Abstract
Spermatogenesis is an intricately regulated process of cellular differentiation transforming spermatogonial stem cells to spermatozoa. Elimination of the transcription factor EGR4 generates subfertile male mice yet the expression and function of EGR4 in the mammalian testis has yet to be fully investigated. We performed in situ hybridization and immunofluorescence to identify Egr4 transcript and protein localization in the developing murine testis. EGR4 was detected in both germ and somatic cells in the neonatal testis but was specific to germ cells inside the seminiferous epithelium from juvenile development onward. EGR4 also displayed distinct intracellular localization patterns within specific cell populations of the testis. In addition, Egr4-deficient testis tubules regress from relatively normal to Sertoli cell and undifferentiated spermatogonia only over time. Taken together, these data suggest that Egr4 may regulate spermatogenesis at multiple steps, with roles in the dividing Sertoli cells, peritubular myoid cells, and the meiotic and elongating haploid germ cell populations.
Keywords: spermatogenesis, EGR4, testis, germ cells
INTRODUCTION
Spermatogenesis is an intricately regulated process of cellular differentiation transforming primordial germ cells to spermatozoa. Aberrant gene expression during male germ cell development can lead to abnormal sperm production and infertility. Spermatogonial differentiation and meiotic initiation are essential to spermatogenesis as these events ensure that correct numbers of healthy haploid male gametes are released from the seminiferous epithelium.
In the mouse, the first wave of spermatogenesis begins directly after birth when gonocytes, which entered the G0/G1 mitotic cell cycle phase in utero, reinitiate mitosis, migrate to the basement membrane of the seminiferous epithelium, and differentiate to spermatogonial stem cells (SSCs; de Rooij and Russell, 2000). The SSCs then divide to either replenish the stem cell population or start a series of mitotic cell divisions which first result in paired A spermatogonia, connected by an intracellular bridge, and then create chains of up to 32 A aligned spermatogonia (Aal; de Rooij and Russell, 2000). Spermatogonia from the SSCs to the Aal represent the undifferentiated population. The Aal then differentiate, without dividing, to produce A1 spermatogonia, which in turn undergo five mitotic divisions to produce the B spermatogonia. The B spermatogonia will then divide to become spermatocytes and initiate meiosis (Russell et al., 1990).
The complex series of cell divisions during spermatogenesis relies on the precisely regulated expression of a vast number of genes. Several transcription factors are known to play key roles in the differentiation of spermatogonia and meiotic initiation. SOX3, a member of the high mobility group (HMG) family of transcription factors, is required for the transition from undifferentiated to differentiated spermatogonia as the seminiferous tubules of the Sox3 null male mouse testis are agametic by 14 days post partum (dpp; Raverot et al., 2005). Loss of the germ-cell specific factor SOHLH1, a basic helix–loop–helix transcription factor, blocks the differentiation of spermatogonia to spermatocyte, also resulting in male infertility (Ballow et al., 2006). In addition, retinoic acid (RA), an active metabolite of vitamin A, is crucial to the initiation of meiosis in both females and males (Anderson et al., 2008) and can directly induce the transition from undifferentiated to differentiated spermatogonia in culture through stimulating the expression of Stra8 and Kit (Zhou et al., 2008). However, there is still much to be learned about how spermatogonial differentiation and meiotic initiation are controlled.
The early growth response factor (Egr) family consists of four different transcription factors which were initially identified as genes whose expression is induced quickly in response to growth factors (reviewed in O’Donovan et al., 1999). All four family members share a highly conserved DNA binding domain, composed of three zinc-finger motifs (Chavrier et al., 1988; Gashler and Sukhatme, 1995). Transcription factors EGR1, 2, and 3 also share an R1 domain, missing in EGR4, which can be bound by NAB proteins to prevent expression of EGR target genes (Svaren et al., 1998). The EGR zinc finger motifs are designed to specifically recognize a 9 base pair consensus sequence (5′ GCG TGG GCG 3′), with each finger spanning three nucleotides (Christy and Nathans, 1989; Nardelli et al., 1991; Pavletich and Pabo, 1991).
Initially, it was thought that the EGR proteins only regulated changes in gene expression controlling neuronal plasticity; however, functional analysis of the individual EGRs using knockout mice has revealed importance for these genes in other systems. For example, mice which lack Egr3 display severe motor abnormalities due to a lack of muscle spindle fiber development (Tourtellotte and Milbrandt, 1998), and EGR1 is a direct transcriptional regulator of luteinizing hormone beta (LHβ). The Egr1-null mouse displays a significant reduction in LHβ expression, leading to female infertility (Lee et al., 1996). In contrast, male mice deficient in Egr1 exhibit no identifiable defects in spermatogenesis, a result of partial redundancy between Egr1 and Egr4 as the dual Egr1/Egr4-deficient male displays very low levels of serum LH and male infertility (Tourtellotte et al., 2000).
Our current understanding of EGR4 function suggests that it acts as a transcriptional repressor in vitro (Zipfel et al., 1997) and the fertility phenotype of the global Egr4 knockout mouse demonstrates that it is essential for normal spermatogenesis. Early expression data suggested Egr4 was specific to the central nervous system (Crosby et al., 1992); however, the Egr4-deficient mouse is viable and displays no obvious phenotypic abnormalities except that males, and not females, are subfertile (Tourtellotte et al., 1999). In situ hybridization has localized Egr4 transcript to primary and secondary spermatocytes in the adult mouse testis and reverse transcriptase PCR confirmed that Egr4 transcripts were only present in the germ cell fraction in the adult animal (Tourtellotte et al., 1999). A loss of Egr4 results in a block in spermatogenesis at the early-mid pachytene spermatocyte stage leading to a significant reduction in the number of spermatozoa produced. Additionally, Erg4-null spermatozoa show abnormal morphology such as separated heads and fragmented flagellum. The block also occurred during the first wave of spermatogenesis as apoptotic germ cells were present from 14 dpp onward.
The importance of EGR4 for fertility in mammals has also been demonstrated in human males with cryptorchidism (Hadziselimovic et al., 2009). Immunohistochemistry localized EGR4 to germ and Leydig cells in testes of 3-year-old boys. Microarray analyses of undescended testes revealed a down-regulation of Egr4 in patients at a high risk of infertility, i.e., had testes that did not contain Adark spermatogonia, the stem cells of the human testis. In these patients, no EGR4 could be detected within the seminiferous epithelium; however, the Leydig cells still displayed positive signal. Hence, this study links EGR4 to the development of stem cells in the human testis, and taken together with the data generated by the Egr4-null mouse, indicates that EGR4 may be important to germ cell differentiation at several different stages during spermatogenesis.
Even with the current published EGR4 expression studies in the testis, there are still gaps in our understanding of wild-type Egr4 biology. The published studies provide expression information for the adult mouse testis and the neonatal human testis. Given that the first apoptotic germ cells in the Egr4-deficient testis are present at 14 dpp, an expression analysis encompassing the first wave of spermatogenesis is necessary to understand how Egr4 regulates male germ cell development and bridge the gap between the two published studies. To complete the analysis, in situ hybridization and immunofluoresence were used to investigate the histological expression and localization of Egr4 transcript and protein throughout the first wave of spermatogenesis. We also extend the morphological analysis of the Egr4-null testis phenotype by quantitatively counting the regression of the seminiferous epithelium in these animals as they age.
RESULTS
Egr4 Expression Is Developmentally Regulated During Testis Development
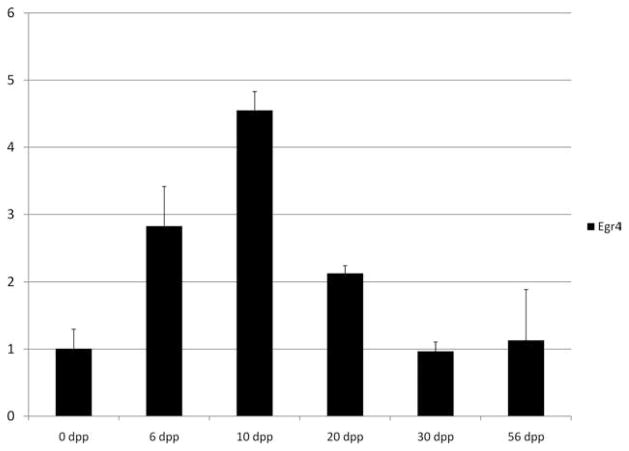
Our laboratory generated a publicly available global microarray expression profile database for a developmental time course of embryonic ovary and embryonic and postnatal testis tissues (Shima et al., 2004; Small et al., 2005). These data sets allow a quick and simple analysis of time points in the testis and embryonic ovary at which our genes of interest are highly expressed. These array data sets were mined to graph an expression profile for Egr4; however, in each of the three arrays, the Erg4 probe set was flagged as ‘‘absent’’ in every sample. In addition, a search of the GEO Profiles database on the NCBI Web site (http://www.ncbi.nlm.nih.gov) for Egr4 expression profiles in testis samples also yielded mostly data sets with ‘‘absent’’ detection calls for the Egr4 probe (transcript) and a previous study also found it difficult to generate a mouse array expression for Egr4 (Hamra et al., 2004). Hence, we were not able to generate a microarray expression profile for Egr4. Instead, we performed quantitative real-time reverse transcriptase-polymerase chain reaction (RT-PCR) to generate an expression profile for Egr4 using the RNA samples that were analyzed with microarrays. Egr4 transcript was maximally expressed during the neonatal and juvenile periods of mouse testis development, with a peak of expression present at 10 dpp (Fig. 1).
Fig. 1.
Egr4 expression profile during testis development. Real-time polymerase chain reaction used to determine the levels of Egr4 transcript in total testis RNA samples from mice collected at 0, 6, 10, 20, 30, and 56 days post partum (dpp; X axis). Data are presented as the relative fold change (Y axis) compared with the 56 dpp testis sample. Error bars represent mean ± standard deviation, n = 2.
Egr4 Transcript Is Present in Both Germ and Somatic Cells in the Developing Murine Testis
Egr4 has been reported as being germ cell specific in the adult mouse testis, as no transcript could be detected in somatic cells using either in situ hybridization or RT-PCR (Tourtellotte et al., 1999). However, protein has been detected in both germ and somatic cells in the neonatal human testis (Hadziselimovic et al., 2009) and the testes of Egr4-deficient mice begin to display apoptotic cells from 14 dpp onward (Tourtellotte et al., 1999). Hence, we performed in situ hybridization on a developmental time course of mouse testis sections to visualize the wild-type cellular expression pattern of Egr4 to bridge the gap between the two published studies and before and after morphological differences in the knockout animal are apparent.
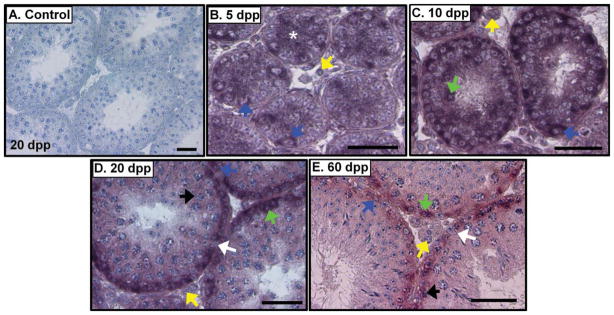
Egr4 transcript was present in both germ and somatic cells in the neonatal testis (Fig. 2). Spermatogonia display a positive signal for Egr4 transcript at 5, 10, 20, 30, and 60 dpp (Fig. 2). Egr4 message was also present in preleptotene, leptotene, and zygotene spermatocytes at 10, 20, and 60 dpp (Fig. 2). Signal was also faintly detectable in the pachytene spermatocytes at 20 and 60 dpp (Fig. 2D,E); however, no signal was detected in the haploid germ cells at any of the ages tested. In terms of the somatic cells of the testis, Sertoli cells also contained Egr4 transcript at 5 dpp (Fig. 2B) and signal was present in Leydig cells at 5 and 10 dpp (Fig. 2B,C); however, both of these cell types were negative at 20 and 60 dpp. Egr4 transcript was also not detectable in the peritubular myoid cells (PTMs). We used the Egr4 sense probe hybridized to wild-type mouse testis sections as negative controls for our in situ hybridization experiments (Fig. 2A), and signal was never detected on these samples.
Fig. 2.
Egr4 transcript localizes to specific populations of both germ and somatic cells in the developing testis. A–E: In situ hybridization of 5, 10, 20, and 60 days post partum (dpp) mouse testes with Egr4 antisense cRNAs. Sense cRNA control shown in A. Scale bars = 50 μm. Blue arrows, spermatogonia; green, preleptotene spermatocytes; black, pachytene spermatocytes; yellow, Leydig cells; white arrow, Sertoli cell nucleus; white asterisk, Sertoli cell cytoplasm.
EGR4 Localizes to Both Testicular Germ and Somatic Cells and Has an Intracellular-Specific Localization Pattern
Hadziselimovic et al. (2009) used a commercially available EGR4 antibody to localize this protein in the human testis, and we used this same reagent to perform immunofluorescence to determine which testis cell types contain EGR4 during the first wave of spermatogenesis.
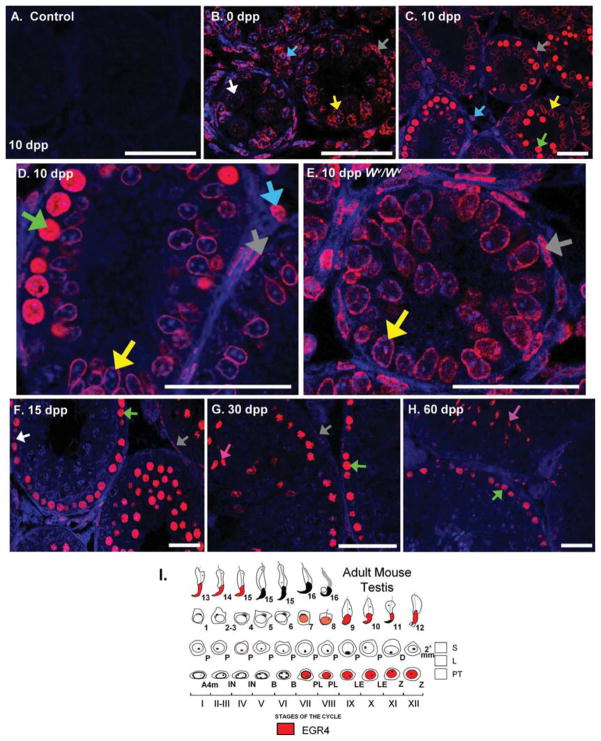
Both a cell- and intracellular-specific localization pattern was apparent for EGR4 in the developing mouse testis (Fig. 3). EGR4 was present in spermatogonia at 0 dpp (Fig. 3B), in preleptotene and leptotene spermatocytes from 10 dpp onward (Fig. 3C,D,F–H) and in elongating spermatids at 30 and 60 dpp (Fig. 3G,H). Stage specificity was also evident in the adult mouse testis staining pattern from 30 dpp onward (Fig. 3I). At stages VII–IV, EGR4 protein was present in the nuclei of round and elongating spermatids, whereas the preleptotene, leptotene, and zygotene spermatocyte nuclei stain positively for EGR4 at stages VII through XI (Fig. 3H,I). In addition to the staining pattern seen in the germ cell population, EGR4 was also detected in the PTMs from 0 through 30 dpp (Fig. 3B–D,F,G), and in the Sertoli and Leydig cells at 0, 5, and 10 dpp (Fig. 3B,C, 5 dpp data not shown).
Fig. 3.
EGR4 protein localizes to germ and somatic cells in the developing testis. A–H: Immunofluorescence localizing EGR4 in sections of 0, 10, 20, 30, and 60 days post partum (dpp) wild-type mouse testes and to the 10 dpp Wv/Wv mouse testis. No primary antibody control shown in A. I: Mouse testis staging diagram detailing the cell expression and localization of EGR4 in the adult mouse testis. Scale bars = 20 μm. White arrows, spermatogonia; green, preleptotene/leptotene spermatocytes; pink, elongating spermatids; light blue, Leydig cells; yellow, Sertoli cells; gray, PTMs.
Of interest, EGR4 displayed a distinct pattern of localization in germ cells and PTMs versus Sertoli cells. At 0 and 5 dpp, EGR4 was present in the cytoplasm and nucleus of gonocytes and spermatogonia (Fig. 3B); however, at 10 dpp, EGR4 localizes throughout the nucleus of germ cells and PTMs (Fig. 3C,D), whereas Sertoli cells appear to contain a ring of EGR4 protein localized to the perinuclear rim (Fig. 3C,D). We also analyzed cross-sections of a testis from a 10 dpp Wν/Wν mouse, a naturally occurring mutation in the c-kit gene which leads to the production of a Sertoli-cell only testis phenotype (Motro et al., 1991), and observed that the perinuclear rim localization was conserved in these animals as was EGR4 staining throughout the PTM nucleus (Fig. 3E).
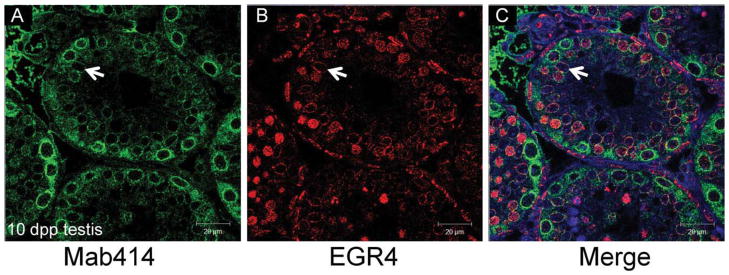
To further investigate the distinct perinuclear localization of EGR4 in the 10 dpp Sertoli cells, colocalization of EGR4 and Mab414 was performed (Fig. 4). Mab414 is a monoclonal antibody raised against the FXFG repeats in multiple nuclear pore complex proteins and has been successfully used to highlight the nuclear envelope (Lopez-Soler et al., 2001). In the 10 dpp testis, EGR4 and Mab414 colocalize to the nuclear envelope of the Sertoli cells (Fig. 4C); however, they do not colocalize in any other testicular cell type at 10 dpp.
Fig. 4.
Colocalization of nucleoporins with EGR4 in Sertoli cells. A,B: Immunofluorescence colocalizing EGR4 with the nucleoporin marker Mab414 in sections of 10 days post partum (dpp) testis. The staining patterns for Mab414 (green) and for EGR4 on the same testis tubule are shown individually in panels A and B, respectively. C: The merged image. The white arrow denotes a Sertoli cell nuclear envelope. Scale bars = 20 μm.
Egr4 Deficiency Results in Gradual Tubule Regression
It is clear that a loss of Egr4 disrupts the normal progression of spermatogenesis. In the absence of EGR4, germ cell differentiation is blocked during meiosis and the cells which escape this block develop as morphologically abnormal spermatozoa (Tourtellotte et al., 1999). However, not all knockout males were completely infertile, as two null males were able to sire litters. We have generated our own colony of these Egr4-deficient mice and found that some null males never produce litters and those that do only sire 2–3 litters before they are also infertile at approximately 5 months of age. To begin to assess why the older Egr4-deficient males were losing their fertility, testes from heterozygous and homozygous mice were collected at 120 and 365 dpp, fixed and cross-sections were prepared so the testes could be morphologically analyzed. The testes at both ages were found to contain an assortment of normal, partially deficient or completely deficient tubules (Fig. 5). Partially deficient tubules contained germ cells which appeared morphologically apoptotic, were missing layers of germ cells and/ or contained vacuoles and are represented by Figure 5E. The completely deficient tubules are represented by Figure 5F. These tubules contained Sertoli cells but were devoid of almost all germ cells. All stages of the seminiferous epithelium were morphologically normal and represented among the wild-type looking tubules (represented by Fig. 5D). Immunofluorescence staining for the germ cell marker GCNA at 180 and 365 dpp was used to determine whether germ cells remained in the completely deficient tubules at these ages. At 180 dpp, GCNA is clearly present throughout all tubules of a wild-type control testis (Fig. 5G). GCNA-positive cells were also present in almost all completely deficient tubules at 180 dpp (Fig. 5H); however, at 365 dpp, GCNA negative, completely deficient tubules were present (Fig. 5I).
Fig. 5.
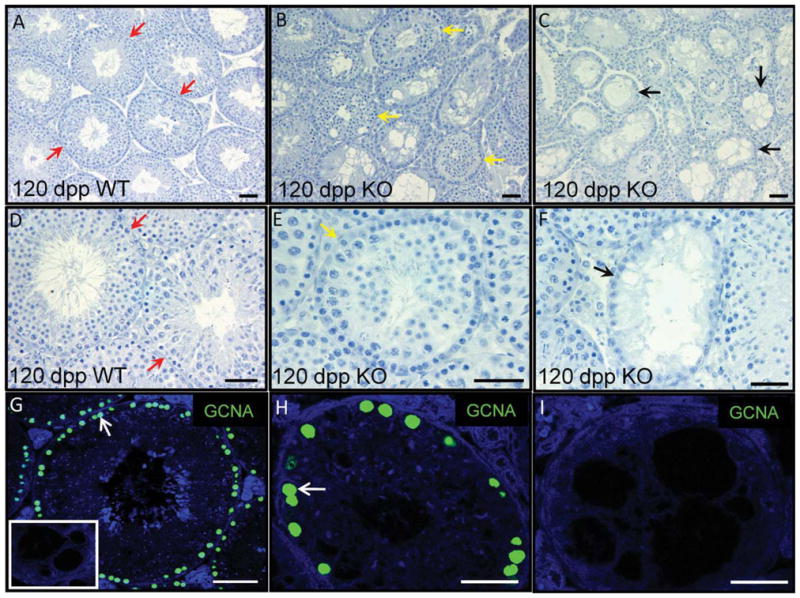
Egr4-null adult testis tubules regress with age. A–F: Hematoxylin stained sections of a 120 days post partum (dpp) wild-type (A,D) and Egr4-deficient (B,C,E,F) mouse testes. Red arrows denote normal tubules (A,D), yellow denotes partially regressed (B,E), and black denotes completely regressed tubules (C,F). G–I: Immunofluorescence localizing GCNA (green) to sections of Egr4 wild-type and null testes isolated from 1-year-old mice. A illustrates GCNA positive cells in tubules of a wild-type testis. Panel B depicts an Egr4-deficient testis tubule with multiple GCNA-positive cells at the basement membrane; however, C shows a tubule containing no GCNA-positive cells from the testis of the same animal. No primary antibody control on knockout tissue is shown as an insert in A.
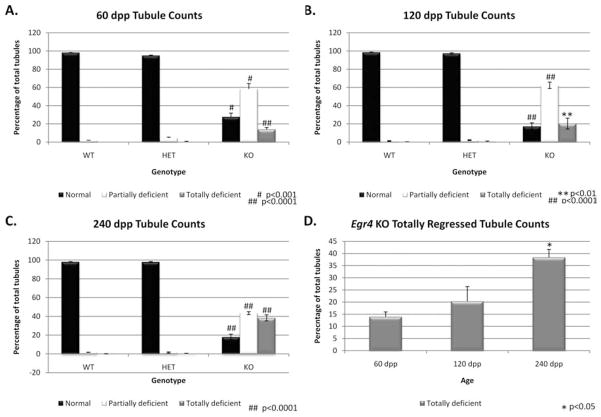
To quantitate the germ cell regression observed in the older Egr4-deficient males, testes were collected from wild-type, heterozygous and knockout males aged 60, 120, and 240 dpp. These testes were fixed, sectioned, and the testis tubules scored as either normal, partially deficient, or completely deficient. For each age, testes were collected from 3 animals of each genotype and at least 200 tubules were scored for each animal. No significant differences were observed between the wild-type and heterozygous animals in any of the tubule scoring categories. At all three ages, the knockout animals contained significantly less normal tubules and significantly more partially and completely deficient tubules (Fig. 6A–C). A comparison of the tubule scoring data only for the knockout animals at all three ages revealed that there were no significant differences between any of the tubule scoring categories at 60 and 120 dpp. At 240 dpp, the numbers of normal tubules were not significantly different from the other two ages; however, there were significantly less partially deficient tubules and significantly more completely deficient tubules at this age.
Fig. 6.
Egr4-deficient mouse testis tubule scoring. A–C: Column graphs depicting the percentage of normal (black), partially regressed (white) and totally regressed (gray) tubules in Egr4 wild-type (WT), heterozygous (HET), and knockout (KO) testes at 60 dpp (A), 120 dpp (B), and 240 dpp (C). D: Column graph depicting only the numbers of totally regressed tubules in Egr4 deficient testes at 60, 120, and 240 dpp. Statistical significance was determined using a Student’s t-test with JMP-7 software and the representative P values are given on each graph.
DISCUSSION
It has been known for 10 years that Egr4 is important for the normal progression of spermatogenesis; however, there remains to be an extensive investigation of its expression and function throughout the first wave of spermatogenesis. Two previous publications (Tourtellotte et al., 1999; Hadziselimovic et al., 2009) provide Egr4 mRNA and protein expression data in the testis for the neonatal and adult mammal and this study now bridges the gap. This study describes, for the first time, a complete Egr4 mRNA and protein expression pattern in the neonatal, juvenile, and adult murine testis and also extends the morphological analysis of the Egr4-deficient mouse testis.
Previous studies have localized Egr4 transcript to primary and secondary spermatocytes and spermatids in the adult mouse testis and to germ and Leydig cells in the neonatal human testis (Tourtellotte et al., 1999; Hadziselimovic et al., 2009). Our data build on these previous expression patterns to provide a complete histological analysis of EGR4 cellular expression and localization in the murine testis. Egr4 mRNA and protein is present throughout the first wave of spermatogenesis, and we have localized EGR4 to germ cells, Sertoli cells, Leydig cells, and PTMs. As the animal ages, Egr4 expression becomes restricted to specific subpopulations of germ cells and the PTMs with protein only detectable in the pre-pachytene spermatocytes and elongated spermatids from 60 dpp. In agreement with the published localization pattern in the human neonatal testis, we detected EGR4 in both germ and Leydig cells in the 5 and 10 dpp mouse testis. However, we also observed EGR4 in Sertoli cells, a cell type that was not positive in the Hadziselimovic study. In addition, our study localized Egr4 transcript to spermatogonia and spermatocytes in the adult mouse testis with no signal detectable in the haploid germ cells or PTMs and, therefore, does not completely agree with the previously published result (Tourtellotte et al., 1999). The most likely explanation for these slight differences in data is the variation which occurs in experiments which rely on fixed tissues. A different fixative has been used in each of the three studies, and so the extent to which transcripts and proteins are preserved and consequently can be detected in each study will be varied. In addition, the observation that EGR4 is present in PTMs but the mRNA was not detected in this cell type in this study could be due to the level of Egr4 transcript being below the level of detection in our system. Nevertheless, taken together, the three studies provide a more complete picture of the expression and localization of EGR4 in the mammalian testis.
The use of knockout tissue is the ideal negative for cellular expression experiments such as in situ hybridization and immunofluorescence. However, Egr4-deficient tissue was not used as the negative control for these studies as the knockout construct only deletes the 3′ region of the transcript, specifically the zinc-finger domain. The Egr4-null mouse expresses a dominant negative form of the protein, lacking the DNA binding domain; hence, both transcript and protein identical to the 5′ region of Egr4 will be present in genotypically knockout tissue. There is a high level of sequence conservation amongst the EGR family, however, EGR4 is distinct from the other three family members as it lacks the 5′ located R1 domain. Additionally, Egr4 contains only one intron, located at the 5′ end; hence, the 5′ region of the Egr4 transcript was the most appropriate area for in situ hybridization probe design.
Our immunofluorescence data illustrate a cell- and intracellular-specific localization patterns for EGR4 in the postnatal testis. A previous study detected EGR4 protein in both the cytoplasm and nucleus of germ cells in the neonatal human testis and our results agree with this observation. EGR4 was also localized throughout the nucleus of germ cells and PTMs, which fits its predicted function as a DNA binding protein. The perinuclear ring of stain seen in neonatal Sertoli cells is intriguing. The colocalization of EGR4 with Mab414 indicates that EGR4 is present either in or at the nuclear envelope in Sertoli cells at 10 dpp. A role for EGR4 outside of regulating transcription by binding to its consensus sequence has never been described, so this localization pattern intimates that EGR4 may serve a different function in these cells. Further investigation of the binding partners of EGR4 in Sertoli cells will be crucial to understanding whether the nuclear envelope localization of this protein affects its function. The observation that EGR4 is also only detected in Sertoli cells which are actively dividing suggests a possible role for this transcription factor in Sertoli cell differentiation. Sertoli cells are mitotically active in the mouse until approximately 15 dpp when they enter G1/G0 and terminally differentiate. Given that EGR4 has been shown to act as a transcriptional repressor (Zipfel et al., 1997), it will be interesting to investigate whether EGR4 is responsible for directly repressing the expression of genes responsible for driving Sertoli cell quiescence.
Our analysis of Egr4-deficient testis morphology presented here extends the previously published data and suggests that EGR4 may regulate multiple events in spermatogenesis. A loss of EGR4 in male mice results in subfertility of these animals due to germ cell apoptosis during early-mid pachytene stage of meiosis (Tourtellotte et al., 1999). EGR4 also appears to be important for the correct progression of spermiogenesis as germ cells which escape the meiotic block in the deficient male display an abnormal morphology. Our expression data support these conclusions as we detect protein in wild-type spermatocytes and elongating spermatids. However, our analysis of this knockout animal also suggests that the reduced fertility over time in these animals is a result of the progressive loss of germ cells. Most Egr4-deficient males were able to sire two to three litters before becoming infertile. Our morphological analysis of an adult testis age series demonstrated that over time, the seminiferous epithelium in these mice could potentially regress to only Sertoli cells and undifferentiated spermatogonia and then could become completely devoid of all germ cells by one year of age. The testes of the Egr4-deficient animals contained significantly more partial and regressed tubules at each of the ages tested and examination of the knockout animals alone illustrates that over time, the numbers of partially and completely deficient tubules in these animals increases. The counting data, combined with our observation that the knockout males become infertile over time, suggest that there may be a loss of SSCs as the Egr4-deficient males age. The conclusion that EGR4 may aid in maintaining the stemness of mammalian SSCs is supported by microarray data generated from cultures of rat SSCs (Hamra et al., 2004). Both Egr4 and Egr3 are transcripts with initially high expression patterns in cultures of rat SSCs but then decrease rapidly as these cells began to differentiate. This observation suggests that a possible redundancy between EGR3 and EGR4 should be investigated and may be the reason why the regression of the seminiferous epithelium in the Egr4-deficient male happens over quite a few months, rather than over several weeks. In addition, the published observation that Egr4 is not detectable in human testes which do not contain Adark spermatogonia also suggests a role for this protein in the development of the stem cell population in the mammalian gonad. Taken together, these studies demonstrate that further investigation into the regulation of SSC differentiation by EGR4 is required.
The observations presented in this study demonstrate that EGR4 displays both a cell- and intracellular-specific localization pattern in the neonatal, juvenile, and adult mouse testis. The progressive loss of germ cells in the testis of the Egr4-deficient male animals suggests a novel role for this protein in maintaining the SSCs population at a level which permits fertility.
EXPERIMENTAL PROCEDURES
Animals and Tissues
All animal experiments were approved by Washington State University Animal Care and Use Committees and were conducted in accordance with the guiding principles for the care and use of research animals of the National Institutes of Health. A BL/6-129 mouse colony was maintained in a temperature- and humidity-controlled environment with food and water provided ad libitum. BL/6-129 mice ranging from birth to adulthood (35–90 dpp) used in these studies were collected from this colony. The animals were killed by decapitation (0 dpp–10 dpp) or asphyxiation before cervical dissociation (10 dpp–adult) and their testes or ovaries dissected. Tissue samples for RNA preparation were snap frozen immediately after collection and stored at −80°C until use. Tissues for in situ hybridization and immunofluorescence were placed in Bouin’s fixative for 5–8 hr immediately after collection, then dehydrated through a graded ethanol series and embedded in paraffin. Sections of tissue 3–5 μm were placed on Superfrost Plus slides (Menzel-Glaser, Braunschweig, Germany).
Real-Time RT-PCR
Total RNA was extracted from mouse testes using the TRIzolTM reagent as per the manufacturer’s instructions (Invitrogen, CA). Reverse transcription was performed to generate cDNA samples using the iScript kit (Bio-Rad, CA) as per the manufacturer’s instructions with 1,000 μg of each total RNA as template. A two-step real-time RT-PCR was used to measure the expression of Egr4 in a testis developmental time course as previously described (Zhou et al., 2005). The RNA samples were analyzed in triplicate with primers specific for the appropriate genes. Egr4 primers amplified a 200 bp product (primers: forward, 5′CTCCACCTGAGCGACTT CTC3′ and reverse, 5′ TCCAGGAAGC AGGAGTCTGT3′) and the control Rps2 primers amplified a 112 bp product (primers: forward, 5′CTGACTCCC GACCTCTGGAAA3′ and reverse, 5′ G AGCCTGGGTCCTCTGAACA3′). Expression of Egr4 was normalized to Rps2 expression and analyzed using the δδCT method with comparisons made to the 0 dpp testis. Data represent mean fold change ± SD. Duplicate testis samples from each age were prepared for this analysis and replicate real time RT-PCR reactions were performed for each sample with consistent results.
In Situ Hybridization
In situ hybridization was used to localize Egr4 transcript on mouse testis and ovary sections as previously described (Hogarth et al., 2006). PCR products were derived from adult mouse testis cDNA using the following primer sets: F: 5′ CTCCACCTGAGC GACTTCTC and R: 5′ TCCAGGAAG CAGGAGTCTGT, ligated into pGEM TEasy (Promega, Madison, WI) and transformed into cells of the DH10β host strain. Positive colonies were identified by means of PCR, verified by sequencing and the plasmids used as PCR templates to derive products using M13 forward and reverse primers (Sigma-Aldrich, T) with the Egr4 probe region flanked by the T7 and Sp6 promoter sequences. These PCR products were used as template for PCR-based production of DIG-cRNAs from the T7 and Sp6 promoters. Hybridization and washing were performed at 55°C. Both antisense and sense (negative control) cRNAs were used on each sample, in every experiment, for each set of conditions tested.
Immunofluorescence
Immunofluorescence with an antibody raised against an EGR4 (Santa Cruz, CA; sc-19868), the nuclear envelope marker Mab414 (Covance, NJ; MMS-120R), or with the germ cell marker GCNA (kindly supplied by Prof. George Enders, University of Kansas Medical Centre) was performed essentially as previously described (Loveland et al., 2006). Antigen retrieval was performed in 0.01 M citrate (pH6; ≥90°C maintained for 5 min), and primary antibody was applied at 0.5–1.0 μg/ml for overnight incubation at room temperature in 0.1% bovine serum albumin/phosphate buffered saline (PBS). For the co-immunofluorescence experiments, both anti-EGR4 and the Mab414 antibody were applied at the same time. Control sections were incubated without primary antibody. Subsequent steps were performed at room temperature, with PBS washes between incubations. EGR4 primary antibody binding was detected using the AlexaFluor 568 donkey anti-goat secondary antibody (Invitrogen, CA; A-11057); the Mab414 and GCNA were visualized using the AlexaFluor 488 goat anti-rabbit secondary antibody (Invitrogen; A-11008). All secondary antibodies were applied to sections for 1 hr and diluted at 1:500 in 0.1% bovine serum albumin/PBS. The sections were mounted under cover-slips using Vectashield with DAPI (4′,6-diamidine-2-phenylidole-dihydrochlor-ide. Vector Laboratories, CA), and cells were analyzed and imaged using a Leica confocal microscope at the Washington State University Imaging facility. Germ and somatic cell types were identified on the basis of their nuclear morphology and position within the developing gonad (Russell et al., 1990). In the postnatal testis, sections from at least three BL/6-129 animals were analyzed for protein localization and each immunofluorescence experiment was repeated with consistent results obtained.
Tubule Quantitation
Cross-sections of testes from Egr4-deficient mice and mice heterozygous for the mutant Egr4 allele, aged 60, 120, and 240 dpp, were stained with Harris Hematoxylin (Sigma-Aldrich) for morphological analysis. For each age, at least 600 tubule cross-sections, minimum of 200 tubules from three different animals, were scored as either wild-type, partially deficient (containing apoptotic nuclei and vacuoles and missing layers of germ cells), or completely deficient (Sertoli cell and undifferentiated spermatogonia only). An unpaired Student’s t-test was used to determine whether there were statistically different numbers of normal, partial and completely regressed tubules at the three ages analyzed.
Acknowledgments
We thank Prof. George Enders for supplying us with the GCNA antibody. This research was supported by a Contraceptive Center Grant U54 42454 and by HD 10808 from NIH.
Grant sponsor: NIH; Grant sponsor: Contraceptive Center; Grant number: U54 42454; Grant number: HD R01 10808
ABBREVIATIONS
- Aal
A aligned spermatogonia
- dpp
days post partum
- Egr
early growth response factor
- E
embryonic day
- LH
luteinizing hormone
- kb
kilobases
- PTM
peritubular myoid cell
- RA
retinoic acid
- SSCs
spermatogonial stem cells
References
- Anderson EL, Baltus AE, Roepers-Gajadien HL, Hassold TJ, de Rooij DG, van Pelt AM, Page DC. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc Natl Acad Sci U S A. 2008;105:14976–14980. doi: 10.1073/pnas.0807297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballow D, Meistrich ML, Matzuk M, Rajkovic A. Sohlh1 is essential for spermatogonial differentiation. Dev Biol. 2006;294:161–167. doi: 10.1016/j.ydbio.2006.02.027. [DOI] [PubMed] [Google Scholar]
- Chavrier P, Lemaire P, Revelant O, Bravo R, Charnay P. Characterization of a mouse multigene family that encodes zinc finger structures. Mol Cell Biol. 1988;8:1319–1326. doi: 10.1128/mcb.8.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christy B, Nathans D. DNA binding site of the growth factor-inducible protein Zif268. Proc Natl Acad Sci U S A. 1989;86:8737–8741. doi: 10.1073/pnas.86.22.8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby SD, Veile RA, Donis-Keller H, Baraban JM, Bhat RV, Simburger KS, Milbrandt J. Neural-specific expression, genomic structure, and chromosomal localization of the gene encoding the zinc-finger transcription factor NGFI-C. Proc Natl Acad Sci U S A. 1992;89:6663. doi: 10.1073/pnas.89.14.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij DG, Russell LD. All you wanted to know about spermatogonia but were afraid to ask. J Androl. 2000;21:776–798. [PubMed] [Google Scholar]
- Gashler A, Sukhatme VP. Early growth response protein 1 (Egr-1): prototype of a zinc-finger family of transcription factors. Prog Nucleic Acid Res Mol Biol. 1995;50:191–224. doi: 10.1016/s0079-6603(08)60815-6. [DOI] [PubMed] [Google Scholar]
- Hadziselimovic F, Hadziselimovic NO, Demougin P, Krey G, Hoecht B, Oakeley EJ. EGR4 is a master gene responsible for fertility in cryptorchidism. Sex Dev. 2009;3:253–263. doi: 10.1159/000249147. [DOI] [PubMed] [Google Scholar]
- Hamra FK, Schultz N, Chapman KM, Grellhesl DM, Cronkhite JT, Hammer RE, Garbers DL. Defining the spermatogonial stem cell. Dev Biol. 2004;269:393–410. doi: 10.1016/j.ydbio.2004.01.027. [DOI] [PubMed] [Google Scholar]
- Hogarth CA, Calanni S, Jans DA, Loveland KL. Importin alpha mRNAs have distinct expression profiles during spermatogenesis. Dev Dyn. 2006;235:253–262. doi: 10.1002/dvdy.20569. [DOI] [PubMed] [Google Scholar]
- Lee SL, Sadovsky Y, Swirnoff AH, Polish JA, Goda P, Gavrilina G, Milbrandt J. Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A (Egr-1) Science. 1996;273:1219–1221. doi: 10.1126/science.273.5279.1219. [DOI] [PubMed] [Google Scholar]
- Lopez-Soler RI, Moir RD, Spann TP, Stick R, Goldman RD. A role for nuclear lamins in nuclear envelope assembly. J Cell Biol. 2001;154:61–70. doi: 10.1083/jcb.200101025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveland KL, Hogarth C, Szczepny A, Prabhu SM, Jans DA. Expression of nuclear transport importins beta 1 and beta 3 is regulated during rodent spermatogenesis. Biol Reprod. 2006;74:67–74. doi: 10.1095/biolreprod.105.042341. [DOI] [PubMed] [Google Scholar]
- Motro B, van der Kooy D, Rossant J, Reith A, Bernstein A. Contiguous patterns of c-kit and steel expression: analysis of mutations at the W and Sl loci. Development. 1991;113:1207–1221. doi: 10.1242/dev.113.4.1207. [DOI] [PubMed] [Google Scholar]
- Nardelli J, Gibson TJ, Vesque C, Charnay P. Base sequence discrimination by zinc-finger DNA-binding domains. Nature. 1991;349:175–178. doi: 10.1038/349175a0. [DOI] [PubMed] [Google Scholar]
- O’Donovan KJ, Tourtellotte WG, Millbrandt J, Baraban JM. The EGR family of transcription-regulatory factors: progress at the interface of molecular and systems neuroscience. Trends Neurosci. 1999;22:167–173. doi: 10.1016/s0166-2236(98)01343-5. [DOI] [PubMed] [Google Scholar]
- Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Raverot G, Weiss J, Park SY, Hurley L, Jameson JL. Sox3 expression in undifferentiated spermatogonia is required for the progression of spermatogenesis. Dev Biol. 2005;283:215–225. doi: 10.1016/j.ydbio.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Russell LD, Ettlin RA, Sinha Hikim AD, Clegg EP. Histological and histo-pathological evaluation of the testis. St. Louis, MO: Cache River Press; 1990. [Google Scholar]
- Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- Small CL, Shima JE, Uzumcu M, Skinner MK, Griswold MD. Profiling gene expression during the differentiation and development of the murine embryonic gonad. Biol Reprod. 2005;72:492–501. doi: 10.1095/biolreprod.104.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svaren J, Sevetson BR, Golda T, Stanton JJ, Swirnoff AH, Milbrandt J. Novel mutants of NAB corepressors enhance activation by Egr transactivators. EMBO J. 1998;17:6010–6019. doi: 10.1093/emboj/17.20.6010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourtellotte WG, Milbrandt J. Sensory ataxia and muscle spindle agenesis in mice lacking the transcription factor Egr3. Nat Genet. 1998;20:87–91. doi: 10.1038/1757. [DOI] [PubMed] [Google Scholar]
- Tourtellotte WG, Nagarajan R, Auyeung A, Mueller C, Milbrandt J. Infertility associated with incomplete spermatogenic arrest and oligozoospermia in Egr4-deficient mice. Development. 1999;126:5061–5071. doi: 10.1242/dev.126.22.5061. [DOI] [PubMed] [Google Scholar]
- Tourtellotte WG, Nagarajan R, Bartke A, Milbrandt J. Functional compensation by Egr4 in Egr1-dependent luteinizing hormone regulation and Leydig cell steroidogenesis. Mol Cell Biol. 2000;20:5261–5268. doi: 10.1128/mcb.20.14.5261-5268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Shima JE, Nie R, Friel PJ, Gris-wold MD. Androgen-regulated transcripts in the neonatal mouse testis as determined through microarray analysis. Biol Reprod. 2005;72:1010–1019. doi: 10.1095/biolreprod.104.035915. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Li Y, Nie R, Friel P, Mitchell D, Evanoff RM, Pouchnik D, Banasik B, McCarrey JR, Small C, Griswold MD. Expression of stimulated by retinoic acid gene 8 (Stra8) and maturation of murine gonocytes and spermatogonia induced by retinoic acid in vitro. Biol Reprod. 2008;78:537–545. doi: 10.1095/biolreprod.107.064337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel PF, Decker EL, Holst C, Skerka C. The human zinc finger protein EGR-4 acts as autoregulatory transcriptional repressor. Biochim Biophys Acta. 1997;1354:134–144. doi: 10.1016/s0167-4781(97)00084-5. [DOI] [PubMed] [Google Scholar]