Abstract
The DNA replicative gene, thymidylate synthase (TYMS), is inhibited upon treatment with the anticancer drug 5-fluorouracil (5-FU). TYMS has 28-bp tandem repeat sequences or VNTR (variable numbers of tandem repeats) in the 5′-untranslated region (5′-UTR). The number of these repeats is variable in any given population, but the most prevalent are double (2R) and triple (3R) repeat sequences. A single G/C nucleotide polymorphism in the triple repeat sequence gives rise to a 3Rc or a 3Rg triple repeat structure. A widely cited literature used plasmid constructs of the 5′-UTR and proposed that genotyping the TYMS UTRs would predict the efficiency of Tyms protein translation, justifying altered therapeutic dosage of 5-FU. Prior studies had unusual features in experimental design, such as using the firefly Kozak sequence in place of the native human TYMS Kozak sequence to determine the ribosomal translational efficiency of TYMS mRNA. Our results using transient transfection, antibiotic-selected pools of transfected cells, and stably transfected clones, while using plasmids having native human Kozak sequence, refute the earlier results.
Key words: thymidylate synthase, 5-fluorouracil, VNTR, single nucleotide polymorphism (SNP), Kozak sequence
Introduction
5-fluorouracil (5-FU), a classical thymidylate synthase (Tyms) inhibitor developed more than five decades ago,1 is widely used2 against malignancies of the gastrointestinal tract, breast, head, neck and other cancers.3–5 5-FU has been extensively studied and accounts for 48,000 publications in the PubMed. It causes cell death by incorporating the fluorinated nucleotides into DNA and RNA, by covalent binding of its metabolites with Tyms protein, and perhaps by inhibiting the DNA replication enzyme primase.6–8
The accumulated literature on the Tyms-5-FU interaction, 25 years old now, contains analyses of both transcriptional9–12 and post-transcriptional (translational) activities9,13–16 associated with the TYMS VNTR (variable number of tandem repeats). The earliest report by Kaneda et al.13 summarizes a prior observation that expression of TYMS gene during cell cycle in normal human fibroblasts is mainly controlled by post-transcriptional events.17 Over time, the possible relationship between TYMS 5′-untranslated region (5′-UTR) repeats and Tyms protein expression gained more impact and citation in the field. Horie et al. in 1995 (319 citations) concluded that expression efficiency could be associated with the TYMS VNTR and suggested either a transcriptional or post-transcriptional mechanism. This observation was followed by three highly cited reports in literature: two of which supported a strong (3- to 4-fold) translational influence for the TYMS VNTR [Kawakami et al. 2001 (164 citations); Kawakami and Watanabe, 2003 (160 citations)], while one supported a transcriptional influence [Mandola et al. 2003 (219 citations)]. These mechanistic studies implicating the VNTR as an important determinant of Tyms protein levels in tumors and normal tissues led to several clinical studies of the concept that genotyping the 5′-UTR of the TYMS gene in blood could be used as a predictor of 5-FU-related toxicity,18–21 although blood DNA can differ significantly from the genotype obtained from aneuploid tumor DNA.22
Although not the subject of current study, the clinical reports produced varying odds ratios and were largely inconsistent. A recent Blue Cross Blue Shield (BCBS) opinion (BCBS report, Volume 24, No. 13, August, 2010) refuted these toxicity studies and concluded that the determination of the VNTR alone could not reliably predict toxicity from 5-FU. Therefore, several questions remain regarding the mechanistic links between Tyms expression and 5-FU treatment. Publications on a given side of this topic are often refuted by others.22 Other variables are reported to also affect 5-FU responses.7,23–29 In addition, it was reported that the colorectal cancer patients with microsatellite instability (MSI) tend to not respond to 5-FU-based therapy.30 Thus the compelling nature of the clinical results, based on a translational activity of the TYMS VNTR affecting Tyms protein expression, urged that continued mechanistic explorations be pursued in vitro. The “in vitro” subset of publications that received maximal clinical attention had individually supported TYMS tandem repeats as having a mechanistic role in determining Tyms protein expression levels, but to us remained unconvincing. Although also outside the scope of current work, reports concerning transcriptional effects produced conflicting results,31 causing one recent author to discuss potential transcriptional regulators “despite these discrepancies”.12
Many mechanistic reports had drawbacks in their experimental design. Most used the firefly luciferase Kozak (which is a “strong” Kozak), instead of a human TYMS Kozak (which is a “weak” Kozak) to model TYMS translation. The efficiency of protein translation depends in large part upon the efficiency of initiating translation. Use of the firefly gene's Kozak instead of the human TYMS gene's Kozak sequence might invalidate such studies, as one presumes that the human 5′-UTR of TYMS gene might interact functionally with the Kozak so as to regulate translation initiation. Other difficulties included assaying the function of human sequences in mouse cells in the original paper by Kaneda et al. 1987.13 Some reports measured RNA levels so as to normalize the reporter activity (i.e., to assess translation efficiency), but contaminating plasmid DNA used in transient transfections can invalidate some of these methods. Complete statistical techniques and replication of experiments were not reported in all the papers. One report in reference 16, demonstrated sensitivity of several established cancer cell lines having a 2R or a 3R genotype to 5-FU and to FUdR (5-fluoro-2′-deoxyuridine), but did not distinguish between the 3Rc and the 3Rg genotypes. Arguably, the proposal to use TYMS genotyping with measurements of Tyms protein levels to predict 5-FU dosage and patient response to 5-FU therapy did not have a strong ‘in vitro’ experimental basis.
TYMS expression is known to be variable in primary tumors and considerably difficult to estimate accurately as the tumor tissue can be heavily contaminated with non-neoplastic stromal cells. In addition, the tumor cells might experience allelic deletion,32–34 chromosomal copy number variation,35 or gene amplification as a mechanism of 5-FU resistance.36 Tyms protein expression depends rigorously upon the phase of the cell division cycle.17 None of these parameters are expected to remain constant across all regions of an aneuploid and potentially metastatic malignancy, but would be rather predictable in normal tissues, where the side effects of chemotherapy manifest.
We re-examined the subject, using the native human TYMS Kozak consensus sequence in luciferase reporter constructs in place of the previously employed Kozak sequences to determine whether translation efficiency was robustly dependent on variation in the TYMS 5′-UTR. In order to address the problem of high variation in transient transfection results, we generated stably transfected, pooled, and individual antibiotic-selected cell lines. We measured RNA levels of the reporter gene to obtain normalized reporter activity values. Experimental replicates and standard statistical techniques were employed to provide a perspective on discrepancies with prior literature.
Results
Failure to confirm a direct association between TYMS repeats and Tyms protein expression: Using firefly Kozak in transient transfection studies.
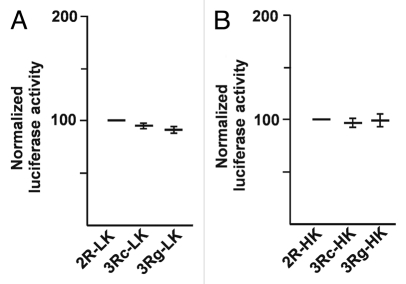
We re-visited the inferred effect of TYMS 5′-UTR tandem repeats or VNTR on TYMS expression, using luciferase reporter constructs having the firefly Kozak sequence, with attention to different sources of statistical variation. Plasmids 2R-LK, 3Rc-LK and 3Rg-LK (Fig. 1) containing the firefly Kozak sequence were used in transient transfection assays in seven independent experiments. We tested seven parallel plasmid DNA preparations and repeated the transient transfection assay in DLD-1 cells, with each particular DNA preparation at least twice, to determine the mean Renilla-normalized luciferase activity of each plasmid preparation (Fig. 2A). We found that the reporter activity was indistinguishable in the 2R-LK, 3Rc-LK and 3Rg-LK firefly Kozak constructs, in contrast to the previously published results. We confirmed our observation in two additional cell lines, namely, RKO and HCT116 (data not shown).
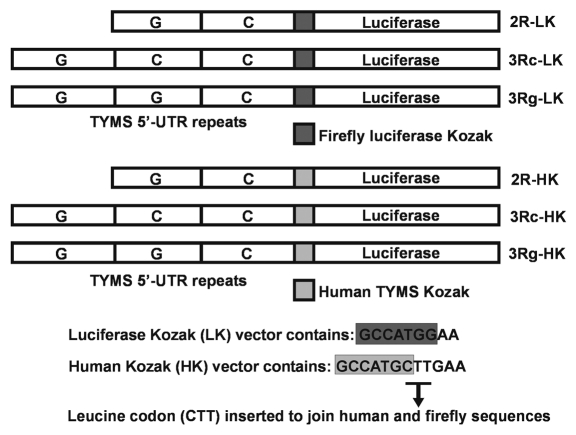
Figure 1.
Structure of TYMS 5′-UTR tandem repeats-luciferase reporter constructs having Kozak sequences from firefly luciferase (LK) and human TYMS (HK) are shown. Leucine (CTT) was inserted by site mutagenesis between methionine (ATG) and glutamine (GAA) to generate constructs with native TYMS Kozak sequence GCCATGC in place of the firefly Kozak GCCATGG.
Figure 2.
(A) Transient transfection results using reporter constructs having the firefly luciferase Kozak (LK). Reporter activities from seven plasmid DNA preparations were used in transient transfection assays, each constituting two replicates of 2R-LK, 3Rc-LK and 3Rg-LK. In each experiment, the 2R-LK activity was arbitrarily defined as “100”. No meaningful difference was found between reporter activities using the three VNTR structures. The luciferase reporter values were normalized by the renilla readings of the same well. The normalized luciferase value for each plasmid construct is presented as a mean of the seven experiments ± standard error (bars). (B) Transient transfection results using reporter constructs having the human Kozak (HK). Reporter activities from seven plasmid preparations were used in transient transfection assays, each constituting two replicates of 2R-HK, 3Rc-HK and 3Rg-HK. In each experiment, the 2R-HK activity was arbitrarily defined as 100. No meaningful difference was found between the reporter activities using the three VNTR structures. The normalized luciferase value for each plasmid construct is presented as a mean of the seven experiments ± standard error (bars).
Failure to confirm a direct association between TYMS tandem repeats and Tyms protein expression: Using the human TYMS Kozak in transient transfection assays.
We examined the effect of TYMS 5′-UTR tandem repeats on TYMS translation efficiency using reporter constructs having the native human TYMS Kozak sequence in transient transfection assays using seven independent experiments with independent plasmid preparations. The Renilla-normalized reporter activity from plasmids 2R-HK, 3Rc-HK and 3Rg-HK (Fig. 1), having the native human TYMS Kozak sequence, were indistinguishable from each other (Fig. 2B). Using the human Kozak constructs, we found similar results in RKO and HCT116 (data not shown).
We also tried to normalize the reporter values using measurements of luciferase RNA, but owing to the RNA following transient transfection being heavily contaminated with plasmid DNA, this proved challenging. This plasmid DNA could not be removed completely after prolonged treatment with DNaseI or with other common techniques. This residual plasmid DNA interfered with the RNA quantitative assays. In contrast, the genomic DNA of the non-transfected parental DLD-1 cells (experimental control) could be productively reduced in RNA samples by treatment with DNaseI for 30 min at 37°C (data not shown).
Absence of a direct relationship between TYMS tandem repeats and Tyms protein expression: Using geneticin-selected cultures after transfection.
Because the reporter values from transient transfections could not be properly normalized to levels of plasmid transcribed RNA, we introduced a temporal delay before RNA isolation. We established geneticin-selected pooled cultures by co-transfecting each of the plasmids 2R-LK, 3Rc-LK, 3Rg-LK, 2R-HK, 3Rc-HK and 3Rg-HK, having either the firefly luciferase or human TYMS Kozak sequences with pcDNA3.1, encoding the selectable marker geneticin. The stably integrated plasmid DNA was readily eliminated using DNaseI after RNA isolation. The pooled resistant cells expressing luciferase both from the firefly Kozak and TYMS Kozak were used for luciferase assays. Cells were taken for luciferase assay and RNA isolation separately, followed by quantitative real-time PCR (qPCR) quantification of luciferase RNA levels. The luciferase activity was divided by the luciferase RNA quantity from each cell line to obtain the final normalized results (Fig. 3). We determined the luciferase and RNA values for each of the six stable transfectants on three different days so as to obtain the three data points shown in Figure 3. The literature had indicated the 3Rg variant would have 3- to 4-fold more expression.15 Among each trio of constructs in stable transfected pools of cells having either firefly Kozak or the human TYMS Kozak sequence, no systematic difference of the expected direction were seen. As compared with the 3R clones, we instead observed a higher luciferase activity for the 2C-LK and the 2C-HK clones (containing the luciferase and human Kozaks, respectively).
Figure 3.
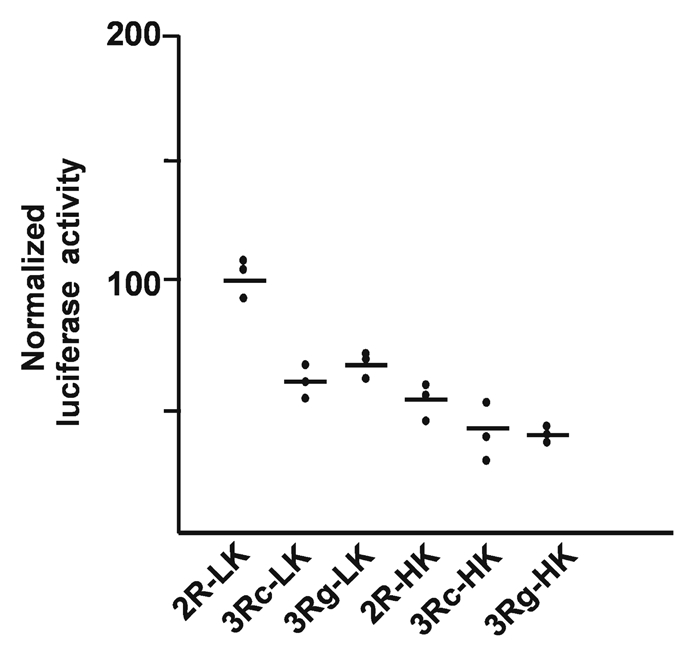
Normalized expression of luciferase reporter values in stably transfected, selected pools of cells having firefly Kozak and human TYMS Kozak constructs. For each construct, one transfection was performed to generate a stable clone. Each of the six clones was evaluated in three independent experiments, plotted as three dots, and the mean for each. The normalized luciferase activity was derived from the luciferase enzymatic activity divided by the luciferase RNA values measured in the corresponding cell population by qPCR. The expected systematic difference in the desired direction was not found between the different stable lines having different VNTR structures.
Absence of a direct relationship between TYMS tandem repeats and Tyms protein expression: Using stably transfected clones.
The pools of selected cells could have represented a heterogenous population of several geneticin-resistant clones, potentially masking an underlying relationship. We thereby generated individual selected clones that would permit normalization to integrated plasmid-derived mRNA levels in order to examine more closely the long-standing claims9,14–16 regarding the TYMS 5′-UTR polymorphism and translational efficiency, but using an improved experimental control.
We established stably transfected cell lines. Luciferase results were examined from two clones for each specific construct, except that only a single clone was obtained for one of the constructs. Five independent measurements were done for each construct (generally three with one clone and two with the other). Our experimental variation among “replicate” clones far exceeded the systematic variation attributed to the TYMS polymorphisms from earlier publications. We observed that for the same construct, most of the experimental variance was contributed by clonal variation among the cell lines isolated (data not shown).
Discussion
Prior studies had not directly challenged the published experimental evidence that the TYMS VNTR strongly affected translation efficiency. We argue here that the experimental basis of this proposal (from the “in vitro” literature) lacked sufficient rigor. The Kozak sequence in a gene is known to dictate the translational efficiency of any given site of translational initiation37 but previous investigators have largely used firefly Kozak instead of human TYMS Kozak to infer the efficiency of TYMS translation. Also, the influence of unintended experimental variation and uncontrolled variables were not adequately explored in the mechanistic papers. Our results from the present study refute the prior experimental results concerning the functional effects of the 5′-UTR polymorphism on reporter gene translation. We designed our experiments with a number of controls, performed several replicates of an experiment, included numerous preparations of DNA samples, examined parallel clones, and presented the variation among results. We used stably transfected cells to normalize the reporter values to a quantification of luciferase mRNA, permitting examination of the specific proposal that repeats in 5′-UTR govern translation efficiency of each mRNA molecule. Our results, however, failed to confirm the reported relations in which 3R or 3Rg TYMS tandem repeats strongly augment reporter protein expression in transient and stable transfection reporter assays using constitutive promoters. Based on our previous work in reference 35, and work from others,36 we should emphasize that genotyping of constitutional DNA would not reliably reveal the tumor genotypes, as the tumor cells might experience copy number changes due to LOH or gene amplification (a mechanism of 5-FU resistance). Determining the TYMS copy number in gastrointestinal cancers may help in 5-FU-based therapy, but this may prove difficult in a metastatic disease.
One can re-examine the prior “in vitro” literature with a brief but critical eye. The original paper by Kaneda et al.13 identified a triple, tandemly repeated, sequence in the 5′-UTR of the human TYMS mRNA. They did not, however, identify the 2R and 3R alleles and therefore did not compare in their assays the natural forms of then-undiscovered polymorphic alleles. The authors appropriately used the native human TYMS Kozak consensus sequence in the constructs, but measured the Tyms expression for their plasmids in mouse cells rather than in human cells. Mice do not have these repeats.
Eight years later, Horie et al.9 identified the presence of the double and triple repeat polymorphisms and a G/C single nucleotide polymorphism (SNP) in the triple repeat structure. They used deletions to mimic 2R and 3R variants in transient transfection assays, finding the double repeat construct to have reduced translation of a chloramphenicol acetyl transferase (CAT) coding sequence. Most of the deletion constructs lacked the native human TYMS Kozak; instead, the CAT gene Kozak sequence was used. The CAT activity values were not normalized to trans-fection-related variation.
Kawakami et al.14 concluded that the TYMS mRNA with the three-repeat sequence had 3.5-fold greater translational efficiency than with the two-repeat sequence when tested using transiently transfected luciferase plasmids. Although the luciferase reporter results were normalized using an RNase protection assay to control for variation in plasmid transcription and four replicate experiments were reported, no controls were performed to rule out a high assay background as would be expected from the large excess of biologically inactive plasmid DNA.38
Kawakami and Watanabe15 examined further a G/C polymorphism (a SNP) in the triple repeat sequence of TYMS giving rise to either a 3Rc or a 3Rg triple repeat. The 3Rg construct was concluded to have 3–4 times higher translational efficiency than the 2R and 3Rc constructs, again after normalizing for RNA quantity and using a firefly Kozak sequence. Although they reported four replicate experiments for each plasmid, the error bars were not defined, and replicate plasmid preparations were not reported.
Yawata et al.16 reported, using transient transfection, that the 3Rg repeat construct had a 2-fold increased luciferase activity as compared with the 2R allele. Their use of a strong constitutive promoter precluded the study of transcriptional enhancers for the native TYMS promoter. The reporter values were normalized using a co-transfected Renilla luciferase construct. The definition of the error bars was unstated. p values were provided, but the numbers of replicates of experiments or replicate plasmid preparations were not reported.
In addition to the TYMS VNTR, some rare polymorphisms have been identified in the TYMS 5′-UTR.11,12,15 The twelfth nucleotide in each 28-bp VNTR is a G or a C such that the wild type sequence is 2RGC (2R, in this study) for the two-repeat VNTR, and 3RGGC (3Rg, in this study), for the three-repeat VNTR. We have identified the 3RGCC polymorphism in several individuals that we genotyped (3Rc, in this study), but failed to identify 2RCC, 2RGG and 3RCCC polymorphisms (data not shown). These polymorphisms do not show any significant difference in luciferase activity as demonstrated by deBock et al. 2010,12 and are not suspected to be clinically important.
A broader perspective also remains essential. Showalter et al.22 analyzed prior studies related to Tyms expression levels and response to 5-FU chemotherapy. Little difference was found among response rates between tumors having low- and high-Tyms expression upon analyzing the grouped data. Although admittedly some articles had used greater sophistication in Tyms protein determinations, the authors expressed doubt that a compelling clinical utility had been found in these reports. Several other variables may affect Tyms expression and 5-FU therapy besides the proposed TYMS genetic polymorphisms. Patients with deficiency in dihydropyrimidine dehydrogenase, the rate-limiting enzyme of pyrimidine catabolism in the 5-FU metabolic pathway to the inactive 5-fluoro-5,6-dihydrouracil, suffer from severe toxicity in response to 5-FU treatment.23 Another candidate gene for explaining variable 5-FU toxicity is methylenetetrahydrofolate reductase, which forms the reduced folate cofactor essential for inhibiting Tyms.21 Orotate phosphoribosyl-transferase (OPRT), the enzyme necessary for stabilization and formation of the ternary complex39 following 5-FU treatment, is also a potential predictor of 5-FU effects.27 The G213A polymorphism in OPRT is reported to be associated with grade 3–4 toxicity in response to 5-FU therapy.20 In addition to Tyms levels, a combination of enzymes in the 5-FU metabolic pathway could affect 5-FU response.
Based on a conflicting clinical literature, conflicting studies of the TYMS native promoter, and our reexamination of the evidence in favor of translational regulation by varying the TYMS 5′-UTR's VNTR, we see neither an empiric nor mechanistic basis for the genotyping of germline patterns of TYMS repeats in patients as a guide to 5-FU-therapy.
Materials and Methods
Cell and culture conditions.
DLD-1 cells previously used to study the TYMS 5′-UTR repeats16 were obtained from the American Type Culture Collection and subsequently cultured in RPMI medium supplemented with 10% FBS and 1% penicillin/streptomycin in the presence of 5% CO2 at 37°C.
Plasmid construction.
The TYMS 5′-UTR sequence was obtained from cells having different TYMS genotypes by PCR using the forward primer TS 33, modified to contain a HindIII site, and reverse primer TS 28, containing an NcoI site.15 The amplified sequence was digested with HindIII and NcoI and inserted into the pGL3-Control vector (Promega) having been cut with the same pair of enzymes, to create three constructs having 2R, 3Rc and 3Rg TYMS 5′-UTR repeats upstream of the firefly luciferase reporter gene, for use in transient assays. These plasmids having the TYMS 5′-UTR repeats and firefly luciferase Kozak sequence were termed 2R-LK, 3Rc-LK and 3Rg-LK (Fig. 1) and mimicked published plasmids.9,14–16
The firefly luciferase Kozak consensus sequence of GCC ATG G was thereafter replaced by site mutagenesis (Quikchange, Stratagene) by inserting an amino acid leucine (CTT) between the luciferase start codon ATG and second codon GAA, giving rise to the native human TYMS Kozak (a non-consensus or weak Kozak) sequence of GCC ATG C (Fig. 1). We introduced the new sequence in all the three constructs to yield human Kozakluciferase reporter constructs having 2R, 3Rc and 3Rg repeats from TYMS 5′-UTR region. These constructs were termed 2R-HK, 3Rc-HK and 3Rg-HK (Fig. 1). The above plasmids were confirmed by full plasmid DNA sequencing.
Transient transfection.
Transient transfections were done using Lipofectamine (Invitrogen). Cells (2.5 to 4 × 105) were plated in six-well plates 24 h before transfection. The next day, cells were transfected using appropriate quantities (detailed below) of plasmid constructs and incubated for 5 h before the transfection medium was exchanged.
Generation of stably transfected antibiotic-selected pools and individual clones.
The pooled population was generated by co-transfecting the luciferase reporter constructs and pcDNA3.1 (Invitrogen) vector at a ratio of 6:1 in DLD-1 cells, followed by selection for a week in geneticin (200 mg/ml). Separately, individual stable clones were obtained by limiting dilution of transfected cells; serum-free transfection medium was changed 6 h following transfection. Cells were allowed to grow for 18 h in complete medium lacking geneticin. Cells were thereafter distributed at serial dilutions to 96-well plates containing complete medium plus geneticin (200 mg/ml), and grown for two weeks. The 96-well plates were then screened by PCR using luciferase-based primers. This PCR did not distinguish between stable transfectants and non-integrated plasmids. We identified stable integration by expanding the positive clones and demonstrating easily detectable luciferase reporter activity in them. Clones with varying levels of active luciferase (un-normalized) were obtained.
Luciferase assay.
The luciferase assay was done 24 h and 48 h after co-transfecting cells with 0.2 µg of each VNTR plasmid and 20 ng of pRL-SV40 renilla vector. Results of 24 and 48 h were similar, and 24 h was used subsequently for all reported experiments. Cells were washed twice with PBS and lysed by passive lysis buffer (300 µl/well). Each sample was measured in duplicate. Twenty µl of cell lysate was mixed with 100 µl of luciferase reagent II (LARII). 100 µl of Stop and Glo reagent was used after recording the luciferase luminescent signals in order to record the renilla luminescence readings in a PerkinElmer Microbeta Trilux plate reader. The luciferase reporter values were normalized by the renilla readings of the same well. Normalized luciferase activity was measured from duplicate wells for each plasmid.
Quantitative real-time PCR (qPCR) assay for analysis of firefly luciferase RNA levels.
RNA was extracted from stably transfected pooled cultures or individual clones using the RNeasy Mini Kit (Qiagen). RNA (1 µg) was treated with DNaseI (Invitrogen) and converted to cDNA for real-time PCR using SuperScript III Reverse Transcriptase (Invitrogen). qPCR was done on the iCycler (Bio-Rad) with luciferase and actin-specific primer sets (available upon request). Standard curves, melting curves and PCR efficiencies were determined for each primer set. At least three experiments were done for each data point. The RNA quantity was used to normalize the luciferase expression values. Despite considerable effort, we could not eliminate signals from plasmid DNA when assaying transiently transfected cells, and we found no verified method in the technical literature to do so.
Statistical representation.
Luciferase values were expressed as normalized means and standard error for transient transfection experiments using seven plasmids each, and as means with individual data plotted for pooled stable transfections, having three readings each.
Acknowledgments
This work has been supported by NIH grant CA62924 and the Everett and Marjorie Kovler Professorship in Pancreas Cancer Research.
Abbreviations
- TYMS
thymidylate synthase
- 5-FU
5-fluorouracil
- VNTR
variable number of tandem repeats
- UTR
untranslated region
- mRNA
messenger ribonucleic acid
- DNA
deoxyribonucleic acid
- BCBS
Blue Cross Blue Shield
- MSI
microsatellite instability
- FUdR
5-fluoro-2′-deoxyuridine
- SNP
single nucleotide polymorphism
- CAT
chloramphenicol acetyl transferase
- qPCR
quantitative polymerase chain reaction
- LOH
loss of heterozygosity
- OPRT
orotate phosphoribosyl-transferase
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Heidelberger C, Chaudhuri NK, Danneberg P, Mooren D, Griesbach L, Duschinsky R, et al. Fluorinated pyrimidines, a new class of tumour-inhibitory compounds. Nature. 1957;179:663–666. doi: 10.1038/179663a0. [DOI] [PubMed] [Google Scholar]
- 2.Lokich J. Infusional 5-FU: historical evolution, rationale and clinical experience. [Williston Park] Oncology. 1998;12:19–22. [PubMed] [Google Scholar]
- 3.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 4.Rustum YM, Harstrick A, Cao S, Vanhoefer U, Yin MB, Wilke H, et al. Thymidylate synthase inhibitors in cancer therapy: direct and indirect inhibitors. J Clin Oncol. 1997;15:389–400. doi: 10.1200/JCO.1997.15.1.389. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham D, Allum WH, Stenning SP, Thompson JN, Van de Velde CJ, Nicolson M, et al. Perioperative chemotherapy versus surgery alone for resectable gastroesophageal cancer. N Engl J Med. 2006;355:11–20. doi: 10.1056/NEJMoa055531. [DOI] [PubMed] [Google Scholar]
- 6.Noordhuis P, Holwerda U, Van der Wilt CL, Van Groeningen CJ, Smid K, Meijer S, et al. 5-Fluorouracil incorporation into RNA and DNA in relation to thymidylate synthase inhibition of human colorectal cancers. Ann Oncol. 2004;15:1025–1032. doi: 10.1093/annonc/mdh264. [DOI] [PubMed] [Google Scholar]
- 7.Brody JR, Hucl T, Costantino CL, Eshleman JR, Gallmeier E, Zhu H, et al. Limits to thymidylate synthase and Tp53 genes as predictive determinants for fluoropyrimidine sensitivity and further evidence for RNA-based toxicity as a major influence. Cancer Res. 2009;69:984–991. doi: 10.1158/0008-5472.CAN-08-3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parker WB, Cheng YC. Metabolism and mechanism of action of 5-fluorouracil. Pharmacol Ther. 1990;48:381–395. doi: 10.1016/01637258(90)90056-8. [DOI] [PubMed] [Google Scholar]
- 9.Horie N, Aiba H, Oguro K, Hojo H, Takeishi K. Functional analysis and DNA polymorphism of the tandemly repeated sequences in the 5′-terminal regulatory region of the human gene for thymidylate synthase. Cell Struct Funct. 1995;20:191–197. doi: 10.1247/csf.20.191. [DOI] [PubMed] [Google Scholar]
- 10.Luo HR, Lu XM, Yao YG, Horie N, Takeishi K, Jorde LB, et al. Length polymorphism of thymidylate synthase regulatory region in Chinese populations and evolution of the novel alleles. Biochem Genet. 2002;40:41–51. doi: 10.1023/A:1014589105977. [DOI] [PubMed] [Google Scholar]
- 11.Mandola MV, Stoehlmacher J, Muller-Weeks S, Cesarone G, Yu MC, Lenz HJ, et al. A novel single nucleotide polymorphism within the 5′ tandem repeat polymorphism of the thymidylate synthase gene abolishes USF-1 binding and alters transcriptional activity. Cancer Res. 2003;63:2898–2904. [PubMed] [Google Scholar]
- 12.de Bock CE, Garg MB, Scott N, Sakoff JA, Scorgie FE, Ackland SP, et al. Association of thymidylate synthase enhancer region polymorphisms with thymidylate synthase activity in vivo. Pharmacogenomics J. doi: 10.1038/tpj.2010.43. [DOI] [PubMed] [Google Scholar]
- 13.Kaneda S, Takeishi K, Ayusawa D, Shimizu K, Seno T, Altman S. Role in translation of a triple tandemly repeated sequence in the 5′-untranslated region of human thymidylate synthase mRNA. Nucleic Acids Res. 1987;15:1259–1270. doi: 10.1093/nar/15.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawakami K, Salonga D, Park JM, Danenberg KD, Uetake H, Brabender J, et al. Different lengths of a polymorphic repeat sequence in the thymidylate synthase gene affect translational efficiency but not its gene expression. Clin Cancer Res. 2001;7:4096–4101. [PubMed] [Google Scholar]
- 15.Kawakami K, Watanabe G. Identification and functional analysis of single nucleotide polymorphism in the tandem repeat sequence of thymidylate synthase gene. Cancer Res. 2003;63:6004–6007. [PubMed] [Google Scholar]
- 16.Yawata A, Kim SR, Miyajima A, Kubo T, Ishida S, Saito Y, et al. Polymorphic tandem repeat sequences of the thymidylate synthase gene correlates with cellular-based sensitivity to fluoropyrimidine antitumor agents. Cancer Chemother Pharmacol. 2005;56:465–472. doi: 10.1007/s00280-005-1018-z. [DOI] [PubMed] [Google Scholar]
- 17.Ayusawa D, Shimizu K, Koyama H, Kaneda S, Takeishi K, Seno T. Cell cycle-directed regulation of thymidylate synthase messenger RNA in human diploid fibroblasts stimulated to proliferate. J Mol Biol. 1986;190:559–567. doi: 10.1016/0022-2836(86)90241-X. [DOI] [PubMed] [Google Scholar]
- 18.Pullarkat ST, Stoehlmacher J, Ghaderi V, Xiong YP, Ingles SA, Sherrod A, et al. Thymidylate synthase gene polymorphism determines response and toxicity of 5-FU chemotherapy. Pharmacogenomics J. 2001;1:65–70. doi: 10.1038/sj.tpj.6500012. [DOI] [PubMed] [Google Scholar]
- 19.Lecomte T, Ferraz JM, Zinzindohoue F, Loriot MA, Tregouet DA, Landi B, et al. Thymidylate synthase gene polymorphism predicts toxicity in colorectal cancer patients receiving 5-fluorouracil-based chemotherapy. Clin Cancer Res. 2004;10:5880–5888. doi: 10.1158/1078-0432.CCR-04-0169. [DOI] [PubMed] [Google Scholar]
- 20.Ichikawa W, Takahashi T, Suto K, Sasaki Y, Hirayama R. Orotate phosphoribosyltransferase gene polymorphism predicts toxicity in patients treated with bolus 5-fluorouracil regimen. Clin Cancer Res. 2006;12:3928–3934. doi: 10.1158/10780432.CCR-05-2665. [DOI] [PubMed] [Google Scholar]
- 21.Schwab M, Zanger UM, Marx C, Schaeffeler E, Klein K, Dippon J, et al. Role of genetic and nongenetic factors for fluorouracil treatment-related severe toxicity: a prospective clinical trial by the German 5-FU Toxicity Study Group. J Clin Oncol. 2008;26:2131–2138. doi: 10.1200/JCO.2006.10.4182. [DOI] [PubMed] [Google Scholar]
- 22.Showalter SL, Showalter TN, Witkiewicz A, Havens R, Kennedy EP, Hucl T, et al. Evaluating the drug-target relationship between thymidylate synthase expression and tumor response to 5-fluorouracil. Is it time to move forward? Cancer Biol Ther. 2008;7:986–994. doi: 10.4161/cbt.7.7.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyss AP, Lilenbaum RC, Harris BE, Diasio RB. Severe 5-fluorouracil toxicity in a patient with decreased dihydropyrimidine dehydrogenase activity. Cancer Invest. 1993;11:239–240. doi: 10.3109/07357909309024846. [DOI] [PubMed] [Google Scholar]
- 24.Metzger R, Danenberg K, Leichman CG, Salonga D, Schwartz EL, Wadler S, et al. High basal level gene expression of thymidine phosphorylase (platelet-derived endothelial cell growth factor) in colorectal tumors is associated with nonresponse to 5-fluorouracil. Clin Cancer Res. 1998;4:2371–2376. [PubMed] [Google Scholar]
- 25.Grem JL. 5-Fluorouracil: forty-plus and still ticking. A review of its preclinical and clinical development. Invest New Drugs. 2000;18:299–313. doi: 10.1023/A:1006416410198. [DOI] [PubMed] [Google Scholar]
- 26.Salonga D, Danenberg KD, Johnson M, Metzger R, Groshen S, Tsao-Wei DD, et al. Colorectal tumors responding to 5-fluorouracil have low gene expression levels of dihydropyrimidine dehydrogenase, thymidylate synthase and thymidine phosphorylase. Clin Cancer Res. 2000;6:1322–1327. [PubMed] [Google Scholar]
- 27.Fujii R, Seshimo A, Kameoka S. Relationships between the expression of thymidylate synthase, dihydropyrimidine dehydrogenase and orotate phosphoribosyl-transferase and cell proliferative activity and 5-fluorouracil sensitivity in colorectal carcinoma. Int J Clin Oncol. 2003;8:72–78. doi: 10.1007/s101470300013. [DOI] [PubMed] [Google Scholar]
- 28.Kornmann M, Schwabe W, Sander S, Kron M, Strater J, Polat S, et al. Thymidylate synthase and dihydropyrimidine dehydrogenase mRNA expression levels: predictors for survival in colorectal cancer patients receiving adjuvant 5-fluorouracil. Clin Cancer Res. 2003;9:4116–4124. [PubMed] [Google Scholar]
- 29.Kakimoto M, Uetake H, Osanai T, Shirota Y, Takagi Y, Takeshita E, et al. Thymidylate synthase and dihydropyrimidine dehydrogenase gene expression in breast cancer predicts 5-FU sensitivity by a histocultural drug sensitivity test. Cancer Lett. 2005;223:103–111. doi: 10.1016/j.canlet.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 30.Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.BioMed Crit Comm. 2011;4:2417. [Google Scholar]
- 32.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 33.Kawakami K, Ishida Y, Danenberg KD, Omura K, Watanabe G, Danenberg PV. Functional polymorphism of the thymidylate synthase gene in colorectal cancer accompanied by frequent loss of heterozygosity. Jpn J Cancer Res. 2002;93:1221–1229. doi: 10.1111/j.1349-7006.2002.tb01227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Villafranca E, Okruzhnov Y, Dominguez MA, Garcia-Foncillas J, Azinovic I, Martinez E, et al. Polymorphisms of the repeated sequences in the enhancer region of the thymidylate synthase gene promoter may predict downstaging after preoperative chemoradiation in rectal cancer. J Clin Oncol. 2001;19:1779–1786. doi: 10.1200/JCO.2001.19.6.1779. [DOI] [PubMed] [Google Scholar]
- 35.Brody JR, Hucl T, Gallmeier E, Winter JM, Kern SE, Murphy KM. Genomic copy number changes affecting the thymidylate synthase (TYMS) gene in cancer: a model for patient classification to aid fluoropyrimidine therapy. Cancer Res. 2006;66:9369–9373. doi: 10.1158/0008-5472.CAN06-2165. [DOI] [PubMed] [Google Scholar]
- 36.Wang TL, Diaz L, Jr, Romans K, Bardelli A, Saha S, Galizia G, et al. Digital karyotyping identifies thymidylate synthase amplification as a mechanism of resistance to 5-fluorouracil in metastatic colorectal cancer patients. Proc Natl Acad Sci USA. 2004;101:3089–3094. doi: 10.1073/pnas.0308716101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes and organelles. Microbiol Rev. 1983;47:1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coonrod A, Li FQ, Horwitz M. On the mechanism of DNA transfection: efficient gene transfer without viruses. Gene Ther. 1997;4:1313–1321. doi: 10.1038/sj.gt.3300536. [DOI] [PubMed] [Google Scholar]
- 39.Santi DV, McHenry CS. 5-Fluoro-2′-deoxyuridylate: covalent complex with thymidylate synthetase. Proc Natl Acad Sci USA. 1972;69:1855–1857. doi: 10.1073/pnas.69.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]