Abstract
Polyadenylation (poly(A)) of mRNA plays a critical role in regulating gene expression. Identifying the sequence, structural and epigenomic determinants of poly(A) site usage is an important long-term goal. Several cis elements that mediate poly(A) regulation have been identified. Highly used poly(A) sites are also known to have a greater nucleosome occupancy in the immediate downstream. However, a detailed exploration of additional epigenomic and mRNA structural correlates of poly(A) site usage has not been reported. Importantly, functional interaction between sequence, structure and the epigenome in determining the poly(A) site usage is not known. We show that highly used poly(A) sites are positively associated with an mRNA structure that is energetically more favorable and one that better exposes a critical polyadenylation cis element. In exploring potential interplay between RNA and chromatin structure, we found that a stronger nucleosome occupancy downstream of poly(A) site strongly correlated with (1) a more favorable mRNA structure and (2) a greater accumulation of RNA Polymerase II (PolII) at the poly(A) site. Further analysis suggested a causal relationship pointing from PolII accumulation to a stable RNA structure. Additionally, we found that distinct patterns of histone modifications characterize poly(A) sites and these epigenetic patterns alone can distinguish true poly(A) sites with ∼76% accuracy and also discriminate between high and low usage poly(A) sites with ∼74% accuracy. Our results suggest a causative link between chromatin structure and mRNA structure whereby a compacted chromatin downstream of the poly(A) site slows down the elongating transcript, thus facilitating the folding of nascent mRNA in a favorable structure at poly(A) site during transcription. Additionally we report hitherto unknown epigenomic correlates for poly(A) site usage.
Key words: polyadenylation, RNA structure, epigenomics, histone modifications
Introduction
Polyadenylation [poly(A)] is a two phase process that involves endonucleolytic cleavage at a poly(A) site of nascent mRNA followed by addition of poly(A) tail.1 Nearly all mature mRNAs are polyadenylated and poly(A) is critical for many aspects of mRNA metabolism, including mRNA stability, translation and transport.2,3 Much like alternative splicing and alternative usage of transcription start sites, alternative usage among multiple poly(A) sites is a vital mechanism by which the multitude of gene isoforms are regulated.4,5 Characterizing the factors that mediate the usage of a specific poly(A) site is an important long term goal in biology.
The sequence flanking a poly(A) site harbors several cis elements that are specifically recognized by trans-acting factors, which then carry out the cleavage and the addition of poly(A) tail at the 3′ end of the transcript.6 One of the most important of these cis elements is the hexamer A(A/U)UAAA, or a close variant, usually referred to as the polyadenylation signal (PAS), and is located 10–35 nt upstream of most human poly(A) sites.7 Additional cis elements include TGTA, TATA, G-rich and C-rich elements.8 The PAS acts as a substrate for Cleavage and Polyadenylation Specificity factor (CPSF) which is critical for poly(A) recognition.1 In addition to the primary sequence elements, mRNA structure plays an important role in poly(A) site selection.9,10 For instance, in the case of human immunodeficiency virus type 1 mRNA, occlusion of PAS by the stem region of a hairpin structure can interfere with its binding to CPSF, thus inhibiting polyadenylation.11 Besides enabling proper presentation of PAS, mRNA structure may also facilitate bringing PAS in close proximity to other enhancer elements important for correct identification of PAS by CPSF.12 Finally, epigenomic features and chromatin structure has been shown to be an important determinant of poly(A) site usage. For instance, CpG methylation in the region separating consecutive poly(A) sites in murine H13 gene influences the relative usage of poly(A) sites.13 Furthermore, even though the genomic regions near poly(A) sites are generally depleted of nucleosomes, highly used poly(A) sites have a significantly greater nucleosome occupancy downstream of the site than the rarely used poly(A) sites.14 Despite progress in identifying sequence, structural and epigenomic determinants of poly(A) site selection, additional factors remain to be identified and, more importantly it is not clear how these various properties interact with each other in ultimately determining the poly(A) site usage.
Here we show that, in addition to the known association between high poly(A) usage and high nucleosome occupancy,14 the highly used poly(A) sites are also associated with a more favorable mRNA structure. A direct comparison between nucleosome occupancy and mRNA structure revealed that greater nucleosome occupancy downstream of a poly(A) site strongly correlates with an mRNA structure at the poly(A) site that is energetically more stable and that better exposes the critical cis element—PAS. In further exploring the interaction between chromatin and RNA structure, we found that the downstream nucleosome occupancy also positively correlated with a greater accumulation of PolII at the poly(A) site. We carried out further analysis to discern a causal relationship between PolII accumulation and RNA structural stability. Our analysis suggests that PolII accumulation facilitates the formation of stable RNA structure. Taken together, our findings are consistent with a mechanism whereby a compacted chromatin immediately downstream of the poly(A) site promotes the slowing down of elongating transcript resulting in greater PolII accumulation, thus facilitating the folding of mRNA in a structure that is energetically and functionally favorable for polyadenylation around the poly(A) sites.
Specific patterns of histone modifications have been previously found to associate with post-transcriptional regulation signals such as splice sites.15 Another study showed that poly(A) sites are marked by a decrease in H3K36 di- and trimethylation.16 Here we examined all available histone modifications in human CD4+ T cell17,18 and found that various modifications occur in distinct patterns surrounding poly(A) sites. These patterns are distinct from the underlying nucleosome occupancy pattern,14 and a support vector machine (SVM) classifier based on the histone modifications can distinguish functional poly(A) sites from background PAS with ∼76% accuracy. Notably, the classification based on epigenomic features alone was superior to classification accuracy based on 15 previously identified cis element features.8 Moreover, using epigenetic patterns we were able to differentiate high usage poly(A) sites from low usage poly(A) sites with ∼74% accuracy.
Overall, our results unfold novel epigenomic and mRNA structural correlates of polyadenylation and suggest a causative link between chromatin structure and mRNA structure favorable for polyadenylation site usage.
Results
Region surrounding the high usage polyadenylation sites exhibit a stable RNA structure that is conducive to its recognition.
Alternative poly(A) sites for a gene are utilized with varying frequency.19 Here we investigated whether the high and the low usage poly(A) sites differ in the mRNA structure of the region surrounding the poly(A) site. Based on EST data, the human poly(A) sites have been classified according to their usage frequency into 5,139 high usage and 21,204 low usage poly(A) sites19 (see Methods). We first quantified the structural stability of the region near poly(A) site by estimating the free energy (FE) of the thermodynamic ensemble of flanking 250 bp region (200 bp upstream and 50 bp downstream) using RNAfold.20 This region was chosen because it encompasses the most critical PAS, which is located 10–30 bp upstream of poly(A) site as well as other cis elements suggested to play a role in polyadenylation.1,8 We found that high usage poly(A) sites have a significantly lower FE than low usage poly(A) sites (Wilcoxon test p value < 2.2e–16). However, FE is known to depend on the GC content of the RNA sequence. Therefore we repeated the comparison of the high and the low usage poly(A) site regions after controlling for GC content using a matched sampling procedure (see Methods). Even with matched GC content, we found that high usage poly(A) sites have a significantly lower FE than low usage poly(A) sites (Wilcoxon test p value = 0.001). Thus our results are suggestive of a more stable RNA structure forming near the highly used poly(A) sites.
However, the FE alone does not immediately provide a mechanistic explanation of the observed differences between high and low usage poly(A) sites. RNA structure that appropriately exposes the critical cis elements is likely to be more favorable.11 The PAS located 10–30 bp upstream of the poly(A) site is critical for its recognition by the CPSF complex.21,22 We quantified the exposure of PAS in a folded mRNA by computing the fraction of nucleotides within the PAS involved in a base pairing; the fewer the base pairing, the greater the exposure of PAS. We found that the high usage sites have significantly fewer PAS nucleotides involved in base pairing (Wilcoxon p-value = 0.02). This result was observed despite the fact that high usage poly(A) regions had significantly more overall base pairing than the low usage poly(A) regions (Wilcoxon p-value = 9.9e–09), as would be expected from a lower FE.
In the above analysis, the poly(A) usage is derived from a global prevalence of ESTs pooled from multiple tissue types. Next, we investigated whether the above relationship is also observed based on cell type specific polyA usage. We obtained RNA-seq data for human CD4+ T-cells23 and used it to estimate poly(A) usage based on previously published method.24 However, the approaches to estimate poly(A) usage from RNA-seq data are in their infancy and not sufficiently accurate. Therefore, to minimize errors, we considered a poly(A) site high (respectively, low) usage if it was deemed so both by the EST-based measure as well as by the RNA-seq based measure (see Methods). This yielded 1,113 high usage poly(A) sites and 7,796 low usage poly(A) sites. Upon controlling for GC content using a matched sampling procedure (see Methods), we confirmed that here too the high usage poly(A) sites have a significantly lower FE than low usage poly(A) sites (Wilcoxon test p value = 0.03).
Taken together, our results suggest that high usage poly(A) sites are associated not only with higher nucleosome occupancy, as shown before,14 but also a more favorable RNA secondary structure near poly(A) sites.
A higher nucleosome occupancy downstream of poly(A) site significantly correlates with a more stable RNA structure around the poly(A) site.
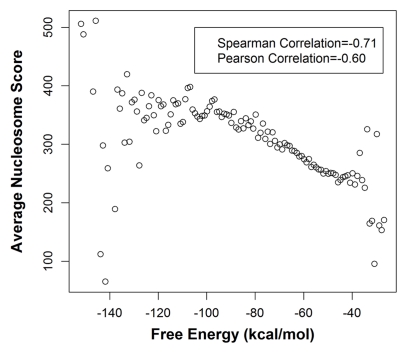
Given that highly used poly(A) sites have a greater nucleosome occupancy downstream, as well as a more stable secondary structure around the poly(A) site, as we show above, next we investigated the direct relationship between the downstream nucleosome occupancy and RNA structure around the poly(A) site, using all 31,871 poly(A) sites in 3′ UTRs (see Methods). For each poly(A) site we computed the FE of the RNA structure near the poly(A) site as above in the 250 bp region, and we also estimated the nucleosome occupancy score in the 600 bp downstream of the site based on ChIP-Seq nucleosome occupancy data from human CD4+ T cells25 (see Methods). As evident in the scatter plot shown in Figure 1, we found a strong negative correlation (Spearman rho = −0.71 p-value < 2.2e–16, Pearson rho = −0.60 p-value = 3.07e–13) between the FE and nucleosome occupancy. Since both nucleosome occupancy26 and FE strongly depend on the GC content of the sequence, we controlled for the GC content using a matched sampling procedure (see Methods). We still found that the poly(A) regions with high nucleosome occupancy (top 50 percentile) have a more stable RNA structure than those with low nucleosome occupancy (bottom 50 percentile) (Wilcoxon p value = 6.01e–14). Thus we conclude that a greater nucleosome occupancy downstream of poly(A) site corresponds to a more stable RNA structure at the poly(A) site.
High nucleosome poly(A) sites are associated with a greater PolII occupancy.
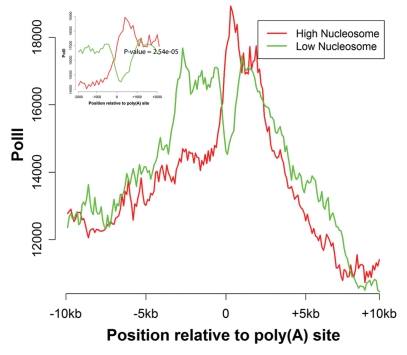
Pausing of PolII is known to associate with poly(A) site recognition and transcription termination.27–29 PolII pausing can enhance cleavage and polyadenylation30,31 possibly via greater recruitment of cleavage/polyadenylation factors at paused PolII.32 For example, it was previously suggested that PolII pausing modulates the poly(A) site usage in Immunoglobulin M pre-mRNA.28 There are several factors that can slow down elongating PolII such as stable RNA structure and compacted chromatin structure.29 Here we investigated whether a greater nucleosome occupancy downstream of a poly(A) site has a bearing on PolII pausing. PolII accumulation at a genomic site for a transcriptionally competent polymerase is indicative of pausing of PolII,32,33 therefore we used the density of PolII ChIP-seq tags in human CD4+ T cells to quantify PolII accumulation.17 We obtained two subsets of poly(A) sites according to downstream nucleosome occupancy score—high nucleosome occupancy (highest 25 percentile scores) and low nucleosome occupancy (smallest 25 percentile scores). Figure 2 reveals distinct patterns of PolII occupancy near the poly(A) sites for the two nucleosome occupancy based classes of poly(A) sites; there is a much greater accumulation of PolII in the high nucleosome class immediately downstream of poly(A) site. We quantified the significance of this difference using the χ2 test by comparing the number of PolII ChIP-seq tags mapped to 500 bp upstream and 500 bp downstream of the poly(A) sites for the two classes (p-value = 2.54e-05). Thus our results suggest that a greater nucleosome occupancy is associated with a greater accumulation of PolII immediately downstream of the poly(A) site which may be a result of PolII pausing,33 in addition to being associated with a more favorable secondary structure around the poly(A) site.
Figure 2.
Differential polII occupancy at Poly(A) sites with high versus low nucleosome occupancy. Poly(A) sites were classified based on the nucleosome occupancy in the 600 bp region downstream of the site. The figure shows the PolII occupancy profiles flanking the poly(A) sites for the high (top 25 percentile) (red) and low (bottom 25 percentile) (green) nucleosome occupancy classes. The profiles are centered around the poly(A) site. The inset shows that a greater PolII pausing immediately after the poly(A) site for high nucleosome poly(A) sites (Chi-square p-value = 2.54e-05).
We verified whether the observed association between PolII and nucleosome occupancy at poly(A) sites was a reflection of a broader relationship between the two or if it was specific to poly(A) sites. We analyzed whether the two quantities tracked each other in an extended region around the poly(A) site. As shown in Supplemental Figure 1, we did not find this to be the case and thus our observed relationship between PolII and nucleosome occupancy applies specifically at the poly(A) sites.
Dissecting the causality between PolII accumulation and RNA structure.
We have shown that high nucleosome occupancy downstream of the poly(A) sites is associated with a more stable RNA structure as well as with a significantly greater accumulation of PolII, possibly a manifestation of PolII pausing.33 However the causality between RNA structure and PolII pausing is not known. For instance, PolII pausing can affect mRNA structure, presumably by affecting transcriptional dynamics at poly(A) site,34 or conversely, a more stable RNA structure as well as the higher nucleosome occupancy downstream of poly(A) sites may jointly affect the transcriptional elongation by slowing down the PolII.29 To assess relative importance of the two possibilities, we probabilistically quantified the influence of PolII accumulation on the RNA structure and conversely, the influence of RNA structure on PolII accumulation in the region surrounding poly(A) sites.
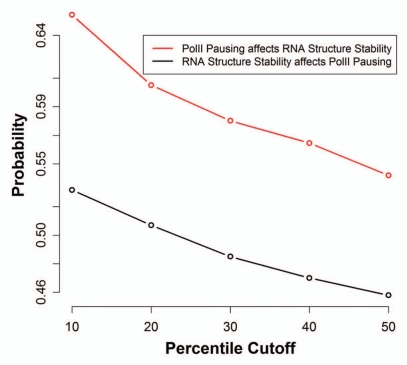
More specifically, we estimated the probability that a poly(A) site has low FE (lowest 50 percentile) given that the site has high PolII occupancy (in top 10, 20, 30, 40 and 50 percentile), and conversely, we estimated the probability that a poly(A) site has high PolII occupancy (greatest 50 percentile) given that the site has low FE (in bottom 10, 20, 30, 40 and 50 percentile). PolII occupancy was computed in the region 1,000 bp downstream of the poly(A) sites and the FE was predicted in the 250 bp region (200 bp upstream and 50 bp downstream) surrounding the poly(A) sites. We observed a consistently higher conditional probability (ranging from 0.54 to 0.65) of stable structure around poly(A) sites given that there was higher PolII accumulation, as compared with the conditional probability of a greater PolII accumulation (ranging from 0.46 to 0.53) given a more stable RNA structure (Fig. 3). Thus, PolII accumulation is more likely to have a causal effect on RNA structural stability of poly(A) region compared with the converse in the context of polyadenylation.
Figure 3.
Putative causality between PolII pausing and RNA structure stability. The figure shows in red the probability (y-axis) that a poly(A) site has low FE (in lowest 50 percentile) given that the site has high PolII occupancy (in top 10, 20, 30, 40 and 50 percentile shown on x-axis). The figure shows in black the probability (y-axis) that a poly(A) site has high PolII occupancy (greatest 50 percentile) given that the site has low FE (in bottom 10, 20, 30, 40 and 50 percentile shown on x-axis). PolII occupancy was computed for 1,000 bp downstream of poly(A) site and minimum free energy of RNA was computed for the 250 bp region (200 bp upstream and 50 bp downstream) surrounding the poly(A) site.
Taken together, our results are consistent with a model whereby a greater nucleosome occupancy downstream of poly(A) site causes slowing down of elongating transcript, as evidenced by a greater PolII accumulation, thus facilitating the formation of appropriate RNA secondary structure using only the already transcribed mRNA sequence in the immediate vicinity of poly(A) sites. This structure is energetically stable and also favorable for recognition of poly(A) sites as shown above.
Distinct patterns of histone modifications demarcate poly(A) site.
While nucleosome occupancy provides a high-level view of the chromatin organization, post-translational histone modifications (HM) provide a much richer mechanism to modulate chromatin structure, with implication for a variety of biological processes.15,35–37 Here we examined in greater detail the patterns of HM surrounding the 31,871 poly(A) sites in 3′ UTRs (see Methods).
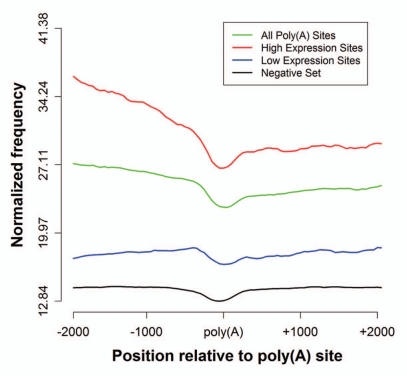
First, we pooled the ChIP-seq mapped tags for 37 HMs as well as for the Histone variant H2A.Z in human CD4+ T cells obtained from reference 17 and 18 and examined the pattern of the pooled data around the poly(A) sites (±2 kb). We observed that the overall distribution indicates a depletion of HMs in a narrow region around the poly(A) sites similar to the previously observed nucleosome occupancy profile around the poly(A) sites14 (Fig. 4). We also examined the pattern of the pooled HMs for the negative set formed of the 87,679 non-poly(A) regions harboring a PAS (See Methods) (Fig. 4). The overall levels of HMs is significantly lower at the negative sites (Wilcoxon p-value < 2.2e–16).
Figure 4.
Distribution of histone modifications around Poly(A) Sites. Profile was obtained by pooling all available HM ChIP-seq tags (see Methods) averaged in each 200 bp positional bin and a sliding window of 50 bp for all 3′ UTR poly(A) sites (green 31,871 sites), only the poly(A) sites belonging to high expression genes (red 7,583 sites), only the poly(A) sites belonging to the low expression genes (blue 7,577 sites) and non-poly(A) positions that form the negative set (black 87,679 sites). Profiles are centered on poly(A) sites.
Next, we examined the profiles of each of the 37 distinct HMs and H2A.Z separately and found several patterns surrounding the poly(A) sites that were distinct from the patterns of pooled histone modifications as well as distinct from the previously reported patterns of nucleosome occupancy.14 Consistent with previous observations,38 we found that the levels of H3K36me3, known to associate with transcription elongation, drops gradually after the poly(A) site (Sup. Fig. 2s). Several modifications such as H2BK5me1, H2BK20ac, H3K4me1, H3K4me2, H3K4me3, H3K9me1, H3K27me1, H3K27me2, H3K36me1, H3K18ac, H4K5ac and H4K91ac were depleted around the poly(A) site, following the pattern of nucleosome occupancy (Sup. Fig. 2d, g, i–l, o, p, r, ab, ah, al). Interestingly, in contrast, a few HMs were enriched around the poly(A) site, such as H3K9me3 and H3R2me2 (Sup. Fig. 2n and x). The levels of all HMs differed considerably for the negative set with the exception of H2AZ and H3K14ac (Sup. Fig. 2c and aa). The above analysis reveals rich and characteristic patterns of various HMs near poly(A).
While the functional relevance of the HM patterns to poly(A) usage cannot be determined from computational analysis alone and will require directed experiments, here we tested whether such patterns can be used to computationally distinguish functional poly(A) sites from other non-functional genomic sites harboring a putative PAS motif. As our set of features, we used the number of HM tags mapped to the six 200 bp bins in ±600 bp region flanking poly(A) site. As the negative control we randomly selected the same number of GC-matched poly(A)-like regions harboring a PAS (see Methods). We trained a SVM and then carried out the testing on independent datasets (see Methods). The classification accuracy for individual marks ranged between 51–71% (Sup. Table 1). We then extended the features by combining the top five marks based on their individual classification accuracy, resulting in 30 features, and re-trained SVM (see Methods). This resulted in an improved accuracy of 75.3%–75.5% (95% confidence interval, Sensitivity = 91.4%–91.5%, Specificity = 59.4%–59.5%). Combining the top ten marks resulted in further improved accuracy of 76.1%–76.2% (95% confidence interval, Sensitivity = 95.2–95.3%, Specificity = 56.9–57%).
As a comparison to classification by sequence elements, we repeated the SVM classification by using the 15 cis-elements surrounding the human poly(A) sites that have been previously shown to enhance polyadenylation8,39 (see Methods). This resulted in classification accuracy of 69.3%–69.5% (95% confidence interval, Sensitivity = 62.8%–62.9%, Specificity = 75.7%–75.8%). Thus, classification accuracy based on epigenomic features alone is superior to the classification based on sequence features. Classification based on epigenomic features resulted in a much greater sensitivity and slightly reduced specificity as compared to that obtained by using sequence features. Furthermore, repeating the classification by combining the 15 cis-elements with the top performing 5 HMs did not result in any improvement in the accuracy (data not shown).
HM levels surrounding transcription start sites have been previously shown to correlate with the level of gene expression.35 Here we examined whether this is also true for HMs around the poly(A) sites. Based on the gene expression data for human CD4+ T cells, we partitioned the poly(A) sites into 7,583 highly expressed (top 25 percentile by overall gene expression) and 7,577 lowly expressed (bottom 25 percentile). We found that although for most HMs the overall pattern of HMs near poly(A) sites is similar between the two classes, the highly expressed genes have higher levels of HMs (Sup. Fig. 2). However, for a few of the marks—H2AZ, H3K9me2, H3K9me3, H3K27me2, H3K27me3, H3K36me1, H3R2me1, H3K14ac, the opposite is true (Sup. Fig. 2c, m, n, p–r, w and aa). Also, the levels of H2AK9ac, H3R2me2, H3K9ac, H3K18ac, H3K36ac and H4R3me2 around poly(A) sites do not vary substantially with expression (Sup. Fig. 2b, x, z, ab, ae and ag).
Patterns of histone modifications differentiate high and low usage poly(A) sites.
We compared the HM patterns between high and low usage poly(A) sites obtained using EST/cDNAs and separately for the subset of these that agreed with the usage based on CD4+ T cell RNA-seq data. The pattern is highly similar for these two sets (Sup. Figs. 3 and 4) and for simplicity we have described the findings using the larger EST/cDNA based usage set of poly(A) sites. We emphasize that the categorization of poly(A) sites by usage is independent of the overall level of gene expression. Although the patterns of histone modification were similar between the high and low usage sites, the level of certain marks was higher in the high usage set, whereas others were higher in the low usage set (Sup. Fig. 3). Interestingly, H3K9me2 and H3K27me1, known to be associated with condensed chromatin and gene silencing,40,41 were more prevalent in the low usage set (Wilcoxon p-value < 2.4e-05). However, H3K27me2/3, which were previously found to be biased towards silent promoters were present at higher levels (Wilcoxon p-value < 1.9e–14) in high usage poly(A) sites.17 On the other hand, H3K4me2/3, which is associated with active chromatin40,42 were more prevalent in high usage poly(A) sites (Wilcoxon p-value < 0.004), whereas other modifications associated with active chromatin—H3K36me3, H3K79me1/2/3, were more prevalent in low usage poly(A) sites (Wilcoxon p-value < 0.0002).17
Next, we tested whether HM patterns can be used to computationally distinguish the high usage from the low usage poly(A) sites using the usage determined from EST/cDNAs and separately for the subset of sites that agreed with the usage based on CD4+ T cell RNA-seq data (Sup. Table 2 and 3). Again, we only discuss the results for the larger set based on EST/cDNAs below as the classification results for both were highly similar. We trained a SVM as above, using each individual modification, resulting in classification accuracy between 52%–66% (95% confidence interval) (Sup. Table 2). Upon combining the top five most discriminating HMs the classification accuracy was 63%–66.1% (95% confidence interval, Sensitivity = 82.9%–88.2%, Specificity = 46.1%–46.2%), and using the ten most discriminating marks resulted in improved classification accuracy to 69.7%–74% (95% confidence interval, Sensitivity = 79%–79.1%, Specificity = 61.1%–61.3%). We repeated the SVM classification using the 15 cis elements surrounding the human poly(A) sites that have been identified to enhance polyadenylation,8,39 resulting in classification accuracy of 76.5–76.6% (95% confidence interval, Sensitivity = 81.1%–81.2%, Specificity = 71.8%–71.9%). Thus, HM patterns can be used to discriminate high and low usage poly(A) sites with a reasonable accuracy and sensitivity, although with a lower specificity as compared to that obtained by classification using cis-elements. No improvement in performance was achieved by combining the cis-elements with the top ten most discriminating HMs (data not shown).
Discussion
A critical aspect of all cellular processes is an accurate and timely decoding of genomic information. One of the first and arguably the most regulated step in this decoding process is transcription, comprised of three separately regulated events—initiation, elongation and termination.43 The last of the three events—termination, involves cleavage and polyadenylation at the 3′ end of the mRNA. Recent genome-wide availability of accurately mapped 3′ end of transcripts has led to an increased appreciation of the role that alternative polyadenylation plays in generating the repertoire of gene isoforms and in differential spatio-temporal regulation of isoforms.5,19
The mechanism by which a poly(A) site is recognized by the cellular machinery for cleavage and polyadenylation is not entirely known.1 Similar to other crucial genomic signals such as transcription initiation sites and splice sites, the identifying features of poly(A) site can be grouped into three broad classes pertaining to sequence, structure and epigenomic marks.8,9,14,16,44 Several sequence elements enriched near poly(A) sites have been shown to facilitate polyadenylation by acting as binding sites for specific cleavage and polyadenylation enzymes.45 In addition, mRNA structure has been shown to be a critical determinant of polyadenylation in vitro.9,10 It has been suggested that structure plays a role in poly(A) site definition and selection through the exposure or occlusion of cis elements and by influencing the distance separating the poly(A) factor binding sites.11,12 Lastly, poly(A) site usage has been shown to correlate with nucleosome occupancy immediately downstream of the poly(A) site.14 Here we provide a possible mechanistic explanation of this observation by showing correlations between nucleosome occupancy with slowing down of PolII elongation perhaps due to PolII pausing and RNA structure. Nucleosomes present a barrier to PolII in vitro.46,47 Our results based on in vivo data suggest a similar effect. While the data show a strong correlation between PolII accumulation and stable RNA structure [both correlated to high nucleosome occupancy and poly(A) usage], the causality relationship between the two is less clear. While on one hand, PolII pausing which causes accumulation of PolII can affect mRNA structure, presumably by affecting transcriptional dynamics at poly(A) site,34 on the other hand, a more stable RNA structure and a compact chromatin downstream of poly(A) sites may jointly affect PolII pausing.29 Our conditional probability analysis favors the former causality link, that is, PolII pausing affects RNA structure. Even though RNA structure depends on the static RNA sequence, with the changes in the PolII elongation rate the formation of local RNA structure varies due the differences in the availability of the RNA sequence. Thus, taken together, our result suggests that high nucleosome occupancy downstream of poly(A) sites affects PolII pausing, which in turn affects the RNA structure stability making it more conducive to recognition of poly(A) sites for cleavage and polyadenylation.30,31 We emphasize that our results based on statistical analysis of the data does not constitute a proof of this causality and it is not inconsistent with potential causal role of RNA structure on PolII pausing.29
Chromatin structure as well as poly(A) site usage is often specific to tissue and developmental stage.5,48,49 Thus, as far as possible, we obtained our datasets for human CD4+ T cells. The nucleosome occupancy, histone modification, PolII occupancy and gene expression data were all measured in vivo in human CD4+ T cells. Although RNA-seq performed on nuclear extracts from CD4+ T cells can, in principle, be used to obtain cell type specific usage of poly(A) sites, the tools needed to accurately quantify poly(A) site usage from RNA-seq data are not sufficiently accurate. In contrast to exon skipping events, quantifying poly(A) site usage requires unambiguously assigning RNA-seq reads to each of the alternative poly(A) sites in a gene, which is non-trivial especially for the tandem alternative 3′ poly(A) sites. We have obtained an estimate of relative usage of the poly(A) sites from RNA-seq data in CD4+ T cells using the relative usage of downstream poly(A) site (RUD) score as described in reference 50. This measure relies on the mappable reads and thus assumes that the lack of reads downstream/upstream of a poly(A) site is due to the poly(A) site usage, which may not be true. Also, as mentioned previously, this method does not make an attempt to probabilistically assign reads to specific poly(A) sites. Therefore we also obtained the poly(A) site usage using EST/cDNA data, as done previously in reference 14. To be relevant for most cell types we focused only on the highly used (≥75%) and rarely used (≤25%) poly(A) sites. Furthermore, we used the poly(A) sites that were discovered to be high/low usage using both the datasets as our high confidence set to verify the findings.
Chromatin can affect both transcriptional and post-transcriptional event by different mechanisms. Nucleosome occupancy and epigenomic modifications can affect transcription initiation by mediating the interactions between transcription factors and the chromatin.51,52 Whereas, due to coupling of transcription and mRNA processing, chromatin structure is likely to affect mRNA processing via control of transcriptional dynamics.53,54 Spies et al. found that Human poly(A) sites were strongly depleted of nucleosomes, whereas exons contained distinct peaks in nucleosome occupancy as well as peaks of all methylated forms of histone except H3K9me3.14 They also found an inverse correlation between splice site strength and nucleosome occupancy and a positive correlation between poly(A) site usage and nucleosome occupancy and suggested an interdependence of splicing and polyadenylation mechanism on the chromatin structure, possibly mediated through the interactions between nucleosome-associated proteins with RNA splicing43 and cleavage/polyadenylation factors.2 Our findings are consistent with those of Spies et al. and suggest that nucleosome occupancy affects recognition of mRNA processing signals [splice sites or poly(A) sites] by affecting transcriptional dynamics. Thus, our results provide an additional causative link between the epigenome and a post-transcriptional event.
We found distinct histone modification patterns surrounding polyA sites which not only distinguish functional poly(A) sites from putative poly(A) sites but can also discriminate between frequently used poly(A) sites from rarely used poly(A) sites with an accuracy at least as good as that obtained by using known cis elements. There is some loss of accuracy when we used the HMs to classify poly(A) site usage as compared to using the cis elements. This happens due to the fact that the region around the poly(A) site used for discrimination is much larger in the case of HMs, since it was selected to encompass at least six surrounding nucleosomes, and thus overlaps in many cases between the high and low usage poly(A) sites. Whereas the cis elements come from the surrounding ±100 nt of the poly(A) sites.8 Thus, epigenomic features surrounding poly(A) sites appear to encode a substantial portion of the regulatory information. Our analysis revealed that some HMs-H2BK5me1, H3K4me1, H3K4me2, H3K4me3, H3K9me1, H3K27me1, H3K27me2, have a profile similar to the observed pattern of nucleosome depletion around the poly(A) sites.14 On the contrary, H3K9me3 and H3R2me2 were found to be enriched around the poly(A) site. The HM H3K36me3 gradually drops off after the poly(A) site, consistent with previous suggestion that H3K36me3 recognition by chromatin remodeling complexes might affect the rate of PolII elongation.55 The observed diversity in HM patterns, and their ability to distinguish functional poly(A) sites from putative sites and high usage sites from low usage sites, warrants further analysis into their functional role in polyadenylation.
Materials and Methods
Poly(A) site dataset.
The poly(A) sites as well as the locations of the PAS element were obtained from the PolyA_DB2 database19 which includes 54,686 human poly(A) sites. The genome coordinates for these sites were converted to the human genome assembly hg18 using the liftOver tool from UCSC Genome Browser (genome.ucsc.edu). Of the 54,686 poly(A) sites, 31,871 were within the 3′ UTR of the Refseq gene annotations (hg18) downloaded from the UCSC Genome Browser. The PolyA_DB2 also provides the number of polyadenylated EST/cDNA supporting each poly(A) site, which we used to compute the poly(A) site usage, defined as the fraction of times a poly(A) site is used for a given gene. High usage sites are those that are supported by ≥75% EST/cDNAs whereas low usage sites were those supported by ≤25% EST/cDNAs. Thus any gene that has a highly used poly(A) site is guaranteed to also have one or more low usage sites. Only considering the genes with multiple poly(A) sites, we obtained 5,139 high usage sites and 21,204 low usage sites.
Controlling for GC content to compare the RNA structure stability for high and low usage poly(A) site regions.
We obtained 5,139 high usage sites and 21,204 low usage sites as described previously. Based on the GC fraction in the region 250 bp upstream and 50 bp downstream of the poly(A) site, we divided each group into 5 bins of size 0.2 ranging from 0 to 1. For each bin with say, m high usage and n low usage poly(A) sites (m ≤ n), we randomly sampled m out of the n low usage sites. Thus overall, the two classes were matched for GC content.
Poly(A) site usage in human CD4+ T cell.
Mapped reads from RNA-seq performed on nuclear extracts from CD4+ T cells were obtained from NCBI's Gene Expression Omnibus, accession number GSM501716.23 For all the alternative poly(A) sites in PolyA_DB2,19 we calculated the relative usage of downstream poly(A) site (RUD) score, which is the ratio of the density of downstream reads and density of upstream reads, as described in reference 50. For the alternative poly(A) sites in the 3′ UTR, this score was calculated using the downstream region up to the downstream poly(A) site or 500 bp if there is no downstream poly(A) site, and upstream region up to the previous poly(A) site or up to the last exon start position if no upstream 3′ UTR poly(A) site exists. For the alternative poly(A) sites that do not fall within the 3′ UTR, 100 bp downstream and upstream regions were considered for the RUD score. As in reference 50, the reads mapping to ±10 nt region around the poly(A) sites were not used for RUD calculation because the cleavage sites are not mapped precisely. Poly(A) sites with RUD score ≥1 were considered to be low usage whereas those with RUD score in bottom 25 percentile were considered as high usage sites. Overall there were 5,532 high usage poly(A) sites and 11,767 low usage poly(A) sites.
Nucleosome occupancy, histone modification and PolII occupancy in human CD4+ T cell.
High-resolution genome-wide mapping for 37 HMs and histone variant H2A.Z and PolII in human CD4+ T cells were obtained from reference 17 and 18. The genome wide maps of nucleosome occupancy for resting human CD4+ T cells were obtained from reference 25. All genomic coordinates are from the human genome assembly hg18.
Gene expression in human CD4+ T cell.
The microarray gene expression dataset for resting CD4+ T cells was obtained from NCBI's Gene Expression Omnibus repository under accession number GSE10437.25 We performed gcRMA normalization56 on the raw cel data files using the software R (www.r-project.org/) and Bioconductor (www.bioconductor.org). Probes were mapped to all the Refseq Genes from the human genome assembly hg18. Multiple probes mapping to the same gene were summarized by mean expression.
Controlling GC content to examine the correlation between nucleosome occupancy and RNA structure.
Downstream nucleosome occupancy score was computed as the aggregate of the scores in the 600 bp region downstream of the poly(A) site using the nucelosome score profiles obtained from reference 25. These nucleosome score profiles were previously obtained by applying a sliding window of 10 bp across all chromosomes. All reads mapping to the sense strand 80 bp upstream and reads mapping to the antisense strand 80 bp downstream of the window contributed equally to the score of the window.25 The poly(A) sites were divided into two groups based on the median nucleosome occupancy score in the region 600 bp downstream: High nucleosome (score in top 50 percentile), Low nucleosome (score in bottom 50 percentile). Based on the GC content fraction in the region 250 bp upstream and 50 bp downstream of the poly(A) site, we divided each of the groups into bins of size 0.2 ranging from 0 to 1. For each bin with say, m high nucleosome occupancy and n low nucleosome occupancy sites (m ≤ n), we randomly sampled m out of n low nucleosome occupancy sites. Thus overall, the two classes were matched for GC content.
SVM classification based on epigenetic and sequence information.
SVM classification was carried out using the software LIBSVM (www.csie.ntu.edu.tw/∼cjlin/libsvm/), using the radial basis function kernel with parameters: C = 32 and gamma = 0.5, obtained using cross-validation. We also performed the classification using the linear kernel as well as polynomial kernel and found the classification accuracy to be the highest with the radial basis function kernel (data not shown) and hence the results discussed are those obtained using the radial basis function kernel.
To test the classification of the authentic poly(A) sites from poly(A)-like locations, we first identified all genomic locations (genic as well as intergenic) that contain the hexamer AATAAA (best representative PAS). To avoid selecting undetected poly(A) sites from within the gene in our negative set, we further required these decoy sites to lie >50kb away from the end of any gene. In all we obtained 87,679 such positions across the genome. These non-poly(A) positions were used as negative set. All the 31,871 3′ UTR poly(A) sites made up the positive set. For each position the HMs were mapped in the surrounding ±600 bp flanking region in 6 bins of 200 bp which corresponded to the six features used to train the SVM. For training set, we randomly selected 3,000 positions from the positive set and 3,000 positions from the negative set that differ not more that 10% from each other in the GC content of the 1,200 bp region used for mapping the histone modifications. The test set was similarly constructed using 5,000 positive and 5,000 negative positions.
Similarly, for the classification between high usage poly(A) sites and low usage poly(A) sites, the HM data was mapped in the ±600 bp of the poly(A) sites. The training set comprised of randomly selected 3,000 poly(A) sites of each type and the test set comprised of 4,000 randomly selected poly(A) sites of each type.
The classification features of the 5 (respectively 10) HMs that resulted in the best individual accuracies were combined together, resulting in 30 (respectively 60) features for each poly(A) site. While doing so we selected the top performing marks that were not highly correlated with one another in the region ±1,000 bp of the poly(A) sites in order to avoid saturation (average pair-wise correlation ≤0.6). SVM classification was then repeated on this combined dataset as above.
For classification using sequence-based information, we used the 15 cis elements surrounding the poly(A) sites (100 bp upstream and 100 bp downstream) suggested to enhance polyadenylation.8 Position-specific scoring matrices (PSSMs) for these elements were obtained8 and the scores were calculated as in reference 39. The resulting 15 scores were used to train a SVM. The same training and test sets constructed above were used.
The above procedures were each repeated on 100 different instances of training and test datasets created as described above in order to calculate the 95% classification accuracy range.
Figure 1.
mRNA structure stability surrounding poly(A) site correlates with the downstream nucleosome occupancy. The poly(A) sites were grouped according to the free energy (FE) of the thermodynamic ensemble for the 250 bp region surrounding the site and nucleosome occupancy was averaged within each FE bin of 1 kcal/mol. The figure shows the scatter plot of average nucleosome occupancy (600 bp downstream of poly(A) site) and FE. Note: Very few poly(A) sites fall within the bins corresponding to the extreme values of FE which is the reason for the variation in the pattern at both ends of the plot.
Acknowledgements
We thank Logan Everett and Russ Carstens for providing comments on the manuscript. We thank Larry Singh for help with obtaining the gene expression data. We thank the anonymous reviewers for providing constructive comments on the manuscript. This work was supported by the NIH grant R01GM085226.
Supplementary Material
References
- 1.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 2.Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wickens M, Anderson P, Jackson RJ. Life and death in the cytoplasm: messages from the 3′ end. Curr Opin Genet Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- 4.Early P, Rogers J, Davis M, Calame K, Bond M, Wall R, Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980;20:313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- 5.Zhang H, Lee JY, Tian B. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6:100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loke JC, Stahlberg EA, Strenski DG, Haas BJ, Wood PC, Li QQ. Compilation of mRNA polyadenylation signals in Arabidopsis revealed a new signal element and potential secondary structures. Plant Physiol. 2005;138:1457–1468. doi: 10.1104/pp.105.060541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu J, Lutz CS, Wilusz J, Tian B. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 2005;11:1485–1493. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graveley BR, Fleming ES, Gilmartin GM. RNA structure is a critical determinant of poly(A) site recognition by cleavage and polyadenylation specificity factor. Mol Cell Biol. 1996;16:4942–4951. doi: 10.1128/mcb.16.9.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das AT, Klaver B, Berkhout B. A hairpin structure in the R region of the human immunodeficiency virus type 1 RNA genome is instrumental in polyadenylation site selection. J Virol. 1999;73:81–91. doi: 10.1128/jvi.73.1.81-91.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klasens BI, Thiesen M, Virtanen A, Berkhout B. The ability of the HIV-1 AAUAAA signal to bind polyadenylation factors is controlled by local RNA structure. Nucleic Acids Res. 1999;27:446–454. doi: 10.1093/nar/27.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bar-Shira A, Panet A, Honigman A. An RNA secondary structure juxtaposes two remote genetic signals for human T-cell leukemia virus type I RNA 3′-end processing. J Virol. 1991;65:5165–5173. doi: 10.1128/jvi.65.10.5165-5173.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wood AJ, Schulz R, Woodfine K, Koltowska K, Beechey CV, Peters J, et al. Regulation of alternative polyadenylation by genomic imprinting. Genes Dev. 2008;22:1141–1146. doi: 10.1101/gad.473408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lian Z, Karpikov A, Lian J, Mahajan MC, Hartman S, Gerstein M, et al. A genomic analysis of RNA polymerase II modification and chromatin architecture related to 3′ end RNA polyadenylation. Genome Res. 2008;18:1224–1237. doi: 10.1101/gr.075804.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JY, Yeh I, Park JY, Tian B. PolyA_DB 2: mRNA polyadenylation sites in vertebrate genes. Nucleic Acids Res. 2007;35:165–168. doi: 10.1093/nar/gkl870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilmartin GM, Nevins JR. An ordered pathway of assembly of components required for polyadenylation site recognition and processing. Genes Dev. 1989;3:2180–2190. doi: 10.1101/gad.3.12b.2180. [DOI] [PubMed] [Google Scholar]
- 22.MacDonald CC, Wilusz J, Shenk T. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol Cell Biol. 1994;14:6647–6654. doi: 10.1128/mcb.14.10.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nunes NM, Li W, Tian B, Furger A. A functional human Poly(A) site requires only a potent DSE and an A-rich upstream sequence. EMBO. 29:1523–1536. doi: 10.1038/emboj.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tillo D, Hughes TR. G + C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics. 2009;10:442. doi: 10.1186/1471-2105-10-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plant KE, Dye MJ, Lafaille C, Proudfoot NJ. Strong polyadenylation and weak pausing combine to cause efficient termination of transcription in the human Ggamma-globin gene. Mol Cell Biol. 2005;25:3276–3285. doi: 10.1128/MCB.25.8.3276-3285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson ML, Bertolino S, Davis F. An RNA polymerase pause site is associated with the immunoglobulin mus poly(A) site. Mol Cell Biol. 2002;22:5606–5615. doi: 10.1128/MCB.22.15.5606-5615.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uptain SM, Kane CM, Chamberlin MJ. Basic mechanisms of transcript elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 30.Gromak N, West S, Proudfoot NJ. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol Cell Biol. 2006;26:3986–3996. doi: 10.1128/MCB.26.10.3986-3996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yonaha M, Proudfoot NJ. Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol Cell. 1999;3:593–600. doi: 10.1016/s1097-2765(00)80352-4. [DOI] [PubMed] [Google Scholar]
- 32.Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heilman-Miller SL, Woodson SA. Effect of transcription on folding of the Tetrahymena ribozyme. RNA. 2003;9:722–733. doi: 10.1261/rna.5200903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karlic R, Chung HR, Lasserre J, Vlahovicek K, Vingron M. Histone modification levels are predictive for gene expression. Proc Natl Acad Sci USA. 2010;107:2926–2931. doi: 10.1073/pnas.0909344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 37.Hon G, Wang W, Ren B. Discovery and annotation of functional chromatin signatures in the human genome. PLoS Comput Biol. 2009;5:1000566. doi: 10.1371/journal.pcbi.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng Y, Miura RM, Tian B. Prediction of mRNA polyadenylation sites by support vector machine. Bioinformatics. 2006;22:2320–2325. doi: 10.1093/bioinformatics/btl394. [DOI] [PubMed] [Google Scholar]
- 40.Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci USA. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 42.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 43.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 44.Chen F, MacDonald CC, Wilusz J. Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res. 1995;23:2614–2620. doi: 10.1093/nar/23.14.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Proudfoot N. Poly(A) signals. Cell. 1991;64:671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- 46.Izban MG, Luse DS. Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev. 1991;5:683–696. doi: 10.1101/gad.5.4.683. [DOI] [PubMed] [Google Scholar]
- 47.Bondarenko VA, Steele LM, Ujvari A, Gaykalova DA, Kulaeva OI, Polikanov YS, et al. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol Cell. 2006;24:469–479. doi: 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 48.Beaudoing E, Gautheret D. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Res. 2001;11:1520–1526. doi: 10.1101/gr.190501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nunes NM, Li W, Tian B, Furger A. A functional human Poly(A) site requires only a potent DSE and an A-rich upstream sequence. EMBO J. 2010;29:1523–1536. doi: 10.1038/emboj.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turner BM. Defining an epigenetic code. Nat Cell Biol. 2007;9:2–6. doi: 10.1038/ncb0107-2. [DOI] [PubMed] [Google Scholar]
- 52.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 53.Morel JB, Mourrain P, Beclin C, Vaucheret H. DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr Biol. 2000;10:1591–1594. doi: 10.1016/s0960-9822(00)00862-9. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Griffin K, Mondal N, Parvin JD. Phosphorylation of histone H2A inhibits transcription on chromatin templates. J Biol Chem. 2004;279:21866–21872. doi: 10.1074/jbc.M400099200. [DOI] [PubMed] [Google Scholar]
- 55.Sims RJ, 3rd, Reinberg D. Processing the H3K36me3 signature. Nat Genet. 2009;41:270. doi: 10.1038/ng0309-270. [DOI] [PubMed] [Google Scholar]
- 56.Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A Model-Based Background Adjustment for Oligonucleotide Expression Arrays. Journal of the American Statistical Association. 2004;99:909–917. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.