Abstract
The p14/p19ARF (ARF) tumor suppressor gene is frequently mutated in human cancer. Recently ARF has been shown to localize to mitochondria and to induce autophagy. However the controls that regulate the trafficking of ARF to mitochondria remain unknown. We recently reported that 2-phenylethynesulfonamide (PES) selectively interacts with cytosolic heat shock protein 70 (HSP70) and inhibits its function; we further showed that PE S promotes the death of tumor cells, and that this is associated with an impairment of lysosome function and an inhibition of autophagy. In the present work we used a mass spectrometry-based approach to identify mitochondrial ARF-binding proteins. We report that mitochondrial ARF interacts with HSP70. We show that treatment of cells with PE S blocks the trafficking of ARF to mitochondria, indicating that interaction with HSP70 mediates the mitochondrial localization of ARF. We also show that PE S inhibits the ability of ARF to induce autophagy, supporting the premise that localization to this organelle is critical for ARF-induced autophagy. Finally, we report that cells expressing high levels of ARF are more sensitive to PE S than counterparts with ARF silenced. High levels of ARF are characteristic of tumor cells with enhanced MAP K signaling and advanced stage; therefore, these data support the premise that PE S may show preferential cytotoxicity to advanced stage cancers.
Keywords: ARF, mitochondria, autophagy, HSP70, phenylethynesulfonamide
Introduction
The ARF tumor suppressor (p14ARF in humans, p19ARF in mouse) is expressed at low levels in non-transformed cells, but accumulates in response to oncogenic signals. In murine tumors, high levels of ARF are associated with enhanced MAPK signaling and advanced tumor stage.1,2 Upon accumulation, the majority of ARF localizes to the nucleolus and nucleoplasm, where it binds and inhibits the E3 ubiquitin ligase MDM2, thereby stabilizing p53 protein.3,4 In addition to its p53-dependent tumor suppressor function, ARF also possesses growth suppressive activity that is p53-independent; as one example, enforced expression of ARF is able to inhibit the colony-forming ability of MDM2/p53/ARF-/- cells.5 In addition to its nucleolar localization, approximately 5% of ARF has been found to localize to mitochondria.6 Moreover, a minor fraction of murine ARF is translated from an internal methionine, generating a smaller molecular weight variant (smARF) that lacks the nucleolar localization sequence, and shows preferential localization to mitochondria.7 At the mitochondria, ARF and its internal translational variant have been shown to induce autophagy.6–8
Autophagy is a lysosome mediated self-catabolic pathway that can be used during times of metabolic or other stress to promote cell survival. The sequences that are responsible for mitochondrial localization, and presumably also autophagy induction, are distinct on human and murine ARF, probably reflecting the poor degree of homology of these two proteins. Specifically, highly hydrophobic residues in the N-terminus of human ARF act as a mitochondrial import sequence,9 while a version of murine ARF that lacks the N-terminal domain shows enhanced trafficking to mitochondria.10 Presently unknown are the proteins or controls that regulate mitochondrial trafficking and/or autophagy by ARF. In this study we report that mitochondrial ARF interacts with the major cytosolic stress-induced HSP70 protein (HSP70-A1A, also called HSP72). HSP70 is a molecular chaperone that is expressed at low levels in unstressed non-transformed cells but is highly expressed in tumor cells. In both normal and transformed cells, HSP70 re-folds misfolded proteins, and can direct the trafficking of proteins to organelles such as mitochondria. We report that an HSP70/ARF interaction is detectable in both human and murine cells, and that cells from the HSP70 knockout mouse are defective for the mitochondrial localization of ARF. Further, we show that a small molecule inhibitor of HSP70, termed phenylethynesulfonamide (PES),11 inhibits ARF trafficking to mitochondria, as well as ARF-induced autophagy. These data offer HSP70 as a critical regulator of ARF-mediated autophagy, and reinforce the importance of mitochondrial trafficking of ARF for its autophagy function.
Results
Identification of HSP70 as a mitochondrial ARF-interacting protein.
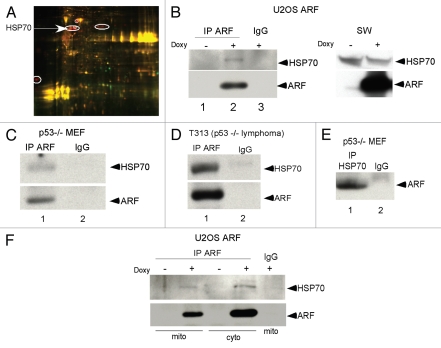
In an attempt to better understand the molecular basis for ARF-induced autophagy, we identified ARF binding partners at the mitochondria using ARF immunoprecipitation followed by 2-Dimensional In-gel Electrophoresis (2D-DIGE). HSP70 was one of the proteins identified as an ARF binding partner using this approach (Fig. 1A). A complete list of ARFinteracting mitochondrial proteins is presented in Table 1. To confirm the interaction between ARF with HSP70 we conducted immunoprecipitation followed by western analysis of extracts obtained from different human and murine cell lines. As shown in Figure 1B, ARF antisera immunoprecipitated HSP70 in U2OS cells containing tetracycline-inducible ARF, as well as in murine p53-/- cells, which express high levels of ARF (Fig. 1B–D). Our quantification of these immunoprecipitations indicates that approximately 30% of total HSP70 appears to complex with ARF in U2OS-ARF cells (data not shown). We also found that HSP70 antisera could specifically immunoprecipitate ARF (Fig. 1E). We next determined whether this interaction exists in purified mitochondria. Mitochondrial and cytoplasmic fractions were isolated on mannitol gradients from inducible U2OS-ARF cells before and after ARF induction with doxycycline, as described.12 Interestingly, we found that ARF antisera could immunoprecipitate the associated HSP70 from both cytosolic as well as mitochondrial extracts (Fig. 1F). The combined data support the premise that endogenous ARF interacts with HSP70, likely at both the mitochondria and in the cytosol.
Figure 1.
ARF interacts with HSP70. (A) Two Dimensional Differential In Gel Electrophoresis (2D-DIGE) of mitochondrial ARF-interacting proteins in mitochondria purified from U2OS-ARF cells. Immunoprecipitates of ARF-induced cells were labeled with CyDye; the circled spots indicate ARF-binding proteins that were excised and sequenced by mass spectrometry. Several spots identified as HSP70 are shown. (B) Co-immunoprecipitation of HSP70 with antisera to ARF (negative control, lane 3) in U2OS-ARF cells treated with doxycycline (Doxy) for 24 h. Normal rabbit IgG is the negative control. IP: immunoprecipitation. On the right, the level of HSP70 and ARF in whole cell lysate from the same samples is depicted. (C) Co-immunoprecipitation of HSP70 with endogenous ARF (negative control IgG in lane 2) in mouse embryo fibroblasts (MEFs) from the p53-/- mouse. (D) Co-immunoprecipitation of HSP70 with endogenous ARF (negative control IgG in lane 2) in a T-cell lymphoma line isolated from the p53-/- mouse (T313). (E) Co-immunoprecipitation of endogenous ARF with HSP70 in p53-/- MEFs. (F) Co-immunoprecipitation of HSP70 with ARF antisera in mitochondrial (Mito) and cytosolic (Cyto) lysates isolated from U2OS-ARF cells treated with doxycycline (doxy) for 24 h.
Table 1.
Mitochondrial ARF-binding proteins
| Mass Spectrometry Results | |||||||
| Top ranked protein name | |||||||
| Accession number | Protein MW | Protein PI | Pep. count | Protein score C. I. % | Total ion score | Total ion C. I. % | |
| Heat shock 70 kDa protein 2 [Homo sapiens] | gi|12804655 | 69977.9 | 5.56 | 12 | 100 | 164 | 100 |
| HYOU1 protein [Homo sapiens] | gi|47938913 | 75296.7 | 5.55 | 13 | 99.969 | ||
| Stress-induced-phosphoprotein 1 | gi|5803181 | 62599.4 | 6.4 | 22 | 100 | 40 | 99.763 |
| Stress-induced-phosphoprotein 1 | gi|5803181 | 62599.4 | 6.4 | 18 | 100 | 27 | 95.802 |
| Chaperonin containing TCP1, subunit 3 isoform c [Homo sapiens] | gi|58761484 | 56395.2 | 6.1 | 16 | 100 | 36 | 99.483 |
| Heat shock 70 kDa protein 9B (mortalin-2) [Homo sapiens] | gi|12653415 | 73681.9 | 6.03 | 11 | 100 | 38 | 99.703 |
| Cationic trypsinogen [Homo sapiens] | gi|11120626 | 9108.8 | 9.24 | 2 | 96.359 | 52 | 99.988 |
| Bcl-xl [Homo sapiens] | gi|510901 | 25086.2 | 4.83 | 3 | 100 | 58 | 99.988 |
| Annexin A2 [Homo sapiens] | gi|18645167 | 38551.8 | 7.57 | 21 | 100 | 372 | 100 |
| Chain C, crystal structure of human P32 [Homo sapiens] | gi|4930075 | 23786.5 | 4.32 | 6 | 100 | 607 | 100 |
Inhibition of HSP70 blocks ARF localization to mitochondria.
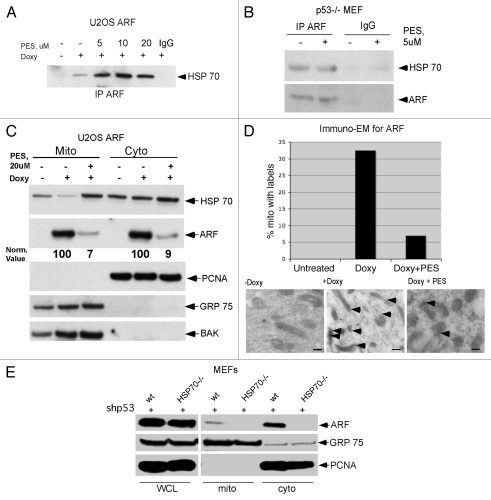
We recently reported that 2-phenylethynesulfonamide (PES) is a selective and specific inhibitor of the major cytosolic, stress-inducible form of HSP70; treatment of cells with PES leads to disruption of the association between HSP70 and some of its co-chaperone proteins. We also showed that PES induces cell death that is associated with an impairment of autophagy.11 To test the hypothesis that interaction with HSP70 plays a role in ARF-dependent autophagy, we inhibited HSP70 function with PES and analyzed the ARF/HSP70 interaction, as well as the ability of ARF to induce autophagy. Toward this end, U2OS-ARF cells and p53-/- mouse embryo fibroblasts (MEFs) were treated with PES and analyzed by IP-western for the ARF-HSP70 complex. As shown in Figure 2, incubation with PES did not disrupt the interaction between HSP70 and ARF (Fig. 2A and B). Indeed, treatment with PES appeared to enhance this interaction, though this was likely because treatment with PES causes increased steady-state levels of HSP70.11 Notably, treatment of cells with PES markedly inhibited the ability of ARF to localize to mitochondria (Fig. 2C). This is consistent with a role for HSP70 in the transport of ARF to mitochondria. To confirm the effect of PES on the trafficking of ARF to mitochondria, we used immuno-electron microscopy to analyze ARF localization. This analysis revealed that treatment of U2OS-ARF cells with PES significantly decreased the amount of ARF immuno-staining gold particles at the mitochondria (Fig. 2D). Again, these data are consistent with the hypothesis that interaction with HSP70 may facilitate the trafficking of ARF to mitochondria. In order to further test this hypothesis, we silenced p53 using a p53 short hairpin (sh-p53) in MEFs from wild-type (wt) or HSP70-/- mice. Because p53 normally transcriptionally represses ARF, its silencing is accompanied by strong upregulation of ARF.13,14 We then assessed the level of total ARF, along with the level of ARF localized to mitochondria. As depicted in Figure 2F, sh-p53-treated cells expressed high levels of ARF in both wt and HSP70-/- MEFs. Interestingly, however, while the absence of HSP70 did not affect the steady-state level of ARF, it markedly decreased the level of ARF associated with mitochondria. The combined data indicate that HSP70 may stabilize cytosolic ARF, and in this way facilitate the localization of ARF to mitochondria.
Figure 2.
HSP70 is required for ARF localization to mitochondria. (A) The HSP70 inhibitor phenylethynesulfonamide (PES) does not block HSP70-ARF complex formation. Co-immunoprecipitation of HSP70 with ARF antisera in U2OS-ARF cells following ARF induction with doxycycline for 24 h in the presence and absence of phenylethynesulfonamide (PES) at the indicated concentrations (5, 10, 20 uM). IgG is the negative control. IP: immunoprecipitation. (B) Co-immunoprecipitation of HSP70 with ARF from p53-/- MEFs following 24 h of treatment with 5 uM PE S. (C) Protein gel analysis using antisera for the proteins indicated of cytosolic (Cyto) and mitochondrial (Mito) extracts isolated from U2OS-ARF cells, treated or untreated for 24 h with doxycycline (Doxy, 100 ng/ml) and/or PE S (20 uM). The normalized values (norm. value) for ARF in the cytosol and mitochondria were assessed by densitometry and normalized to the BAK and PCNA controls. In the bottom part, 20 ug of mitochondrial and cytosolic extract were probed for GRP75 and BAK, which are mitochondrial proteins, and PCNA, which is cytosolic and nuclear, to assess the integrity of mitochondrial purification. (D) Immuno-electron microscopy using ARF antisera and Protein G-Gold was assessed for the percent of mitochondria with three or more gold labels in the mitochondria of U2OS-ARF cells following 24 h of treatment with doxycycline and/or PE S (20 uM). The values shown are averaged from three independent experiments. In the lower part, immuno-electron microscopy using ARF antisera followed by protein G-Gold is depicted in U2OS-ARF cells following 24 h of treatment with doxycycline and/or PE S (20 uM). The arrows indicate gold particles localizing with mitochondria. Scale bars are 500 nm. (E) Protein gel analysis using antisera for the proteins indicated from extracts isolated from whole cell lysate (WCL), cytosol (Cyto) and mitochondria (Mito) purified from mouse embryo fibroblasts (MEFs) from a wild-type (wt) or HSP70-/- mouse. In order to upregulate ARF, MEFs were infected for 48 h with short hairpin to silence p53 (shp53). Twenty ug of mitochondrial and cytosolic extract were probed for GRP75, which are mitochondrial proteins, and PCNA, which is cytosolic and nuclear, to assess the integrity of mitochondrial purification.
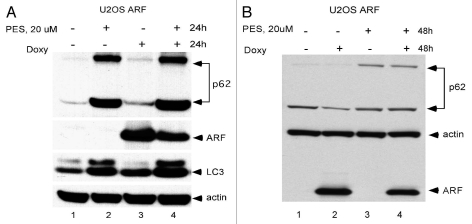
We next chose to determine whether PES could inhibit ARF-mediated autophagy; this would support the contention that mitochondrial localization of ARF is required for autophagy. Toward this end we treated ARF-inducible cells with doxycycline to induce ARF in the presence or absence of incubation with PES, and we assessed autophagy by western analysis for p62SQSTM1, which is degraded by autophagy.15 As depicted in Figure 3A, treatment of U2OS cells with doxycycline and PES for 24 h led to an increase in level of monomeric and oligomeric p62SQSTM1, and in the level of LC3-I, consistent with impaired autophagy. Additionally, 48 h induction of ARF led to reduced levels of monomeric p62SQSTM1, consistent with the induction of autophagy, but concomitant incubation with PES led to an accumulation of p62SQSTM1 (Fig. 3B). These data indicate that inhibition of HSP70 by PES can overcome ARF-mediated autophagy.
Figure 3.
The HSP70 inhibitor phenylethynesolfonamide (PES) blocks ARF-mediated autophagy. (A) Protein gel analysis using antisera for the proteins indicated of whole cell lysate extracts from U2OS ARF cells treated with Doxycycline (Doxy) and/or PE S for 24 h, or left untreated. Shown are the monomeric (bottom) and oligomeric (top band) forms of p62SQSTM1, which accumulate in cells in which autophagy is inhibited, and LC3 (LC3 II), which also accumulates when autophagic flux is inhibited. (B) Protein gel analysis using antisera for the proteins indicated of whole cell lysate extracts from U2OS ARF cells treated with Doxycycline (Doxy) and/or PE S for 48 h, or left untreated. Shown are the monomeric (bottom) and oligomeric (top band) forms of p62SQSTM1, which decrease in autophagic cells, but accumulate in these cells when autophagy is inhibited.
Cell lines that express ARF show increased sensitivity to HSP70 inhibition.
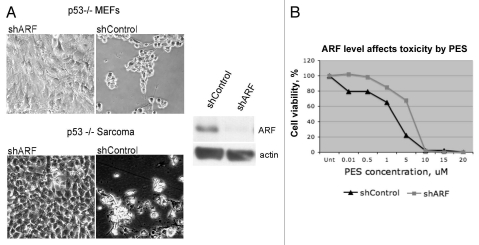
We previously reported that some tumor cell lines utilize ARF-mediated autophagy in order to survive periods of nutrient deprivation.14 These data suggest that cells with high levels of ARF, which would be predicted to utilize the pro-survival functions of ARF-mediated autophagy, might be more sensitive to the autophagy-inhibitory effects of PES. To test this hypothesis, we utilized p53-null MEFs, as well as a sarcoma cell line from the p53-knockout mouse; because loss of p53 de-represses ARF expression,13,14 both of these lines contain high levels of ARF. We infected these cells with a short hairpin to silence ARF (shARF) or a short hairpin control (shControl) in order to create matched cell lines that differ in ARF status. We then tested these matched cell lines for their sensitivity to PES. As shown in Figure 4A, cells infected with shControl were quite sensitive to PES, as judged by the appearance of rounded cells 8 h following PES treatment. In contrast, the same cells in which ARF is silenced appear to be resistant to these effects of PES (Fig. 4A). We then assessed the IC50 for PES in these MEF lines infected with shARF and shControl. As indicated in Figure 4B, cells infected with shARF showed a 3-fold increase in IC50 (2.5–7.5 uM), confirming that cells expressing ARF are more sensitive to the cytotoxic effects of PES.
Figure 4.
ARF is a determinant of PE S-mediated toxicity. (A) Phase-contrast images of MEFs from the p53-/- mouse, as well as a Sarcoma cell line from the p53-/- mouse, infected with shARF to silence ARF or the control retrovirus shControl, in the presence and absence of 5 uM PES after 8 h. Shown on the right is a representative protein gel blot confirming the silencing of ARF in the MEF line. (B) The p53-/- MEF cell line infected with shARF or shControl was treated with the indicated concentrations of PES for 72 h and cell viability was assessed using the sulforhodamine blue assay. Values shown are the averages of three independent experiments, normalized to the viability of the control (DMSO-treated) cells.
Discussion
Several groups have reported that the ARF tumor suppressor can induce autophagy.6,7 However, the controls that mediate the mitochondrial localization of ARF to mitochondria have not been clear. In this report, in order to better elucidate the mechanism underlying this process, we performed 2-dimensional differential in-gel electrophoresis (2D-DIGE) to identify ARF-interacting proteins from purified mitochondria. We identified the major cytosolic heat shock protein 70 (HSP70) as an ARF-interacting protein. HSP70 is known to function in the refolding of misfolded proteins, as well as in the stabilization and trafficking of proteins to mitochondria; indeed, the HSP70 inhibitor we utilized here, PES, was first identified in a chemical screen to identify compounds that inhibit the trafficking of p53 to mitochondria.16 We show that PES abolishes the trafficking of ARF to mitochondria, likely because HSP70 stabilizes cytolsolic ARF, and that it likewise inhibits ARF-mediated autophagy. These data support the relevance of mitochondrial localization and HSP70 function to ARF-mediated autophagy.
We have previously shown that PES is an effective chemotherapeutic agent in a pre-clinical model of murine Burkitt's lymphoma.11 However, the molecular determinants of PES-mediated cytotoxicity (other than HSP70) have not been identified. Herein we show that two cell lines that express high levels ARF are more sensitive to the cytotoxic effects of PES. These data support the contention that high levels of ARF may be a biomarker for treatment success with PES. Recently, ARF was identified as a marker for advanced tumors, as antisera to ARF more frequently stained K-Ras-driven lung adenocarcinomas compared with adenomas.1,2 Therefore, it is possible that PES will prove an effective agent in the most aggressive types of tumors, which bodes well for the use of this compound clinically. Prior to the introduction of PES to clinical use, however, more studies on the efficacy of this compound in pre-clinical models, as well as toxicity studies, need to be done.
Once ARF localizes to the mitochondria, we have shown that it can bind to Bcl-xl and disrupt the Beclin-1/Bcl-xl complex.6 This leads to an increased ability of Beclin-1 to perform its autophagy function(s). In this study, in addition to identifying Bcl-xl, we also identified several stress-induced chaperone proteins, and the protein Annexin A2, as interactors of mitochondrial ARF. Annexin A2 is a calcium-regulated phospholipid binding protein that we have confirmed interacts with mitochondrial ARF and localizes to autophagosomes (Pimkina JS, unpublished data). Therefore, this unbiased proteomic study has set the stage to begin to elucidate the mechanism(s) whereby ARF traffics to mitochondria and initiates autophagy.
Material and Methods
Cell culture, infections.
The p53 null MEF cell line, the p53-/- mouse sarcoma cell line, and the mouse T-cell lymphoma cell line T313 were cultured in DMEM supplemented with 10% fetal bovine serum, 100 units/ml penicillin and 100 µg/ml streptomycin sulfate at 37°C in a humidified atmosphere containing 5% CO2. The U2OS/Tet-On/p19ARF-inducible cell line (U2OS-ARF) was obtained from Pradip Raychaudhuri (University of Illinois at Chicago) cultured in presence of 0.1 µg/ml doxycycline (Sigma) to induce ARF. For retroviral infection with shARF or shControl, Phoenix cells were co-transfected with retroviral vectors and helper plasmid using Fugene 6; supernatants were filtered and used subsequently for retroviral infections as described in reference 17.
Mitochondria isolation, immunoprecipitation-protein gel analysis, 2D-DIGE.
Mitochondria were purified from p53-/- MEF, U2OS ARF cell lines using mannitol gradients as previously described in reference 12. Pellets were placed at −80°C overnight and protein concentration was determined using the DC assay (Bio-Rad Laboratories, Inc., Hercules, CA). Protein gel blotting was performed as described on 100 ug whole cell lysate and 20 ug mitochondrial lysate; antisera used were anti-p19ARF (Abcam), anti-Mdm2 (Ab-1; Ab-2, Calbiochem) anti-actin (AC15, Sigma), anti-GRP75 (C19, Santa Cruz), anti-PCNA (PC10, Santa Cruz), anti-Bak (NT, Upstate). For IP-westerns equivalent amounts of protein (500 µg total cellular or 100 µg mitochondrial fraction) were immunoprecipitated with 1 µg of anti-p19ARF (Abcam) or HSP70 (Cell Signaling). Two dimensional differential in-gel electrophoresis (2D-DIGE) was performed by Applied Biomics (Hayward, CA); briefly, 1,000 ug of purified mitochondria was lysed by freeze-thaw in Nonidet P-40 buffer and immunoprecipitated with 1 ug of antip19ARF antibody (GeneTex) from induced and uninduced U2OS-ARF cells. These samples were covalently linked to CyDye and run with 100 ug of total mitochondrial lysate from induced cells on first dimension isoelectric focusing and second dimension SDS-PAGE. Image analysis was performed using DeCyder software and spots were picked and analyzed by mass spectrometry (MALDI/TOF/TOF).
Electron microscopy.
Cells were trypsinized and prepared for cryosectioning. In brief, cell pellets were fixed in 6% formaldehyde, washed, cryoprotected with 2.1 M sucrose and frozen in liquid nitrogen. Thin sections were cut at −120°C, collected on EM grids and labeled with primary antibody which was subsequently visualized by secondary labeling with colloidal gold conjugated to Protein G. All samples were viewed in a FEI Tecnai 12 TEM operated at 80 kV, the images were recorded using AMT digital camera.
Cell viability (SRB, sulforhodamine B) assay.
Cells (2 × 104) were plated into 96 well plates in 200 ul media, allowed to attach overnight and treated with PES (10 uM) for 24 h. After a 72-h incubation period, the cells were fixed with 100 ul cold 10% TCA and incubated for 1 h at 4°C. The plates were then washed four times with slow-running tap water and air-dried at room temperature. 100 ul 0.057% SRB solution was added to the cells and the cells were incubated for 30 min at room temperature. The cells were then quickly rinsed four times with 1% acetic acid to remove any unbound dye. The protein-bound dye was solubilized using 200 ul 10 mM Tris base solution (pH 10.5) per well, and the resulting colored solutions were spectrophotometrically quantified at an absorbance of 540 nm, as described in reference 18.
Acknowledgments
This work was supported by funding from the National Institutes of Health. This project was also funded in part under a grant with the Pennsylvania Department of Health. The Department specifically disclaims responsibility for any analyses, interpretations or conclusions. The authors declare they have no financial interests in this work.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Feldser DM, Kostova KK, Winslow MM, Taylor SE, Cashman C, Whittaker CA, et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature. 2010;468:572–575. doi: 10.1038/nature09535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Junttila MR, Karnezis AN, Garcia D, Madriles F, Kortlever RM, Rostker F, et al. Selective activation of p53-mediated tumour suppression in high-grade tumours. Nature. 2010;468:567–571. doi: 10.1038/nature09526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/S0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/S0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 5.Weber JD, Jeffers JR, Rehg JE, Randle DH, Lozano G, Roussel MF, et al. p53-independent functions of the p19(ARF) tumor suppressor. Genes Dev. 2000;14:2358–2365. doi: 10.1101/gad.827300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pimkina J, Humbey O, Zilfou JT, Jarnik M, Murphy ME. ARF induces autophagy by virtue of interaction with Bcl-xl. J Biol Chem. 2009;284:2803–2810. doi: 10.1074/jbc.M804705200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reef S, Zalckvar E, Shifman O, Bialik S, Sabanay H, Oren M, et al. A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol Cell. 2006;22:463–475. doi: 10.1016/j.molcel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 8.Balaburski GM, Hontz RD, Murphy ME. p53 and ARF: unexpected players in autophagy. Trends Cell Biol. 2010;20:363–369. doi: 10.1016/j.tcb.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Irvine M, Philipsz S, Frausto M, Mijatov B, Gallagher SJ, Fung C, et al. Amino terminal hydrophobic import signals target the p14(ARF) tumor suppressor to the mitochondria. Cell Cycle. 2010;9:829–839. doi: 10.4161/cc.9.4.10785. [DOI] [PubMed] [Google Scholar]
- 10.Itahana K, Zhang Y. Mitochondrial p32 is a critical mediator of ARF-induced apoptosis. Cancer Cell. 2008;13:542–553. doi: 10.1016/j.ccr.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leu JI, Pimkina J, Frank A, Murphy ME, George DL. A small molecule inhibitor of inducible heat shock protein 70. Mol Cell. 2009;36:15–27. doi: 10.1016/j.molcel.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pietsch EC, Leu JI, Frank A, Dumont P, George DL, Murphy ME. The tetramerization domain of p53 is required for efficient BAK oligomerization. Cancer Biol Ther. 2007;6:1576–1583. doi: 10.4161/cbt.6.10.4719. [DOI] [PubMed] [Google Scholar]
- 13.Robertson KD, Jones PA. The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and downregulated by wild-type p53. Mol Cell Biol. 1998;18:6457–6473. doi: 10.1128/mcb.18.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humbey O, Pimkina J, Zilfou JT, Jarnik M, Dominguez-Brauer C, Burgess DJ, et al. The ARF tumor suppressor can promote the progression of some tumors. Cancer Res. 2008;68:9608–9613. doi: 10.1158/0008-5472.CAN-08-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strom E, Sathe S, Komarov PG, Chernova OB, Pavlovska I, Shyshynova I, et al. Small-molecule inhibitor of p53 binding to mitochondria protects mice from gamma radiation. Nat Chem Biol. 2006;2:474–479. doi: 10.1038/nchembio809. [DOI] [PubMed] [Google Scholar]
- 17.Hemann MT, Zilfou JT, Zhao Z, Burgess DJ, Hannon GJ, Lowe SW. Suppression of tumorigenesis by the p53 target PUMA. Proc Natl Acad Sci USA. 2004;101:9333–9338. doi: 10.1073/pnas.0403286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]