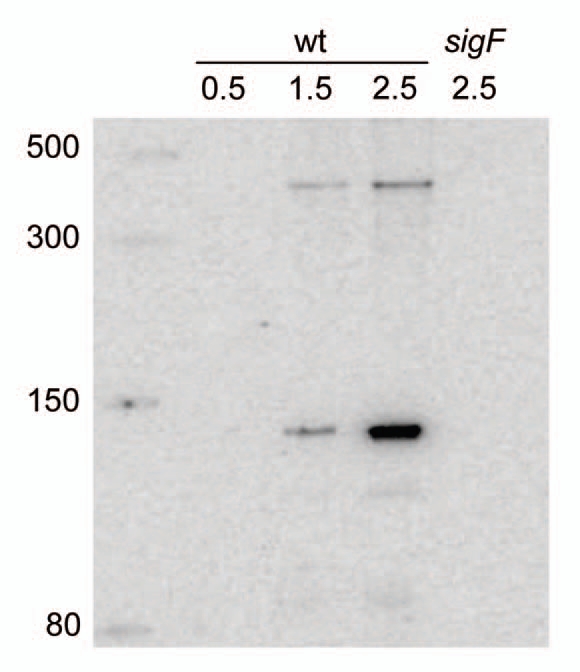
Northern blot detection of CsfG. RNA was extracted from B. subtilis cells (either wild type or deleted for the sigF gene) grown in DSM medium and harvested at various times after entry into sporulation. Hours of sporulation are indicated. Total RNA was separated by electrophoresis on an 8% polyacrylamide gel, transferred to a nylon membrane, and CsfG RNA was detected by hybridization with a complementary oligonucleotide. samples were run alongside ssRNA markers (NEB) with the indicated sizes.
A 11-codon reading frame is present in the four RNAs encoded by the orthologous DNA regions shown in Figure 1B, starting with a UUG triplet at position 22 in the B. subtilis RNA. However, the corresponding ribosome binding site (GAGG) is conspicuously absent in the sequences from the three other Bacilli, making it unlikely that this small open reading frame is actually translated. Moreover, this reading frame is missing in orthologs of this RNA from more distant organisms discussed hereafter. Potential base pairing is identical in these Bacilli RNAs and comprises four stem-loop structures identified by specific colors in Figure 1B. We note that the second loop is 100% conserved between the four Bacilli, whereas the three other loops are highly variable. Also, 14 out of 15 nucleotides between the second and the third stem-loop are fully conserved.
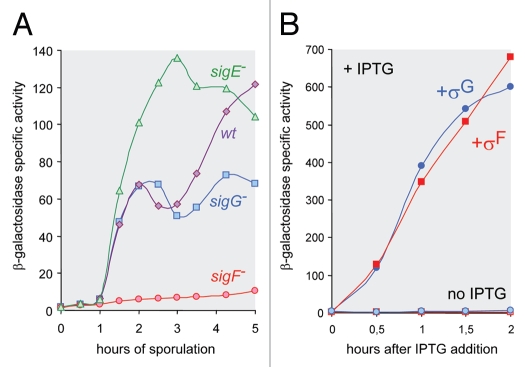
In order to assess quantitatively the synthesis of this sRNA in B. subtilis we fused its promoter region to the Escherichia coli lacZ gene and integrated the resulting transcriptional fusion into the B. subtilis chromosome at the non-essential amyE locus. The cloned fragment contains 132 bp upstream of the predicted transcription start as well as the first 35 bp from the transcribed region. As shown in Figure 3A, β-galactosidase synthesis starts 1-hour after the initiation of the sporulation process and is abolished in the absence of σF. A second wave of β-galactosidase synthesis is observed 2 hours later in a wild-type strain but is missing in the absence of σG. Finally, in a mutant strain where the absence of σE leads to the formation of two forespore compartments, the level of β-galactosidase accumulation is roughly double of what is found in a wild-type strain. Altogether, these results fit perfectly with the presence of a promoter recognized by both σF and σG, and activated exclusively during sporulation, in the forespore. This was further confirmed by inducing expression of the lacZ fusion in exponentially growing cells that were engineered to produce σF or σG in response to the addition of IPTG. The results shown in Figure 3B indicate that both sigma factors are equally efficient in activating transcription of the fusion. From now on, the gene encoding this sRNA will be called csfG (controlled by sigma-F and sigma-G).
Figure 3.
Forespore-specific transcription of the csfG gene. (A) Time course expression of a transcriptional csfG-lacZ fusion integrated into the B. subtilis chromosome in various mutant back-grounds. Cells were grown in DSM. (B) Induction of the same fusion in cells growing exponentially in 2x YT medium and harboring a plasmid allowing synthesis of σF or σG in the presence of IPTG. A control experiment was carried out in the absence of IPTG.
Most of the csfG gene was replaced by a kanamycin-resistance cassette, eliminating 9 bp from the promoter region and 123 bp from the RNA encoding region. A B. subtilis strain harboring this mutation produces as much heat-resistant spores as the wild-type strain and does not show any obvious growth defect (data not shown). To investigate more subtle phenotypes, we subjected the mutant strain to successive cycles of growth, sporulation and germination, in competition with a wild-type strain. After 10 such cycles the ratio of mutant vs. wild-type bacteria shifted from 0.5-0.5 to 0.02–0.98, indicating a loss of about 29% of the mutant strain at each cycle, presumably as a consequence of a slight germination delay of the mutant spores (data not shown).
A highly conserved motif.
As expected from its belonging to a cluster of genes co-occuring in several endospore formers and strongly enriched in sporulation genes csfG is present in a wide range of Bacilli and Clostridia. It is always found immediately upstream of and diverging from a ylbH ortholog. We identified a csfG ortholog in the 21 Bacillaceae that we analyzed, in Alicyclobacillaceae and in Paenibacillaceae, but not in Planococcaceae (e.g., Lysinibacillus sphaericus). We also found a csfG ortholog in various Clostridia, all of them (but Dethiobacter alkaliphilus) being thermophiles. Conversely, csfG is absent from bacteria that do not form endospores.
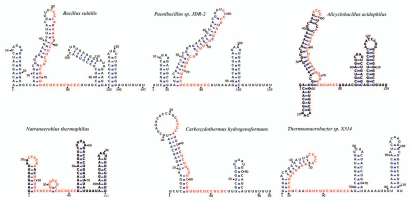
A multiple sequence alignment of 24 CsfG orthologs is shown in Figure 4 where potential base pairing is indicated. Whereas CsfG orthologs from closely related Bacilli are conserved throughout their whole length (Fig. 1B) this extended alignment pinpoints the striking conservation of a central sequence that constitutes the second half of a stem-loop structure and its adjacent unpaired region. All CsfG sequences are predicted to also contain a rho-independent transcription terminator. The remaining predicted secondary structures are variable, with fewer stem loops in sequences originating from Clostridia in accordance with their shorter size. Examples of the predicted secondary structures of a few CsfG RNAs are shown in Figure 5 where the conserved central motif is highlighted.
Figure 4.
Multiple structural alignment of 24 CsfG orthologs. Bacterial names are indicated by the their first letters and correspond to B. subtilis, B. amyloliquefaciens, B. licheniformis, B. pumilus, Bacillus anthracis, Bacillus cereus, Bacillus thuringiensis, Bacillus weihenstephanensis, Bacillus clausii, Bacillus halodurans, Geobacillus kaustophilus, Geobacillus thermodenitrificans, Oceanobacillus iheyensis, Anoxybacillus flavithermus, Paenibacillus sp. JDR-2, Alicyclobacillus acidocaldarius, Halothermotrix orenii, Natranaerobius thermophilus, Carboxydibrachium pacificum, Carboxydothermus hydrogenoformans, Moorella thermoacetica, Thermoanaerobacter ethanolicus, Thermoanaerobacter tengcongensis, Symbiobacterium thermophilum. Text is highlighted in a turquoise/blue/grey gradient to indicate regions of high/medium/low conservation. Arrowheads indicate potential base pairing.
Figure 5.
Predicted secondary structure of representative CsfG RNAs. A varna view (21) of the predicted secondary structure of CsfG RNAs from six distant endospore formers is shown. Highly conserved nucleotides are indicated in red.
All CsfG sequences from Bacilli share an extended conserved motif consisting of UCCCCAU, followed at 4 nt by UCUUC, and one nt further by a strictly conserved 18-residue, pyrimidine-rich sequence. A shorter subset of this motif is present in the CsfG sequences from Clostridia. The conserved CCCCA block is predicted to be in a loop, the UCUUC block to be included in a stem, and the long UC-rich sequence to be at least partially unpaired. The remarkable conservation of this sequence and its specific arrangement suggest that this part of CsfG could be interacting with an RNA target, following a strand displacement mechanism similar to what has been described for DsrA in E. coli.17 It could also be the recognition motif for an RNA binding protein as previously described for some other highly conserved RNAs.8
Discussion
CsfG is a small, non-coding RNA synthesized exclusively in the forespore all along the sporulation process, first by σF then by σG. Its presence in all Bacillaceae and in several other endospore formers, including bacteria that diverged from B. subtilis billions of years ago, indicates that CsfG was present in the earliest endospore formers and that its preservation is somehow beneficial for the organism. Our competition experiments point to a potential germination defect of spores lacking CsfG. Interestingly, in several Clostridia where csfG is missing, it is replaced upstream of ylbH by gpr, a gene that is also transcribed exclusively in the forespore by σF- and σG-associated RNA polymerase. In some Clostridia containing csfG the gpr gene is located only a few kb downstream of csfG, next to a small cluster of σG-controlled ssp genes. These genes encode small proteins stored in the mature spore where they become the substrate of the Gpr protease during germination. These genomic relationships might reflect an ancient genetic linkage and they fit with a possible functional involvement of CsfG in germination.
If CsfG plays a negative role on the expression of some genes, as it is often the case with trans-acting sRNAs, potential targets could be genes controlling vegetative functions not required in the forespore, such as replication initiation or septation. However, synthesizing CsfG in exponentially growing bacteria, from an inducible promoter on a replicative plasmid, did not lead to a reduced growth rate nor to filamentation (data not shown). We used several programs to predict potential targets for B. subtilis CsfG. However, its conserved pyrymidine-rich region has the ability to pair with purine-rich ribosome binding sites, leading to a long list of targets with no clear winners (data not shown). We noticed that the highly conserved CsfG motif UCUUCCUCCGGUUU is perfectly complementary to a segment of stem-loop 40 in 16S rRNA whereas the ylbH gene, next to which csfG is always located, encodes a protein belonging to the methyltransferase superfamily, with highest similarity to RsmD, a 16S rRNA methylase. The significance of these observations remains to be investigated.
Whatever its role it is noteworthy that the CsfG sRNA was initially identified by a computational screen for elements showing phylogenetic co-occurrence in endospore formers. Making a spore involves a large number of genes, but only a subset of them lead to a clearcut sporulation defect when disrupted.4 Moreover, despite the conservation among endospore formers of a core of sporulation genes, mostly the main regulators and their principal targets, the presence or absence of a large number of sporulation-expressed genes depend on the specific habitat of the bacterium.7 Phylogenetic profiles used in the NAPP algorithm are particularly fit to capture such complex patterns as shown by the identification of CsfG.
Materials and Methods
Computational analyses.
The complete NAPP procedure is described in Marchais et al. In short, coding sequences (CDS) and conserved non-coding elements (CNE) from a species of interest (here B. subtilis) are sought systematically in 420 bacterial genomes using Blastn,18 producing a profile of normalized Blast scores. We then measure pairwise Pearson's distances between profiles and we cluster profiles using either K-means or hierarchical clustering.15 The “sporulation” cluster was identified by visual inspection of the hierarchical clustering representation, as part of a larger cluster enriched in known non-coding RNAs identified by Marchais et al. Searches for CsfG homologs were performed against the NCBI nr databases using Blastn. The multiple structural alignment of CsfG homologs was performed using the LocARNA web server19 and further manually edited using the Ralee Emacs plugin20 to correct local errors. Drawing and editing of RNA secondary structure was performed with the varna software.21 The CsfG entry was submitted to the RFAM database.22 A pre-release is available at: en.wikipedia.org/wiki/User:Amarchais/CsfG_RNA.
Growing bacteria.
Bacterial growth conditions and genetic methods were as described in reference 23. All B. subtilis strains are derivatives of JH642. The sig mutants are from our laboratory collection. Sporulation was induced by exhaustion in Difco sporulation medium (DSM) in the absence of antibiotics. Induction of σFor σG activity during exponential growth was carried out in 2x YT medium, in strains harboring a plasmid with the sigF or the sigG sequence under the control of the spac promoter, by addition of 1-mM isopropyl β-d-thiogalactoside (IPTG) at OD570 = 0.25. β-galactosidase activity is expressed as nanomoles of 2-nitrophenyl-β-d-galactopyranoside hydrolyzed per minute per milligram of protein.
Cloning csfG.
The csfG region was cloned by PCR using oligonucleotides (sequences available on request) that created an EcoRI site 130 bp upstream of the predicted csfG transcription start. The natural SacII site located at position 32 in CsfG was used to create a transcriptional fusion with lacZ after cloning into the pDG1661 vector that allows integration by a double recombination event at the amyE locus.24 Similarly, a NcoI site and a PstI site were introduced by PCR at position −9 and +123 respectively, according to the predicted csfG transcription start. A kanamycin resistance cassette was cloned between these two restriction sites and transferred onto the chromosome by a double recombination event that led to inactivation of csfG.
Northern blot analysis.
RNA was prepared according to a published protocol.25 Total RNA (10 µg) was separated on a denaturing 8% polyacrylamide-8 M urea gel and transferred to a Hybond-N+ Membrane (GE Healthcare) for 4 h at 60 V in 0.5× TBE. The membrane was pre-hybridized overnight in Ultrahyb buffer (Ambion) and probed with a 32P-labelled oligonucleotide (GGA GAA GGC AAA GGG AGA GGA GAA ACC GGA GG). Hybridization and wash steps were as described in reference 26. Briefly, the membrane was hybridized 4 h at 42°C in Ultrahyb buffer, washed once in 2x SSC, 0.01% SDS for 10 minutes at 42°C and 5 times at room temperature in 0.2x SSC, 0.01% SDS. The signal was revealed using a Typhoon Trio (GE Healthcare).
Acknowledgements
We are grateful to C. Condon for his advice and encouragement. This work was supported by CNRS (IFR 115 and UPR9073).
References
- 1.Errington J. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol. 2003;1:117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- 2.Eichenberger P, Fujita M, Jensen ST, Conlon EM, Rudner DZ, Wang ST, et al. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2004;2:1664–1683. doi: 10.1371/journal.pbio.0020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steil L, Serrano M, Henriques AO, Völker U. Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology. 2005;151:399–420. doi: 10.1099/mic.0.27493-0. [DOI] [PubMed] [Google Scholar]
- 4.Wang ST, Setlow B, Conlon EM, Lyon JL, Imamura D, Sato T, et al. The forespore line of gene expression in Bacillus subtilis. J Mol Biol. 2006;358:16–37. doi: 10.1016/j.jmb.2006.01.059. [DOI] [PubMed] [Google Scholar]
- 5.Hilbert DW, Piggot PJ. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol Mol Biol Rev. 2004;68:234–262. doi: 10.1128/MMBR.68.2.234-262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paredes CJ, Alsaker KV, Papoutsakis ET. A comparative genomic view of clostridial sporulation and physiology. Nat Rev Microbiol. 2005;3:969–978. doi: 10.1038/nrmicro1288. [DOI] [PubMed] [Google Scholar]
- 7.Stragier P. A gene odyssey: exploring the genomes of endospore-forming bacteria. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and its Closest Relatives: From Genes to Cells. Washington, DC: ASM Press; 2002. pp. 519–526. [Google Scholar]
- 8.Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Yu YTN, Yuan X, Velicer GJ. Adaptive evolution of an sRNA that controls Myxococcus development. Science. 2010;328:993. doi: 10.1126/science.1187200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toledo-Arana A, Repoila F, Cossart P. Small non-coding RNAs controlling pathogenesis. Curr Opin Microbiol. 2007;10:182–188. doi: 10.1016/j.mib.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Silvaggi JM, Perkins JB, Losick R. Genes for small, noncoding RNAs under sporulation control in Bacillus subtilis. J Bacteriol. 2006;188:532–541. doi: 10.1128/JB.188.2.532-541.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmalisch M, Maiques E, Nikolov L, Camp AH, Chevreux B, Muffler A, et al. Small genes under sporulation control in the Bacillus subtilis genome. J Bacteriol. 2010;192:5402–5412. doi: 10.1128/JB.00534-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saito S, Kakeshita H, Nakamura K. Novel small RNA-encoding genes in the intergenic regions of Bacillus subtilis. Gene. 2009;428:2–8. doi: 10.1016/j.gene.2008.09.024. [DOI] [PubMed] [Google Scholar]
- 14.Rasmussen S, Nielsen HB, Jarmer H. The transcriptionally active regions in the genome of Bacillus subtilis. Mol Microbiol. 2009;73:1043–1057. doi: 10.1111/j.1365-2958.2009.06830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marchais A, Naville M, Bohn C, Bouloc P, Gautheret D. Single-pass classification of all noncoding sequences in a bacterial genome using phylogenetic profiles. Genome Res. 2009;19:1084–1092. doi: 10.1101/gr.089714.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci USA. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lease RA, Cusick ME, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc Natl Acad Sci USA. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Will S, Reiche K, Hofacker IL, Stadler PF, Backofen R. Inferring noncoding RNA families and classes by means of genome-scale structure-based clustering. PLoS Comput Biol. 2007;3:65. doi: 10.1371/journal.pcbi.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffiths-Jones S. RALEE—RNA ALignment Editor in Emacs. Bioinformatics. 2005;21:257–259. doi: 10.1093/bioinformatics/bth489. [DOI] [PubMed] [Google Scholar]
- 21.Darty K, Denise A, Ponty Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;7:136–140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karmazyn-Campelli C, Rhayat L, Carballido-López R, Duperrier S, Frandsen N, Stragier P. How the early sporulation sigma factor σF delays the switch to late development in Bacillus subtilis. Mol Microbiol. 2008;67:1169–1180. doi: 10.1111/j.1365-2958.2008.06121.x. [DOI] [PubMed] [Google Scholar]
- 24.Guérout-Fleury AM, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- 25.Bechhofer DH, Oussenko I, Yao S, Mathy N, Condon C. Analysis of mRNA decay in Bacillus subtilis. Methods Enzymol. 2008;447:259–276. doi: 10.1016/S0076-6879(08)02214-3. [DOI] [PubMed] [Google Scholar]
- 26.Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]