Abstract
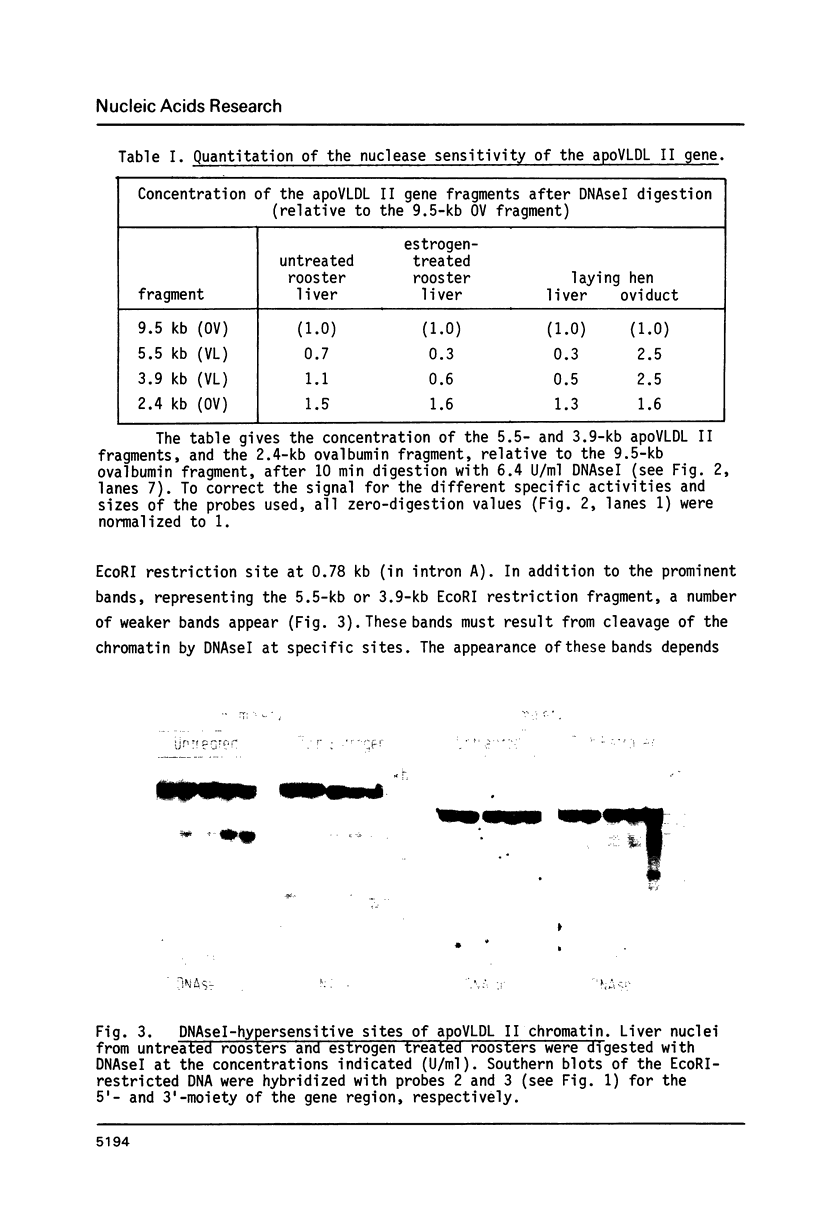
DNAseI-hypersensitive sites were localized in apoVLDL II chromatin from chicken. In the liver two sites at 1.75 and 1.0 kb upstream from the cap-site are present before the gene is activated. After induction by estradiol a number of additional sites appear, three in the promotor region of the gene, one within the coding region and two behind the poly-A signal. These sites disappear when the expression of the gene is shut off upon estradiol withdrawal. All sites appear to be tissue-specific in that they are not found in other tissues of the rooster. However, in oviduct of the laying hen we find a hypersensitive site at 1.6 kb in front of the gene.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson J. N., Vanderbilt J. N., Lawson G. M., Tsai M. J., O'Malley B. W. Chromatin structure of the ovalbumin gene family in the chicken oviduct. Biochemistry. 1983 Jan 4;22(1):21–30. doi: 10.1021/bi00270a004. [DOI] [PubMed] [Google Scholar]
- Balcarek J. M., McMorris F. A. DNase I hypersensitive sites of globin genes of uninduced Friend erythroleukemia cells and changes during induction with dimethyl sulfoxide. J Biol Chem. 1983 Sep 10;258(17):10622–10628. [PubMed] [Google Scholar]
- Becker P., Renkawitz R., Schütz G. Tissue-specific DNaseI hypersensitive sites in the 5'-flanking sequences of the tryptophan oxygenase and the tyrosine aminotransferase genes. EMBO J. 1984 Sep;3(9):2015–2020. doi: 10.1002/j.1460-2075.1984.tb02084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellard M., Kuo M. T., Dretzen G., Chambon P. Differential nuclease sensitivity of the ovalbumin and beta-globin chromatin regions in erythrocytes and oviduct cells of laying hen. Nucleic Acids Res. 1980 Jun 25;8(12):2737–2750. doi: 10.1093/nar/8.12.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgmeyer U., Nowock J., Sippel A. E. The TGGCA-binding protein: a eukaryotic nuclear protein recognizing a symmetrical sequence on double-stranded linear DNA. Nucleic Acids Res. 1984 May 25;12(10):4295–4311. doi: 10.1093/nar/12.10.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan P. N., Olah J., Birnstiel M. L. Major changes in the 5' and 3' chromatin structure of sea urchin histone genes accompany their activation and inactivation in development. Cell. 1983 Jul;33(3):843–848. doi: 10.1016/0092-8674(83)90026-0. [DOI] [PubMed] [Google Scholar]
- Burch J. B. Identification and sequence analysis of the 5' end of the major chicken vitellogenin gene. Nucleic Acids Res. 1984 Jan 25;12(2):1117–1135. doi: 10.1093/nar/12.2.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch J. B., Weintraub H. Temporal order of chromatin structural changes associated with activation of the major chicken vitellogenin gene. Cell. 1983 May;33(1):65–76. doi: 10.1016/0092-8674(83)90335-5. [DOI] [PubMed] [Google Scholar]
- Cartwright I. L., Hertzberg R. P., Dervan P. B., Elgin S. C. Cleavage of chromatin with methidiumpropyl-EDTA . iron(II). Proc Natl Acad Sci U S A. 1983 Jun;80(11):3213–3217. doi: 10.1073/pnas.80.11.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan L., Means A. R., O'Malley B. W. Steroid hormone regulation of specific gene expression. Vitam Horm. 1978;36:259–295. doi: 10.1016/s0083-6729(08)60986-3. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Lomonossoff G. P., Laskey R. A. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 1981 Jun 25;9(12):2659–2673. doi: 10.1093/nar/9.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S. C. DNAase I-hypersensitive sites of chromatin. Cell. 1981 Dec;27(3 Pt 2):413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- Emerson B. M., Felsenfeld G. Specific factor conferring nuclease hypersensitivity at the 5' end of the chicken adult beta-globin gene. Proc Natl Acad Sci U S A. 1984 Jan;81(1):95–99. doi: 10.1073/pnas.81.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritton H. P., Igo-Kemenes T., Nowock J., Strech-Jurk U., Theisen M., Sippel A. E. Alternative sets of DNase I-hypersensitive sites characterize the various functional states of the chicken lysozyme gene. Nature. 1984 Sep 13;311(5982):163–165. doi: 10.1038/311163a0. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Hörz W., Altenburger W. Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res. 1981 Jun 25;9(12):2643–2658. doi: 10.1093/nar/9.12.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A. J., Flavell R. A. A physical map of the DNA regions flanking the rabbit beta-globin gene. Cell. 1977 Oct;12(2):429–439. doi: 10.1016/0092-8674(77)90119-2. [DOI] [PubMed] [Google Scholar]
- Jongstra J., Reudelhuber T. L., Oudet P., Benoist C., Chae C. B., Jeltsch J. M., Mathis D. J., Chambon P. Induction of altered chromatin structures by simian virus 40 enhancer and promoter elements. Nature. 1984 Feb 23;307(5953):708–714. doi: 10.1038/307708a0. [DOI] [PubMed] [Google Scholar]
- Kaye J. S., Bellard M., Dretzen G., Bellard F., Chambon P. A close association between sites of DNase I hypersensitivity and sites of enhanced cleavage by micrococcal nuclease in the 5'-flanking region of the actively transcribed ovalbumin gene. EMBO J. 1984 May;3(5):1137–1144. doi: 10.1002/j.1460-2075.1984.tb01942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene M. A., Corces V., Lowenhaupt K., Elgin S. C. DNase I hypersensitive sites in Drosophila chromatin occur at the 5' ends of regions of transcription. Proc Natl Acad Sci U S A. 1981 Jan;78(1):143–146. doi: 10.1073/pnas.78.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene M. A., Elgin S. C. Micrococcal nuclease as a probe of DNA sequence organization and chromatin structure. Cell. 1981 Nov;27(1 Pt 2):57–64. doi: 10.1016/0092-8674(81)90360-3. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Lawson G. M., Knoll B. J., March C. J., Woo S. L., Tsai M. J., O'Malley B. W. Definition of 5' and 3' structural boundaries of the chromatin domain containing the ovalbumin multigene family. J Biol Chem. 1982 Feb 10;257(3):1501–1507. [PubMed] [Google Scholar]
- Leake R. E., Trench M. E., Barry J. M. Effect of cations on the consideration of hen erythrocyte nuclei and its relation to gene activation. Exp Cell Res. 1972 Mar;71(1):17–26. doi: 10.1016/0014-4827(72)90257-1. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Wood W. I., Dolan M., Engel J. D., Felsenfeld G. A 200 base pair region at the 5' end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981 Nov;27(1 Pt 2):45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- Reeves R. Transcriptionally active chromatin. Biochim Biophys Acta. 1984 Sep 10;782(4):343–393. doi: 10.1016/0167-4781(84)90044-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shermoen A. W., Beckendorf S. K. A complex of interacting DNAase I-hypersensitive sites near the Drosophila glue protein gene, Sgs4. Cell. 1982 Jun;29(2):601–607. doi: 10.1016/0092-8674(82)90176-3. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Hennighausen L., Battey J., Leder P. Chromatin structure and protein binding in the putative regulatory region of the c-myc gene in Burkitt lymphoma. Cell. 1984 Jun;37(2):381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stalder J., Larsen A., Engel J. D., Dolan M., Groudine M., Weintraub H. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell. 1980 Jun;20(2):451–460. doi: 10.1016/0092-8674(80)90631-5. [DOI] [PubMed] [Google Scholar]
- Sweet R. W., Chao M. V., Axel R. The structure of the thymidine kinase gene promoter: nuclease hypersensitivity correlates with expression. Cell. 1982 Dec;31(2 Pt 1):347–353. doi: 10.1016/0092-8674(82)90128-3. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weisbrod S., Weintraub H. Isolation of a subclass of nuclear proteins responsible for conferring a DNase I-sensitive structure on globin chromatin. Proc Natl Acad Sci U S A. 1979 Feb;76(2):630–634. doi: 10.1073/pnas.76.2.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Wu C. Two protein-binding sites in chromatin implicated in the activation of heat-shock genes. Nature. 1984 May 17;309(5965):229–234. doi: 10.1038/309229a0. [DOI] [PubMed] [Google Scholar]
- van den Berg J. A., Kooistra T., Geert A. B., Gruber M. Effect of estradiol on the RNA content and the activity of nucleolar RNA polymerase from rooster liver. Biochem Biophys Res Commun. 1974 Nov 6;61(1):367–374. doi: 10.1016/0006-291x(74)90576-2. [DOI] [PubMed] [Google Scholar]
- van het Schip A. D., Meijlink F. C., Strijker R., Gruber M., van Vliet A. J., van de Klundert J. A., Ab G. The nucleotide sequence of the chicken apo very low density lipoprotein II gene. Nucleic Acids Res. 1983 May 11;11(9):2529–2540. doi: 10.1093/nar/11.9.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]