Abstract
Broadly neutralizing antibodies (bnAbs) against human immunodeficiency virus (HIV)-1 are rare in natural infection and elicitation of HIV-1 bnAbs has not been achieved by any vaccine candidates. We and others have reported that HIV-1 bnAbs are highly diversified from their germline-like predecessors and the germline-like predecessors of bnAbs lack measurable binding to HIV-1 envelope (Env) glycoproteins, suggesting that Env structures containing the epitopes of bnAbs may not initiate somatic maturation pathway, which may partially explain the rarity of HIV-1 bnAbs. To determine the minimum mutations required for converting non-binding germline-like predecessors to Env-binding antibodies, we started with the bnAb b12 as a prototype and generated six “chimeric” scFv b12 variants by sequentially replacing the heavy chain V-segment (HV), D(J)-segment [HD(J)] in the heavy chain variable region (VH), and the whole light chain variable region (VL) in b12 germline-like predecessor with the mature counterparts. We tested the recombinant scFv variants for binding and neutralizing activities. Results showed that a single point mutation in germline D-segment was enough to convert nonbinding germline-like b12 to an Env-binding antibody. Replacement with either mature HV or mature VL also made the germline-like b12 bind to Env, but none of single segment replacements conferred neutralization ability to the germline antibody. Mature VL in combination with mature HD(J) or mature HV, or both conferred increasing neutralization activity to the germline antibody. However, hybrid scFv, mature VH/germline VL, did not neutralize HIV-1, suggesting the importance of mature VL in neutralizing the virus. These results may have implications for vaccine development.
Key words: germline, antibody, immune responses, HIV, vaccine
Introduction
Human immunodeficiency virus/acquired immunodeficiency syndrome (HIV/AIDS) is one of the most devastating infectious diseases in the world. Since the discovery of HIV-1 in 1983, multiple vaccine concepts and vaccination strategies to prevent infection have been tested, but none have proven effective. Broadly neutralizing antibodies (bnAbs) are likely to be a key component of protective immunity conferred by an effective HIV-1 vaccine. However, elicitation of potent bnAbs has not been achieved by any vaccine candidate so far. Potent bnAbs are rare in natural infection, but do exist in a small percentage of infected individuals over a period of years of infection, which may contribute to the long-term cessation of disease progression.1–4 Besides the four known broadly neutralizing human monoclonal anti-bodies (bnmAbs), b12, 2G12, 2F5 and 4E10, recently identified potent bnmAbs, PG9, PG16, HJ16, VRC01, 02 and 03 were also isolated from such long-term nonprogressors (LTNPs).5–11 B12, VRC01, 02 and 03 bind to epitopes overlapping the CD4 binding site (CD4bs) on gp120,12,13 while HJ16 binds an epitope proximal to the CD4bs on gp120.10 PG9 and PG16 bind to a conformational epitope on Env trimer which involves V2 and V3 loops of gp120.9 2F5 and 4E10 recognize linear epitopes located in the membrane proximal external region of gp41.7,8 2G12 is the only bnmAb that binds to sugar moieties on gp120.6 However, immunogens designed to include the structural determinants of some of the known bnmAbs (i.e., b12 and 2F5) failed to elicit the same or similar bnAbs.
We and others have recently reported that HIV-1 bnmAbs are highly divergent from their germline-like predecessors [average percentage amino acid (AA) change in VH: 16%] compared with potent nAbs against SARS CoV and Henipaviruses (average percentage AA change in VH: 5%) and human HIV-1 mAbs with limited cross-reactivity and potency (average percentage AA change in VH: 7%), suggesting that elicitation of HIV-1 bnAbs may take a long time.14 We further found that the germline-like predecessors of known anti-HIV-1 bnmAbs lack measurable binding to HIV-1 envelope glycoprotein (Env), suggesting that Env structures containing the epitopes of bnAbs may not trigger the somatic maturation pathway by binding to the germline-like antibodies. In contrast, the germline-like predecessors of non-neutralizing and weakly neutralizing HIV-1-specific antibodies bind to HIV-1 Env.14 We have hypothesized that somatic maturation of HIV-1 bnAbs may be initiated by primary immunogens that may be unrelated to HIV-1 Env. Intermediate bnAbs generated by the stimulation of primary immunogens may bind Env and further mature to bnAbs upon immunization with Envs or HIV-1 infection.
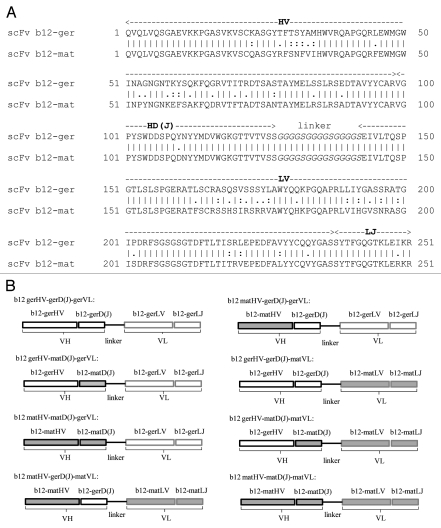
To investigate minimum mutations required for converting the non-binding germline-like predecessors of known bnmAbs to binding antibodies to HIV-1 Env, we used b12 as a model antibody. B12 is one of the most potent bnmAbs reported so far. It binds to a conformationally invariant surface on gp120 that overlaps the distinct subset of the CD4 binding site.13 B12 has a percentage AA change of 13% in both VH and VL. According to JoinSolver sequence analysis,15 b12 VH resulted most likely from recombination of IGHV1-3*01, IGHD3-10*02 and IGHJ6*03 with VD junction of 28 nucleotides insertion (TGG GGC CAT ATA GTT GGG ATG ATT CTC C) and DJ junction of six nucleotides insertion (TATTAT). During somatic maturation to b12, a total of 39 mutations occurred in HV-segment, leading to 20 AA changes; only one point mutation occurred in HD-segment (from CCA GTA CAA T to CCA GGA CAA T), which led to one tyrosine (Y) to aspartic acid (D) change; and one silent mutation occurred in HJ-segment. B12 VL was most likely derived from recombination of IGKV3-20*01 and IGKJ2*01, and somatic maturation led to a total of 19 AA changes in the light chain V-segment (LV) and one AA change in light chain J-segment (LJ) (Fig. 1). Based on the co-crystal structure of Fab b12 with gp120 core, only b12 heavy chain interacted with gp120, with each of the three heavy chain complementarity determining regions (CDRs) making extensive contact. The HCDR2 mimics the phenylalanine interaction of CD4 with gp120, while the HCDR3 also made extensive contact with gp120.13 To localize the important somatic mutations that can convert the non-binding germline-like predecessor of b12 to a binding b12 antibody intermediate, we constructed six germline/mature “chimeric” scFv b12 variants by sequentially replacing one or two segments in germline-like scFv b12 with the mature counterparts. Since b12 light chain did not make contact with gp120 based on the crystal structure, we kept b12 VL as a whole while we dissected b12 VH. Our results showed that a single mutation in HD-segment was enough to make the transition of non-binding germline-like b12 antibody to a binding antibody to HIV-1 Env, but this mutation was not enough to confer neutralization activity to the germline antibody. We found that mature VL was required for “ chimeric” scFv b12 variants to neutralize the virus.
Figure 1.
Amino acid sequence alignment of germline-like scFv b12 and mature scFv b12 (A), and schematic drawing of the constructs of germline-like and mature scFv b12 and six scFv variants (B). V- and D(J)-segments of heavy chain [HV and HD(J)] and V- and J-segments of light chain (LV and LJ) were indicated. A flexible (G4S)3 linker between variable heavy (VH) and light (VL) chains was in italic.
Results
Binding of “chimeric” scFv b12 variants to recombinant HIV-1 Env in ELISA.
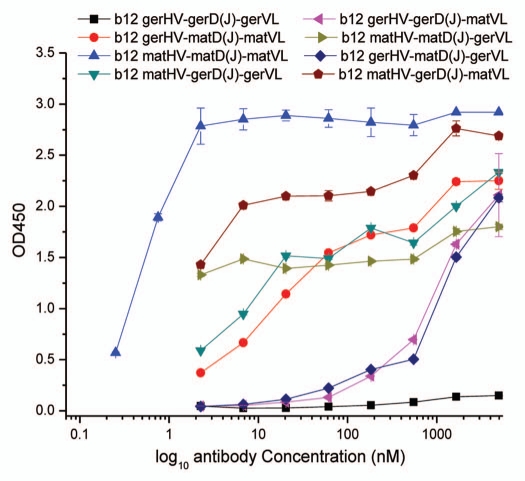
We have previously reported that germline-like predecessor of b12 lacks measurable binding to gp120, while mature scFv b12 has high binding affinity for gp120.14 Based on synthetic germline scFv b12, in which germline VH is covalently linked to germline VL by a flexible (G4S)3 linker, we constructed six “chimeric” scFv b12 variants by sequentially replacing the HV-, HD(J)-segments and the VL in b12 germline-like predecessor with the mature counterparts (Fig. 1). We measured the binding activity of the six “chimeric” scFv b12 variants to gp120Bal in ELISA. Interestingly, all six “chimeric” scFv b12 variants showed measurable binding to gp120Bal (Fig. 2 and Table 1). A single point mutation (from “T” to “G”) in germline D-segment that led to AA change from “Y” to “D” was sufficient to convert the germline b12 to a binding antibody to gp120Bal (EC50 = 1,008.7 nM). Replacement with mature VL alone led to slightly higher binding activity (EC50 = 835.7 nM) compared with this D-mutant, while replacement with mature HV-segment alone led to significantly increased binding affinity for gp120Bal (EC50 = 12.7 nM), suggesting the importance of mature HV in binding to Env, which is in agreement with the result from co-crystal structure of Fab b12 with gp120 core. Based on the crystal structure, HCDR2 of b12 HV is critical for binding to the phenylalanine cavity of gp120.13 In agreement with this, double segment/fragment replacement variants containing mature HV showed even higher binding affinities (EC50 = 2.3 nM for “b12 matHV-matD(J)-gerVL” and 2.4 nM for “b12 matHV-gerD(J)-matVL”) compared with the double segment/fragment replacement variant lacking mature HV (EC50 = 24.1 nM for “b12 gerHV-matD(J)-matVL”). Mature scFv b12 showed the highest binding affinity for gp120 (EC50 = 0.4 nM). These results suggest that somatic mutations in each segment of VH [HV and HD(J)] and VL fragment contribute to the binding of b12, but minimum mutation required for binding to Env can be as low as one point mutation in the D-segment. Although b12 light chain did not make contact with gp120 according to the co-crystal structure of Fab b12 with gp120 core, hybrid scFv, germline VH/mature VL, designated “b12 gerHV-gerD(J)-matVL”, bound to recombinant gp120Bal (EC50 = 853.7 nM) with an affinity similar to that of the D-mutant (Fig. 2 and Table 1).
Figure 2.
Binding of scFv b12 variants to recombinant gp120Bal in ELISA. Three-fold serially diluted recombinant scFvs were added to microwell plates coated with 1 µg/mL gp120Bal. Bound scFvs were revealed by detecting FLAG tag. ger, germline; mat, mature.
Table 1.
Binding and inhibitory activities of scFv b12 variants measured in ELISA and a TZM-bl cell line-based pseudovirus assay, respectively
| scFv b12 variants | EC50 (nM) | IC50 in TZM-bl Pseudovirus Assay (nM) | ||||||
| gp120Bal | JRFL (B) | Bal (B) | ADA (B) | JRCSF (B) | CH1 81.12 (CRF07-BC′) | PCNE1 (B′) | GXC 44 (C) | |
| b12 gerHV-gerD(J)-gerVL | >1,650 | >1,650 | >1,650 | >1,650 | >1,650 | >1,650 | >1,650 | >1,650 |
| b12 matHV-gerD(J)-gerVL | 12.7 | >1,650 | >1,650 | >1,650 | >1,650 | >1,650 | >1,650 | >1,650 |
| b12 gerHV-matD(J)-gerVL | 1,008.7 | >1,650 | >1,650 | >1,650 | >1,650 | >1,650 | >1,650 | >1,650 |
| b12 gerHV-gerD(J)-matVL | 835.7 | >1,650 | >1,650 | >1,650 | >1,650 | >1,650 | >1,650 | >1,650 |
| b12 matHV-matD(J)-gerVL | 2.3 | >1,650 | >1,650 | >1,650 | >1,650 | >1,650 | >1,650 | >1,650 |
| b12 gerHV-matD(J)-matVL | 24.1 | 1,364.7 | >1,650 | >1,650 | >1,650 | >1,650 | >1,650 | >1,650 |
| b12 matHV-gerD(J)-matVL | 2.4 | 15.9 | 55.6 | 1,067.2 | >1,650 | 855.7 | >1,650 | >1,650 |
| b12 matHV-matD(J)-matVL | 0.4 | 8.4 | 13.0 | 170.9 | 77.5 | 281.8 | >1,650 | >1,650 |
EC50s and IC50s were determined using GraphPad prism software. ger, germline; mat, mature.
Binding of “chimeric” scFv b12 variants to cell-associated HIV-1 Env.
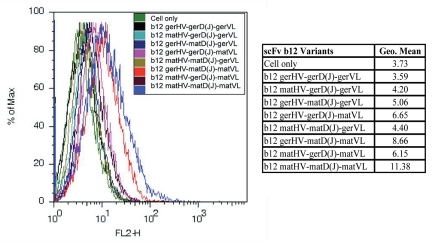
We further tested the binding of the “chimeric” scFv b12 variants to cell-associated HIV-1 gp160 in flow cytometry. As expected, germline-like predecessor of b12 did not bind to cell surface Env. Mature scFv b12 showed high binding affinity for cell-associated gp160Bal. All six “chimeric” scFv b12 variants bound, to various extents, to native Env (Fig. 3). The D-mutant weakly bound to gp160Bal, as well as the other two variants with single segment/fragment replacement and one of the hybrid scFv, mature VH/germline VL. In agreement with the ELISA result, hybrid scFv, germline VH/mature VL showed binding activity to native Env on cell surface.
Figure 3.
Binding of scFv b12 variants to cell-associated HIV-1 gp160Bal in Flow cytometry. 293T cells transfected with plasmid DNA containing gp160Bal gene were used to measure the binding activity of scFv b12 variants in comparison with germline-like predecessor of b12 and mature b12. A fixed concentration (1,650 nM) was used for all recombinant scFvs. Geometry mean values were summarized in the table next to the overlay of the histogram. ger, germline; mat, mature.
Inhibitory activity of “chimeric” scFv b12 variants in TZM-bl pseudovirus assay.
To investigate how somatic maturation of each segment or fragment affects the neutralization activity of b12, we tested six recombinant scFv b12 variants with a panel of HIV-1 isolates in a standardized TZM-bl pseudovirus assay in comparison with the germline-like predecessor and mature scFv b12 (Table 1). None of the variants with single segment/fragment replacement showed neutralization activity at the highest concentration tested (1,650 nM). Two hybrid scFvs, mature VH/germline VL and germline VH/mature VL, also did not neutralize the isolates tested, suggesting that somatic maturation in both heavy and light chains are required for neutralization activity. ScFv b12 variant with mature HD(J) in combination with mature VL weakly neutralized JRFL, while scFv b12 variant with mature HV in combination with mature VL neutralized most of the isolates tested, including JRFL, Bal, ADA and crossclade isolate CH181.12. Mature scFv b12 neutralized all clade B isolates tested, as well as the cross-clade isolate CH181.12. Mature scFv b12 did not neutralize clade B' isolate, PCNE1 and clade C isolate, GXC44.
Discussion
Our study demonstrated that a single mutation in heavy chain D-segment (from “Y” to “D”) can convert a non-binding germline-like predecessor of b12 to a binding antibody to HIV-1 Env, but this point mutation was not sufficient to confer neutralization activity to the antibody. The prediction of germlinelike b12 sequence used in this study was based on the analysis using JoinSolver (joinsolver.niaid.nih.gov). We acknowledged that sequence analysis using IMGT/V-QUEST (imgt.cines.fr/IMGT_vquest/share/textes) resulted in three somatic mutations in HCDR3 and prediction of different D-segment and N-nucleotides in the joint region. Nevertheless, we demonstrated here that a single point mutation can change antibody property. A similar phenomenon was observed with another bnAb 2G12. A few mutations in germline-like 2G12 can cause domain swapping, a major characteristic of mature 2G12.16 The b12 variant with this single mutation had an affinity of about 1 µM for recombinant gp120Bal, and it also weakly bound to cellassociated gp160Bal. This D-mutant, an intermediate antibody of b12 with the least number of somatic mutations, may be used to search for primary immunogen that binds to both germline-like b12 and the intermediate D-mutant, and may trigger the maturation pathway of the germline-like predecessor of b12. We assume that the binding of this D-mutant to Env may be strong enough to initiate the further maturation of b12 upon infection of HIV-1 in vivo as suggested in the literatures.17,18 It would be worth testing in humanized mouse model if B cells bearing the D-mutant gene can mature to b12 upon HIV-1 infection or immunization with HIV-1 Envs. It may be also worthwhile testing other intermediate b12 antibody genes in animal models for possible multiple maturation pathways of b12 in vivo.
Our results showed that somatic maturation in heavy chain V-, D(J)-segments and VL of germline-like predecessor of b12 additively contributed to the binding of b12 to HIV-1 Env. Although b12 light chain did not make contact with gp120 core based on the co-crystal structure of Fab b12 with gp120 core, mature b12 VL in combination with germline VH bound to recombinant Env, as well as cell-associated HIV-1 Env as showed in Flow cytometry analysis. The mechanism for this inconsistency remained to be elucidated. B12 was selected by phage display and its light chain may not be the originally paired one with the heavy chain of b12 in vivo. Nevertheless, we would assume that somatic maturation of the originally paired light chain may also contribute to the binding of b12 to HIV-1 Env as exemplified by bnmAb VRC01 that has the original pair of the heavy and light chains.12 But only the co-crystal structure of this b12 hybrid scFv, germline VH/mature VL, with Env may provide an answer. Mature b12 light chain is important not only for binding to Env, but also for neutralizing HIV-1 because another hybrid scFv, mature VH/germline VL, did not neutralize the virus, suggesting that binding to Env and neutralizing the virus may involve different mechanisms. Note that we used scFv format of germline-like b12, mature b12 and b12 variants in this study. We have shown in our previous study that scFvs and scFv-Fc fusions that mimic the IgG format of antibodies have the same binding characteristics.14 Although we did not use live viruses to measure the neutralization ability of the antibodies in this study, we believe that the results from the standardized TZM-bl cell line-based assay reflect the trend of change in neutralization ability from germline-like b12 to intermediate b12 antibodies and to mature b12.
We often observe that binding profiles of antibodies do not correlate with neutralizing profiles. But for somatic maturation variants derived from the same antibody, in this case b12, the binding profile somewhat correlates with the neutralizing profile (Table 1). During antibody maturation, less mutation, as low as one single nucleotide change, may be needed to turn a non-binding germline-like predecessor of bnAb into a binding antibody intermediate to HIV-1 Env, suggesting the possibility for triggering maturation pathways of bnAbs in vivo. Extensive somatic maturation, however, may be needed to turn a binding antibody intermediate further into a neutralizing antibody, suggesting the existence of binding threshold for neutralization, or a requirement for critical somatic mutations at certain positions for neutralization, or both. Further studies need to be done to elucidate the mechanism. These results may have implications for HIV-1 vaccine development.
Materials and Methods
Cells, viruses, plasmids and proteins.
TZM-bl and 293T were obtained from NIH AIDS Research and Reference Program (ARRP) (Division of AIDS, National Institute of Allergy and Infectious Diseases). Both cell lines were maintained in DMEM containing 10% Fetal Bovine Serum (FBS), 2 mM L-glutamine, 100 U/mL penicillin and 100 ug/mL streptomycin. HIV-1 isolates ADA, JRCSF, JRFL and Bal were obtained from the NIH ARRP. HIV-1 isolate CH181.12 was kindly provided by Dr. Yiming Shao (China CDC, Beijing, China), PCNE1 by Dr. Linqi Zhang (Tsinghua University, Beijing, China) and GXC44 by Dr. Gerald Quinian (USUHS, Bethesda, MD). The pNL4-3.luc.ER-plasmid was obtained from ARRP. Envelope glycoprotein gp120Bal was produced in our laboratory by transient transfection of 293T cells with pSecTag 2 plasmid (Invitrogen, Carlsbad, CA) containing gp120Bal gene amplified from the Bal Env plasmid followed by IMAC affinity purification using Ni-NTA agarose beads (Qiagen, Hilden, Germany).
Generation of germline/mature “chimeric” scFv b12 variants.
Mature and germline-like scFv b12 genes were synthesized by Genescript as described previously in reference 14. Germline-like predecessor of b12 sequence was determined by reverting mutations to the germline sequence while retaining the original CDR3 junctions and terminal deoxynucleotidyl transferase (TdT) N nucleotides. Six germline/mature “chimeric” scFv b12 variants were generated by splice overlap extension PCR (SOE-PCR) to replace either one segment or two segments of germline-like b12 with the mature counterparts and confirmed by DNA sequencing. The pComb3X plasmid (provided by Dennis Burton, Scripps Institute, La Jolla, CA) was used for expression of mature, germline-like scFv b12 and chimeric scFv b12 variants in E. coli HB2151 by following the protocol described previously in reference 19. Recombinant scFvs with his6 and FLAG tags were purified by IMAC affinity purification. FLAG tag was used for detection in ELISA.
ELISA.
One hundred nanograms per well of recombinant gp120Bal in sodium bicarbonate coating buffer (pH 8.3) was coated in high binding 96-well microplates by incubating at 4°C overnight. The wells were then blocked with 2.5% skim milk in PBS buffer (MPBS) and three-fold serially diluted scFvs in MPBS added to the ELISA plate. Bound scFvs were detected using HRP conjugated to mouse anti-FLAG (1:5,000) as second antibody and TMB as substrate. The optical density at 450 nm was determined after color development at RT for 20 min. Antibody concentrations that lead to half-maximum binding (EC50) were determined using GraphPad prism software.
Flow cytometry.
Plasmid DNA containing Bal gp160 were used to transfect 293T cells. Two days post transfection, 293T cells were detached and used in Flow cytometry as follow: 293T cells expressing Bal gp160 were incubated with 1,650 nM of each purified recombinant scFv b12 variant at 4°C for 1 h. The bound antibodies were detected using anti-FLAG mouse mAb M2 (20 µg/mL) by incubation at 4°C for 1 h followed by staining with phycoerythrin conjugated to goat anti-mouse IgG, F(ab')2 (1:500) (ImmunoJackson). A FACSArray plate reader (BD biosciences) was used for flow cytometric analysis and FlowJo software for data interpretation.
Neutralization assay.
Single-round infectious envelope-pseudotyped viruses were prepared by cotransfection of 70–80% confluent HEK 293T cells with pNL4-3.luc.E-R- and HIV-1 Env plasmid using PEI as transfection reagent. Four to six hours post transfection, medium was replaced with DMEM containing 10% FBS (Sigma, St. Louis, MO). Cells were allowed to grow for an additional 40 h. The supernatant was harvested, centrifuged at 16,000 rpm for 5 min at 4°C, and filtered through a 0.45 µm pore filter (Millipore, Bedford, MA). Neutralization assays were carried out in triplicate or, in some cases, in duplicate. Fifty microliters of 3-fold serially diluted scFvs were preincubated with 50 µL of pseudovirus suspension at 37°C for 30 min. Virus-antibody mixtures were then added to TZM-bl cells in 96-well culture plates (Costar, Corning, NY). Plates were incubated at 37°C with 5% CO2 for two days, and then washed with PBS and lysed for 30 min with 60 µL of lysis buffer followed by addition of 25 µL of luciferin (Promega, Madison, WI). Luminescence readings were determined by PE Victor3 luminometer. Antibody concentration that leads to 50% neutralization (IC50) was determined using GraphPad prism software.
Acknowledgments
We thank Dimiter S. Dimitrov, Nancy Longo, Ruth Ruprecht, Peter Kwong, Tongqing Zhou, Xiaodong Xiao for stimulating discussions, Nancy Longo for help with germline sequence analysis and Yiming Shao, Linqi Zhang, Gerald Quinnan, Dennis Burton for providing reagents. This work was supported by the Intramural Research Program of the University of Hong Kong, and by General Research Fund (GRF) of Hong Kong to M.Y.Z.
Abbreviations
- DMEM
Dulbecco's minimal essential medium
- IMAC
immobilized metal affinity chromatography
- HRP
horse radish peroxidase
- TMB
tetramethylbenzidine
- PEI
polyethylenimine
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Dhillon AK, Donners H, Pantophlet R, Johnson WE, Decker JM, Shaw GM, et al. Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1-infected donors. J Virol. 2007;81:6548–6562. doi: 10.1128/JVI.02749-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gray ES, Madiga MC, Moore PL, Mlisana K, Abdool Karim SS, Binley JM, et al. Broad neutralization of human immunodeficiency virus type 1 mediated by plasma antibodies against the gp41 membrane proximal external region. J Virol. 2009;83:11265–11274. doi: 10.1128/JVI.01359-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y, Migueles SA, Welcher B, Svehla K, Phogat A, Louder MK, et al. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13:1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 5.Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 6.Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, et al. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1→2 mannose residues on the outer face of gp120. J Virol. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Purtscher M, Trkola A, Gruber G, Buchacher A, Predl R, Steindl F, et al. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 1994;10:1651–1658. doi: 10.1089/aid.1994.10.1651. [DOI] [PubMed] [Google Scholar]
- 8.Stiegler G, Kunert R, Purtscher M, Wolbank S, Voglauer R, Steindl F, et al. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2001;17:1757–1765. doi: 10.1089/08892220152741450. [DOI] [PubMed] [Google Scholar]
- 9.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009:326–329. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corti D, Langedijk JP, Hinz A, Seaman MS, Vanzetta F, Fernandez-Rodriguez BM, et al. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS ONE. 2010;5:8805. doi: 10.1371/journal.pone.0008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao X, Chen W, Feng Y, Zhu Z, Prabakaran P, Wang Y, et al. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: Implications for evasion of immune responses and design of vaccine immunogens. Biochem Biophys Res Commun. 2009;390:404–409. doi: 10.1016/j.bbrc.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Souto-Carneiro MM, Longo NS, Russ DE, Sun HW, Lipsky PE. Characterization of the human Ig heavy chain antigen binding complementarity determining region 3 using a newly developed software algorithm, JOINSOLVER. J Immunol. 2004;172:6790–6802. doi: 10.4049/jimmunol.172.11.6790. [DOI] [PubMed] [Google Scholar]
- 16.Huber M, Le KM, Doores KJ, Fulton Z, Stanfield RL, Wilson IA, et al. Very few substitutions in a germ line antibody are required to initiate significant domain exchange. J Virol. 2010;84:10700–10707. doi: 10.1128/JVI.01111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Batista FD, Neuberger MS. Affinity dependence of the B cell response to antigen: a threshold, a ceiling and the importance of off-rate. Immunity. 1998;8:751–759. doi: 10.1016/s1074-7613(00)80580-4. [DOI] [PubMed] [Google Scholar]
- 18.Dal Porto JM, Haberman AM, Shlomchik MJ, Kelsoe G. Antigen drives very low affinity B cells to become plasmacytes and enter germinal centers. J Immunol. 1998;161:5373–5381. [PubMed] [Google Scholar]
- 19.Zhang MY, Shu Y, Rudolph D, Prabakaran P, Labrijn AF, Zwick MB, et al. Improved breadth and potency of an HIV-1-neutralizing human single-chain antibody by random mutagenesis and sequential antigen panning. J Mol Biol. 2004;335:209–219. doi: 10.1016/j.jmb.2003.09.055. [DOI] [PubMed] [Google Scholar]