Abstract
The cyclin dependent kinase inhibitor p27Kip1 plays an important role in controlling the eukaryotic cell cycle. The 5′-untranslated region of the p27 mRNA harbors an internal ribosome entry site (IRES) which may facilitate synthesis of p27 in certain conditions. In this study, the RNA-associated protein CUGBP1 was shown to interact with the human p27 5′-untranslated region. Overexpression of CUGBP1 inhibited endogenous p27 expression and reduced translation initiation through the p27 IRES. In contrast, repression of CUGBP1 by siRNA transfection enhanced p27 protein levels and stimulated p27 IRES activity. Addition of recombinant CUGBP1 repressed p27 IRES reporter mRNA translation in vitro. At last, our finding showed that cytosolic form of CUGBP1 binds efficiently to the p27 5′-untranslated region.
Key words: p27Kip1, cell cycle, IRES, CUGBP1, 5′-untranslated region
Background
The tumor suppressor protein p27Kip1 (p27) functions as an inhibitor of G1 cyclin-dependent kinases (CDKs). In many types of human cancer p27 levels are frequently reduced and this is associated with poor prognosis.1 As a tumor suppressor p27 is atypical in that it is rarely mutated in human cancers. Rather, it is aberrant regulation of p27 that is associated with loss of the protein in cancer cells. Cellular levels of p27 are regulated by multiple signaling mechanisms that affect transcription, translation and protein stability and it is these processes that are deregulated in cancer cells that lead to loss of the protein.
Translational control of the p27 mRNA is not completely understood but involves both the 3′ and 5′ untranslated regions (UTRs). The 3′-UTR harbors target sites for miRNAs miR221 and miR222 which repress p27 translation.2 The 5′-UTR of the p27 message harbors an internal ribosome entry site (IRES).3–5 Translation initiation through the p27 IRES is specifically impaired in cells derived from people with X-linked dyskeratosis congenita (X-DC).6 This is a genetic disease caused by a mutation in the DKC1 gene which encodes a pseudouridine synthase that modifies ribosomal RNA.7 One characteristic of this disease is increased susceptibility to cancer, which suggests that loss of p27 IRES activity could be involved in tumorigenesis.8 Recently, Bellodi C, et al. showed that DKC1 gene mutation impairs translation pre-initiation complex formation on p27 IRES element.9 The mechanism of translation initiation through the p27 IRES is not well understood but can be stimulated by polypyrimidine tract binding protein (PTB) and inhibited by HuR.3,4 Ribosome entry appears to occur within a large single-stranded loop that is just upstream of the AUG start codon.10 This single-stranded loop contains a U-rich element that is also the binding site recognized by HuR.11 In this report, a second RNA-associated protein that associates with the U-rich element has been identified. This factor, CUGBP1, represses p27 IRES activity and inhibits expression of endogenous p27 in cultured breast cancer cells. CUGBP1 is localized in both the nucleus and cytoplasm but only the cytoplasmic form of the protein is capable of binding the p27 5′-UTR.
Results
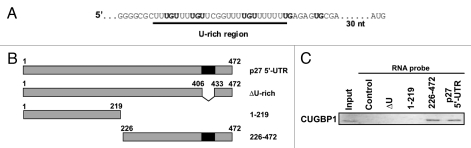
CUGBP1 associates to a U-rich region within the p27 5′-UTR.
The human p27 mRNA has a long 5′-UTR with a high GC content.13 However, it contains an U-rich element located ∼40 nucleotides upstream of the AUG start codon (Fig. 1A). This U-rich element has been shown to be important for translation of the p27 mRNA and to specifically interact with several RNA-binding proteins.4,11 The U-rich region appears to exist as a large single-stranded loop that is important for ribosome entry during cap-independent translation.10 The RNA-binding protein CUGBP1 has been shown to bind with high affinity to UG repeats.14 Marquis et al. defined a high affinity CUGBP1 binding element that consists of multiple UGU sequences within a span of at least 30 nucleotides. The U-rich region of the human p27 5′-UTR is highly similar to this element (Fig. 1A). Therefore, experiments were performed to test if CUGBP1 associates to this region. Biotinylated RNA pulldown assays were performed using various p27 5′-UTR truncations and MCF7 cell extracts (Fig. 1B and C). CUGBP1 was able to associate with the full-length p27 5′-UTR but not with an unrelated control RNA of similar length. The association of CUGBP1 required sequences within the 5′-UTR downstream of nucleotide 219 including the U-rich region. A mutated p27 5′-UTR lacking the U-rich region did not interact with CUGBP1. Thus the U-rich region is required for CUGBP1 to associate with the p27 5′-UTR.
Figure 1.
CUGBP1 associates to a U-rich region of the p27 5′-UTR. (A) Sequence of the U-rich region of the human p27 5′-UTR. (B) Diagram of the human p27 5′-UTR with the U-rich region shown in black. Truncations and deletions used for RNA-binding assays are shown. (C) Binding of CUGBP1 to the p27 5′-UTR. Extracts prepared from MCF7 were used for biotin-labeled RNA pulldown assays. Biotin-labeled probes included a control RNA derived from a plasmid vector, the full-length p27 5′-UTR, and the deletions/truncations shown in (B). RNA-bound CUGBP1 was eluted and assayed by western blotting. For the input lane total MCF7 cell extract was used.
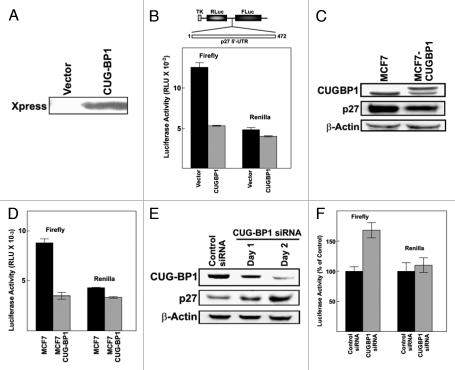
CUGBP1 inhibits p27 IRES activity.
A bicistronic reporter plasmid, pTKLL472, was used as to examine p27 IRES activity.5 An expression vector encoding CUGBP1 (pCDNA3.1-HisB-CUGBP1) or empty vector was transiently cotransfected with pTKLL472 into MCF7 breast cancer cells. The expression of Xpress tagged CUGBP1 in MCF7 was confirmed by western blotting (Fig. 2A). CUGBP1 had almost no effect on expression of Renilla luciferase, which was encoded by the upstream cistron and expected to be translated through a cap-dependent mechanism (Fig. 2B). However, expression of firefly luciferase, which was encoded by the downstream cistron, was strongly repressed by CUGBP1. These results suggest that overexpression of CUGBP1 inhibits cap-independent p27 translation initiation through the p27 5′-UTR.
Figure 2.
CUGBP1 inhibits p27 IRES activity and expression of endogenous p27. (A) MCF7 cells were cotransfected with the bicistronic reporter construct pTKLL472 and either empty vector (pCDNA3.1-HisB) or pCDNA3.1-HisB-CUGBP1 encoding Xpress-tagged CUGBP1. One day after transfection, the expression of CUGBP1 was tested by western blotting. (B) One day after transfection cell lysates were assayed for luciferase activity. Firefly luciferase activity represents p27 5′-UTR IRES-dependent expression and Renilla luciferase activity represents cap-dependent translation. The mean of two independent experiments is shown and error bars represent standard error. (C) An MCF7 cell line constitutively overexpressing CUGBP1 was generated as described in Methods. The levels of CUGBP1, p27 and β-actin in the parental MCF7 cell line and the CUGBP1 overexpressing cell line were compared by western blotting. (D) The bicistronic reporter construct pTKLL472 was transiently transfected into MCF7 cells or MCF7-CUGBP1 cells. Firefly and renilla Luciferase assays were performed as described in (A). (E) MCF7 cells were transfected with control (non-targeting) or CUGBP1-specific siRNAs. CUGBP1 siRNA transfected cells were harvest 24 hours or 48 hours after transfection. The levels of CUGBP1, p27 and β-actin were examined by western blotting. (F) MCF7 cells were transfected with control or CUGBP1-specific siRNAs. Three days later the cells were transfected with the pTKLL472 bicistronic reporter construct. After one additional day the cells were harvested for luciferase assays. Luciferase values were normalized to the levels observed in cells transfected with the control siRNA.
MCF7 cell lines that constitutively overexpress CUGBP1 were generated by stable transfection of pCDNA3.1-HisB-CUGBP1. The stable cell line MCF7-CUGBP1 overexpressed CUGBP1 as shown by western blotting (Fig. 2C). Transient transfection of the pTKLL472 reporter construct into parental MCF7 cells or MCF7-CUGBP1 cells showed that p27 IRES activity was also downregulated by stable expresson of CUGBP1 (Fig. 2D). This line also displayed a lower level of endogenous p27 than the parental MCF7 cell line (Fig. 2C).
To further demonstrate the role of CUGBP1 in p27 IRES activity and expression, siRNA targeted to CUGBP1 was used to knock down endogenous CUGBP1 in MCF7 cells. CUGBP1 siRNA or a control siRNA was transfected into MCF7 cells which were harvested 24 or 48 hours later. Total cell lysates were analyzed by western blotting to examine levels of CUGBP1 and p27 expression (Fig. 2E). Cells transfected with CUGBP1 siRNA had decreased CUGBP1 expression relative to control cells. Decreased CUGBP1 expression correlated with higher levels of p27 protein accumulation, suggesting an inhibitory effect of CUGBP1 on p27 expression. To test whether downregulation of CUGBP1 also affects translation initiation through the p27 IRES, control or CUGBP1 siRNAs were transfected into MCF7 cells. After three days the cells were transfected with the bicistronic reporter construct pTKLL472. Cells that were transfected with the siRNA targeting CUGBP1 showed enhanced expression of firefly luciferase relative to cells transfected with the control siRNA (Fig. 2F). Firefly luciferase was encoded by the downstream cistron that was translated in a p27 5′-UTR-dependent manner. The upstream cistron encoding Renilla luciferase was expressed at nearly equal levels with both control and CUGBP1 siRNAs. This further suggests that CUGBP1 inhibits translation initiated through the p27 5′-UTR.
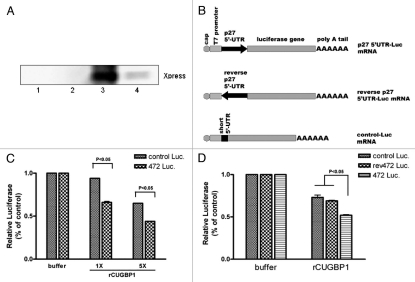
CUGBP1 inhibits p27 IRES reporter mRNA translation in vitro.
Recombinant His-tagged CUGBP1 protein was expressed and purified as described in Methods section. The recombinant CUGBP1 protein was tested by western blotting using anti-Xpress tag antibody (Fig. 3A). The effect of CUGBP1 on p27 IRES activity was tested using the RRL in vitro translation system. Purified recombinant protein was added to the reaction and the efficiency of p27 IRES initiated luciferase mRNA translation was measured. Two control luciferase mRNAs (Control-Luc mRNA and rev472-Luc mRNA) were used to show the specificity of the CUGBP1 effect on p27 IRES activity (Fig. 3B). Control-Luc mRNA contained 50 nucleotide 5′UTR that was derived from the vector and was unrelated to the p27 and a downstream luciferase gene. The translation of the control-Luc mRNA was cap-dependent. Rev472-Luc mRNA had an inverted hp27 5′UTR upstream of the luciferase gene. Previous work in our lab showed that the reverse human p27 5′UTR has low IRES activity (unpublished data). Thus the rev472-Luc mRNA 5′UTR had same length as wild-type 472-Luc mRNA, but low IRES activity. For normalization, the values of luciferase in the absence of CUGBP were set to one. As shown in Figure 3C, rCUGBP1 addition inhibited both control-Luc and 472-Luc mRNA translation in a dose dependent manner. However, CUGBP inhibited 472-Luc mRNA translation to a greater extent, which suggests some specificity for the p27 5′UTR as a target of CUGBP1. Another experiment compared the two control reporter mRNAs to the p27 5′UTR containing mRNA. CUGBP inhibited translation of all reporter mRNAs translation. However, again, CUGBP significantly inhibited p27 IRES activity more than control mRNAs translation.
Figure 3.
CUGBP1 inhibits p27 IRES reporter mRNA translation. (A) Recombinant CUGBP1 protein was expressed and purified as described in Methods. CUGBP1 in each elution fraction was detected by western blotting. The purified protein from the 200 mM imdazole elution fraction (lane 3) was dialyzed and further used for the following experiment. (B) Diagram of reporter mRNAs 472-Luc, rev472-Luc and control-Luc used in the following experiment. (C) Reporter mRNAs were synthesized and used for in vitro translation as described in Methods section. Increasing doses of rCUGBP1 protein (1 × 50 ng, 5 × 250 ng) was added in reaction mixture. The translation efficiency of each sample was measured by luciferase assay. (D) Different reporter mRNAs were used for in vitro translation with or without 250 ng rCUGBP1. The translation efficiency of each sample was measured by luciferase assay.
Overall, the results suggest that CUGBP inhibits p27 IRES activity in a dose dependent manner, and that the p27 5′UTR is a preferred target for CUGBP1.
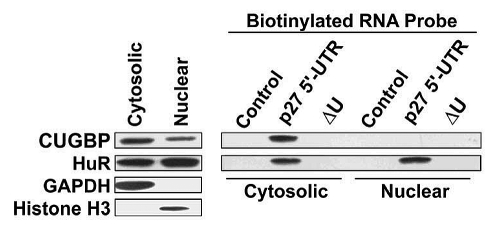
Cytosolic and nuclear CUGBP1 have differential binding activity for the p27 5′-UTR.
CUGBP1 is localized to both the nucleus and cytosol and is known to shuttle between the two compartments.16 In the nucleus CUGBP1 plays a role in regulating alternative splicing events and in the cytosol it functions to control translation or degradaton of specific mRNAs.17 The localization and various biological functions of CUGBP1 appear to be regulated by specific signaling mechanisms. For example, phosphorylation of CUGBP1 changes its binding affinity for various RNA sequences.17–19 It is likely that only a subset of the cellular pool of CUGBP1 is involved in regulating p27 translation. To examine this, MCF7 extracts were separated into nuclear and cytosolic fractions. In agreement with previous findings, CUGBP1 was found in both compartments,16 with somewhat lower levels in nuclei (Fig. 4 and left part). The extracts were used for biotin-labeled RNA pulldown assays to examine binding of nuclear and cytoplasmic CUGBP1 to the p27 5′-UTR (Fig. 4 and right part). Cytosolic CUGBP1 efficiently associated to the p27 5′-UTR but no binding of nuclear CUGBP1 was observed. HuR is another RNA-binding protein that shuttles between the nucleus and cytosol and is known to bind to the U-rich element in the p27 5′-UTR.4,11 In contrast to CUGBP1, both nuclear and cytosolic forms of HuR efficiently bound to the p27 5′-UTR. Deletion of the U-rich elements eliminated binding of both CUGBP1 and HuR. These findings suggest that nuclear and cytosolic forms of CUGBP1 are differentially modified and that it is the cytosolic form of the protein that plays a role in regulating p27 expression.
Figure 4.
Nuclear and cytosolic CUGBP1 have different p27 5′-UTR binding activities. MCF7 cells were fractionated into nuclear and cytosolic fractions. These fractions were then subjected to western blot analysis (left part) for CUGBP1, HuR, GAPDH (cytosolic marker) and histone H3 (nuclear marker). The same cytosolic and nuclear extracts were used for RNA binding assays using the indicated biotinylated probes (right part). RNA-bound proteins were analyzed by western blotting to detect CUGBP1 and HuR.
Discussion
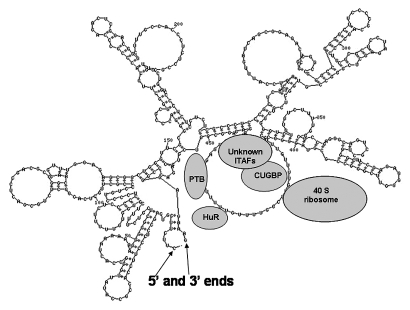
In this study we provide evidence that CUGBP1 associates to the 5′-UTR of human p27 mRNA and inhibits expression of p27 (Fig. 5). Overexpression of CUGBP1 is associated with lower levels of endogenous p27 and also represses p27 IRES activity. In contrast, knockdown of CUGBP1 using siRNAs leads to enhanced expression of p27 that correlates with increased IRES activity. Recombinant CUGBP1 protein addition specifically represses p27 IRES reporter mRNA translation in vitro in a dose dependent manner. These findings suggest that association of CUGBP1 to the p27 5′-UTR interferes with IRES-dependent initiation of translation. The region of the 5′-UTR that is recognized by CUGBP1 is a U-rich element located ∼40 nucleotides upstream of the start codon. This U-rich element is highly similar to the consensus CUGBP1 binding site that was reported by Marquis et al. suggesting a direct interaction between CUGBP1 and the p27 5′-UTR. However, we were unable to detect direct binding of recombinant CUGBP1 to p27 5′-UTR using RNA Electrophoresis Mobility Shift Assay (REMSA) (data not shown). This is probably because that CUGBP1 is indirectly associated to the p27 5′UTR. It may also because that CUGBP1 binding to the mRNA requires other factors. The region of the p27 5′-UTR that includes the U-rich element exists as a large single-stranded loop that functions in ribosome recruitment during initiation of translation.10 CUGBP1 binding may therefore interfere with p27 translation by blocking ribosome recruitment.
Figure 5.
p27 IRES regulation model.
In addition to U-rich or UG-rich sequences, CUGBP1 has been shown to interact with specific GC-rich elements. It has been suggested that phosphorylation acts as a switch that determines whether CUGBP1 binds to U-rich elements or GC-rich elements.18,20 Unphosphorylated CUGBP1 may preferentially bind to U-rich elements while phosphorylated CUGBP1 may preferentially bind to GC-rich target sites.17 CUGBP1 is also subject to phosphorylation by multiple protein kinases which further contributes to its binding specificity. Salisbury et al. showed that CUGBP1 is phosphorylated by Akt at serine 28 and that this enhances binding to GC-islands in the 5′ region of the cyclin D1 mRNA in proliferating myocytes. Skeletal muscles from myotonic dystrophy (DM1), which overexpress CUGBP1 and have higher rates of proliferation than normal skeletal muscles, express elevated levels of cyclin D1 but reduced levels of p27.18 The same group showed that CUGBP1 is phosphorylated at serine 302 by cyclin D3/CDK4. Phosphorylation at this site is associated with differentiation and enhances binding of CUGBP1 to the 5′ regions of p21 and C/EBPβ mRNAs.18 It will be of interest to determine if phosphorylation of CUGBP1 also modulates binding to the p27 mRNA 5′-UTR.
Recently, it was shown that CUGBP1 plays a role in mediating decay of short-lived mRNAs by binding to a GU-rich consensus sequence.21 This sequence was found in the 3′-UTR of the transcripts that are destabilized in a CUGBP1-dependent manner. However, unlike destabilized mRNAs, the CUGBP1 binding site in the p27 message is within the 5′-UTR. Thus it appears that the position of the binding site, as well as the specific RNA sequence, is important for CUGBP1 function. CUGBP1 has been shown to play a role in regulating IRES-dependent translation of the cytoplasmic serine hydroxymethyltransferase (cSHMT) mRNA.22 In this case CUGBP1 is thought to bind to an element in the 3′-UTR and interact with H ferritin to promote initiation of translation. This is in contrast to our finding that CUGBP1 binds to the 5′-UTR of the p27 mRNA and inhibits IRES activity. Again this suggests that position of the binding site is an important factor in determining the consequences of CUGBP1 interactions with mRNAs.
CUGBP1 has both nuclear and cytosolic functions.17 In MCF7 cells significant amounts of CUGBP1 were found in the nucleus but only cytosolic CUGBP1 was able to associate with the p27 5′-UTR. The reason for this difference is not yet known. The subcellular localization of CUGBP1 is thought to be regulated by phosphorylation, with the hypophosphorylated form accumulating in the nucleus.23 It is possible that cytosolic and nuclear CUGBP1 are differentially modified and only cytosolic form of CUGBP1 is able to associate with the p27 5′-UTR. It is also possible that CUGBP1 binding to the p27 5′-UTR requires other factors that are also differentially localized in the cytosol and nucleus. The differential binding may also because that all nuclear CUGBP1 was bound to mRNAs and no free CUGBP1 was available.
How CUGBP1 inhibits cap-independent translation and how CUGBP1 cooperates with other p27 5′-UTR-binding proteins in regulating p27 expression are questions that will require additional experiments. Finally, it will be important to explore the possibility that CUGBP1 plays a role in mediating the observed changes in p27 levels during the normal process of cellular differentiation and in diseases such as cancer and DM1.
Methods
Plasmids construction.
Primers and templates information was summarized in Table 1. Human CUGBP1 cDNA was amplified from plasmid GFP-CUGBP1 (kindly provided by Dr. Lubov Timchenko). The dual luciferase bicistronic plasmid pTKLL472 was previously described in reference 5.
Table 1.
Primer sequences
| Primer | Sequence | Template | Comments |
| 5′CUGBP-PC | AGG ATC CAA TGA ACG GCA CCC TGG A | GFP-CUGBP1 plasmid | primers for subcloning hCUGBP1 cDNA into pCDNA3.1-HisB |
| 3′CUGBP-PC | ACT CGA GTC AGT AGG GCT TGC TGT CA | ||
| 5′CUGBP-PR | AGC TCG AGA ATG AAC GGC ACC CTG GA | pCDNA3.1-HisB-CUGBP1 | primers for subcloning hCUGBP1 cDNA into pRSETB |
| 3′CUGBP-PR | AGA AGC TTT CAG TAG GGC TTG CTG C | ||
| 5′p27UTR | GGA TCC TAA TAC GAC TCA CTA TAG GCT TCT TCG TCA GCC TCC CTT | pTKLL472 | primers for p27 5′UTR DNA amplification |
| 3′p27UTR | CTT TCT CCC GGG CCG TGG CTC GTC GGG G | ||
| 5′p27UTR226 | GGA TCC TAA TAC GAC TCA CTA TAG GCC TCT CCG CCC TCC CGC TCG C | pTKLL472 | primers for p27 5′UTR 226–472 truncation DNA amplification |
| 3′p27UTR | CTT TCT CCC GGG CCG TGG CTC GTC GGG G | ||
| 5′p27UTR | GGA TCC TAA TAC GAC TCA CTA TAG GCT TCT TCG TCA GCC TCC CTT | pTKLL472 | primers for p27 5′UTR 1–219 truncation DNA amplification |
| 3′p27UTR219 | CTT TCT CCC GGG CCG TGG CTC GTC GGG G | ||
| 5′p27UTR | GGA TCC TAA TAC GAC TCA CTA TAG GCT TCT TCG TCA GCC TCC CTT | pTKLL472-ΔU | primers for p27 5′UTR U-rich region deletion DNA amplification |
| 3′p27UTR | CTT TCT CCC GGG CCG TGG CTC GTC GGG G | ||
| 5′-472-Luc-PolA | GGA TCC TAA TAC GAC TCA CTA TAG GCT TCT TCG TCA GCC TCC CTT | pTKLL472 | primers for p27 5′UTR-Luciferase-poly A DNA amplifcation |
| 3′-472-Luc-PolA | TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT AGC TAA GAA TTT CGT CAT CG | ||
| 5′-rev472-Luc-PolA | TAA TAC GAC TC A CT A TAG GAA AGA GGG CCC AGA CGT | pTKLL472R | primers for p27 rev5′UTR-Luciferase-poly A DNA amplifcation |
| 3′-rev472-Luc-PolA | TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT AGC TAA GAA TTT CGT CAT CG | ||
| 5′-ctrl-Luc-PolA | TAA TAC GAC TC A CTA TAG GG | pCDNA3.1-Luc | primers for Luciferase-poly A DNA amplifcation |
| 3′-ctrl-Luc-PolA | TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT AGC TAA GAA TTT CGT CAT CG |
Cell culture, transfection and reporter gene assays.
The human breast cancer cell line MCF7 was cultured in Dulbecco's Modified Eagle's Medium (DMEM) containing 10% fetal calf serum, 100 U/ml penicillin and 100 µg/ml streptomycin at 37°C in a humidified atmosphere containing 5% CO2. Transient transfection of DNAs was performed using DreamFect (Boca Scientific, DF-40500) following the manufacturer's guidelines. For stable transfection, MCF7 cells cultured in 35 mm dishes were transfected with pCDNA3.1-HisB-CUGBP1. One day later, cells were trypsinized and plated in 150 mm dishes. Antibiotic G418 was added to a final concentration of 1 mg/ml and continuously maintained in selection medium. Colonies of cells were isolated, expanded and expression of Xpress-tagged CUGBP1 was tested by western blotting. CUGBP1 siRNA and control siRNA were purchased from Ambion. siRNAs were transfected using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol. Dual luciferase assays were performed using the Dual-Glo Luciferase Assay system (Promega, E2920) as previously described in reference 5.
Preparation of biotinylated RNA probes.
The full-length human p27 5′UTR and its mutants were amplified by PCR as described in Table 1. The PCR products were used as templates for synthesis of biotin-labeled RNAs using AmpliScribe T7-Flash Transcription Kit (Epicentre Biotechnologies, ASF3257) according to the manufacturer's guidelines.
Cell lysis and fractionation.
Cells were harvested and resuspended in five volumes of cell lysis buffer (10 mM Tris pH 8.0, 140 mM NaCl, 1.5 mM MgCl2, 0.1% NP40) and incubated on ice for 5 min. Then the sample was centrifuged at ∼1,000 g to pellet nuclei. The supernatant was used as the cytosolic extract. The nuclear pellet was washed once with NP40-free lysis buffer and resuspended in the same volume of complete lysis buffer as used for the cytosolic fraction. The resuspended nuclei were briefly sonicated on ice. Both nuclear and cytosolic fractions were clarified by centrifugation at 10,000 g and then stored at −80°C until used.
Biotin-labeled RNA pulldown assays.
Biotin-labeled RNA pulldown assays were performed using Dynabeads M-280 Streptavidin (Invitrogen 112-05D). For each sample, 20 µl of resuspended beads were used. Beads were washed twice with solution A [0.1 M NaOH, 0.05 M NaCl prepared in diethylpyrocarbonate (DEPC) treated water], once in solution B (0.1 M NaCl in DEPC treated water) and once in B&W buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA, 2.0 M NaCl). The beads were resuspended in 60 µl B&W buffer and then 1 µl Superase-In (Ambion, AM2694) and 2 µg biotin-labeled RNA were added. The samples were incubated at room temperature for 20 min with occasional mixing. Free biotin-labeled RNAs were removed by collecting the beads using a magnetic stand and then washing the beads twice with B&W buffer. The RNA loaded beads were resuspended with protein samples for a total volume of 200 µl. The samples were incubated at 15°C for 30 min with mixing every 5 min. The beads were then washed five times with cell lysis buffer to remove unbound proteins. After the last washing step, beads were resuspended in 20 µl SDSPAGE sample buffer and the eluted proteins tested by western blotting.
Preparation of capped RNAs for in vitro translation.
The reporter constructs were amplified as described in Table 1. The PCR products were used as templates for capped RNA synthesis using mMESSAGE mMACHINE T7 kit (Ambion, AM1344) following the product instructions.
Recombinant CUGBP expression and purification.
Competent E. coli strain Rossetta cells (Novagen 69450-3) were transformed by pRSETB constructs carrying the CUGBP1 cDNA inserts following the manufacturer's guidelines. Recombinant protein was expressed and purified as previously described in reference 12. Specifically, bacteria cell lysate with target protein was loaded onto a Ni2+ resin column (Novagen). After wash, bound protein was eluted by elution buffer with increasing dose of imidazole (500 mM NaCl, 20 mM Tris, pH 7.9 with 60 mM, 100 mM, 200 mM or 1 M imidazole).
In vitro translation.
Rabbit Reticulocyte Lysate (RRL) was purchased from Promega (L4960). In vitro translation was performed by mixing: 0.25 µl Amino acid mixture minus Leucine, 0.25 µl Amino acid mixture minus methionine, 0.5 µl SuperaseIn, 2 µg reporter mRNA, 50 ng or 250 ng purified rCUGBP1, 17.5 µl RRL and adding water to a final volume of 25 µl. Then the samples were incubated at 30°C for 90 min. The resulting products were analyzed by luciferase assays.
Conclusions
CUGBP1 is a novel regulator of p27 translation. CUGBP1 binds to p27 5′UTR and represses p27 IRES activity. Nuclear and cytosolic CUGBP1 have different affinity to p27 5′UTR. To summarize, our data suggest that CUGBP1 may play a role in modulating levels of p27 protein during differentiation, cellular stress or tumorigenesis.
Acknowledgements and Funding
This work was supported by the National Institutes of Health (grant number R01CA084325).
Authors' Contributions. Y.Z. and W.K.M. initiated the work, designed the experiments and wrote the paper. Y.Z. performed the experiments and statistical analysis.
Abbreviations
- 5′UTR
5′un-translated region
- IRES
internal ribosomal entry site
- CUGBP1
CUG-binding protein 1
References
- 1.Chu IM, Hengst L, Slingerland JM. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat Rev Cancer. 2008;8:253–267. doi: 10.1038/nrc2347. [DOI] [PubMed] [Google Scholar]
- 2.Zhang C, Kang C, You Y, Pu P, Yang W, Zhao P, et al. Co-suppression of miR-221/222 cluster suppresses human glioma cell growth by targeting p27kip1 in vitro and in vivo. Int J Oncol. 2009;34:1653–1660. doi: 10.3892/ijo_00000296. [DOI] [PubMed] [Google Scholar]
- 3.Cho S, Kim JH, Back SH, Jang SK. Polypyrimidine tract-binding protein enhances the internal ribosomal entry site-dependent translation of p27Kip1 mRNA and modulates transition from G1 to S phase. Mol Cell Biol. 2005;25:1283–1297. doi: 10.1128/MCB.25.4.1283-1297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kullmann M, Gopfert U, Siewe B, Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev. 2002;16:3087–3099. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang H, Coleman J, Miskimins R, Srinivasan R, Miskimins WK. Cap-independent translation through the p27 5′-UTR. Nucleic Acids Res. 2007;35:4767–4778. doi: 10.1093/nar/gkm512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, et al. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- 7.Kirwan M, Dokal I. Dyskeratosis congenita: a genetic disorder of many faces. Clin Genet. 2008;73:103–112. doi: 10.1111/j.1399-0004.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- 8.Ruggero D, Grisendi S, Piazza F, Rego E, Mari F, Rao PH, et al. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003;299:259–262. doi: 10.1126/science.1079447. [DOI] [PubMed] [Google Scholar]
- 9.Bellodi C, Krasnykh O, Haynes N, Theodoropoulou M, Peng G, Montanaro L, et al. Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res. 70:6026–6035. doi: 10.1158/0008-5472.CAN-09-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coleman J, Miskimins WK. Structure and activity of the internal ribosome entry site within the human p27 Kip1 5′-untranslated region. RNA Biol. 2009;6:84–89. doi: 10.4161/rna.6.1.7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Millard SS, Vidal A, Markus M, Koff A. A U-rich element in the 5′ untranslated region is necessary for the translation of p27 mRNA. Mol Cell Biol. 2000;20:5947–5959. doi: 10.1128/mcb.20.16.5947-5959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei Q, Eviatar-Ribak T, Miskimins WK, Miskimins R. Galectin-4 is involved in p27-mediated activation of the myelin basic protein promoter. J Neurochem. 2007;101:1214–1223. doi: 10.1111/j.1471-4159.2007.04488.x. [DOI] [PubMed] [Google Scholar]
- 13.Coleman J, Hawkinson M, Miskimins R, Miskimins WK. The major transcription initiation site of the p27Kip1 gene is conserved in human and mouse and produces a long 5′-UTR. BMC Mol Biol. 2001;2:12. doi: 10.1186/1471-2199-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mori D, Sasagawa N, Kino Y, Ishiura S. Quantitative analysis of CUG-BP1 binding to RNA repeats. J Biochem. 2008;143:377–383. doi: 10.1093/jb/mvm230. [DOI] [PubMed] [Google Scholar]
- 15.Marquis J, Paillard L, Audic Y, Cosson B, Danos O, Le Bec C, Osborne HB. CUG-BP1/CELF1 requires UGU-rich sequences for high-affinity binding. The Biochem J. 2006;400:291–301. doi: 10.1042/BJ20060490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujimura K, Kano F, Murata M. Dual localization of the RNA binding protein CUGBP-1 to stress granule and perinucleolar compartment. Exp Cell Res. 2008;314:543–553. doi: 10.1016/j.yexcr.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 17.Barreau C, Paillard L, Mereau A, Osborne HB. Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie. 2006;88:515–525. doi: 10.1016/j.biochi.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 18.Salisbury E, Sakai K, Schoser B, Huichalaf C, Schneider-Gold C, Nguyen H, et al. Ectopic expression of cyclin D3 corrects differentiation of DM1 myoblasts through activation of RNA CUG-binding protein, CUGBP1. Exp Cell Res. 2008;314:2266–2278. doi: 10.1016/j.yexcr.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baldwin BR, Timchenko NA, Zahnow CA. Epidermal growth factor receptor stimulation activates the RNA binding protein CUG-BP1 and increases expression of C/EBPbeta-LIP in mammary epithelial cells. Mol Cell Biol. 2004;24:3682–3691. doi: 10.1128/MCB.24.9.3682-3691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iakova P, Wang GL, Timchenko L, Michalak M, Pereira-Smith OM, Smith JR, Timchenko NA. Competition of CUGBP1 and calreticulin for the regulation of p21 translation determines cell fate. EMBO J. 2004;23:406–417. doi: 10.1038/sj.emboj.7600052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vlasova IA, Tahoe NM, Fan D, Larsson O, Rattenbacher B, Sternjohn JR, et al. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol Cell. 2008;29:263–270. doi: 10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woeller CF, Fox JT, Perry C, Stover PJ. A ferritin-responsive internal ribosome entry site regulates folate metabolism. J Biol Chem. 2007;282:29927–29935. doi: 10.1074/jbc.M706264200. [DOI] [PubMed] [Google Scholar]
- 23.Roberts R, Timchenko NA, Miller JW, Reddy S, Caskey CT, Swanson MS, Timchenko LT. Altered phosphorylation and intracellular distribution of a (CUG)n triplet repeat RNA-binding protein in patients with myotonic dystrophy and in myotonin protein kinase knockout mice. Proc Natl Acad Sci USA. 1997;94:13221–13226. doi: 10.1073/pnas.94.24.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]