Abstract
Esophageal squamous cell carcinoma (ESCC) is among the top ten most frequent malignancies worldwide. In this study, our objective was to identify potential biomarkers for ESCC through a quantitative proteomic approach using the isobaric tags for relative and absolute quantitation (iTRAQ) approach. We compared the protein expression profiles of ESCC tumor tissues with the corresponding adjacent normal tissue from ten patients. LC-MS/MS analysis of strong cation exchange chromatography fractions was performed on an Accurate Mass QTOF mass spectrometer, which led to the identification of 687 proteins. In all, 257 proteins were identified as differentially expressed in ESCC as compared with normal. We found several previously known protein biomarkers to be upregulated in ESCC including thrombospondin 1 (THBS1), periostin 1 (POSTN) and heat shock 70 kDa protein 9 (HSPA9) confirming the validity of our approach. In addition, several novel proteins that had not been reported previously were identified in our screen. These novel biomarker candidates included prosaposin (PSAP), plectin 1 (PLEC1) and protein disulfide isomerase A 4 (PDIA4) that were further validated to be overexpressed by immunohistochemical labeling using tissue microarrays. The success of our study shows that this mass spectrometric strategy can be applied to cancers in general to develop a panel of candidate biomarkers, which can then be validated by other techniques.
Keywords: esophageal carcinoma, progression, metastasis, mass spectrometry, invasion, early detection
Introduction
Gastrointestinal malignancies are among the frequently occurring tumors in humans. Esophageal cancer has been reported to be the eigth most common malignancy worldwide.1 It can be classified into two histological types: esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma. The incidence rate of ESCC is higher as compared with esophageal adenocarcinoma in developing countries being especially common in certain areas of China, Iran and India.2 The incidence and mortality rates are 2- to 3-fold higher in males than females.3
The carcinogenesis of ESCC is a multifactorial and multistep process. It involves various genetic and environmental factors. A number of risk factors have been associated with the development of ESCC. Alcohol and tobacco are known to be major risk factors. Others include diet deficient in vitamins,4 polyaromatic hydrocarbons in smoked foods and consumption of extremely hot beverages.5 Other predisposing factors for ESCC include Plummer-Vinson syndrome, tylosis6 and lye-ingestion. Surgical resection of tumors is the standard mode of treatment in early stages of ESCC. However, ESCC patients are often diagnosed when the cancer is at an advanced stage when surgical intervention is not an option and chemotherapy as well as radiotherapy are the mainstay in the treatment of the majority of cases.
Dysphagia is the most common symptom in patients with ESCC and appears late in the course of the disease. The lack of early clinical symptoms and poor sensitivity of the molecular markers is a major reason for late diagnosis resulting in poor survival. Over the past several years, attempts have been made to identify molecular markers for the diagnosis of ESCC. In a proteomic study, Zhu et al. showed that galectin 7 is upregulated in ESCC tissues.7 Other proteomic studies performed in ESCC have described various molecules that are differentially regulated as potential markers like pituitary tumor transforming gene (PTTG),8 transglutaminase 3 (TGM) 9 and α actinin 4 (ACTN4).10 Some studies have focused on detecting presence of autoantibodies in ESCC patients and found antibodies in ESCC patient sera that were directed against CDC25B11 and HSP70 among others.12
Many of the mass spectrometry-based proteomic studies have been employed to characterize cancer biomarkers.13 Labeling methods allow for multiplexing and relative quantitation of proteins based on the addition of chemical mass tags onto proteins. These labeling strategies have no effect on the analytical or biochemical properties of the labeled proteins or peptides. A number of chemical labeling strategies have been employed which include Isotope Coded Affinity Tags (ICAT) and Isobaric Tags for Relative and Absolute Quantification (iTRAQ). The iTRAQ method has been used as a quantitative approach for identification of biomarkers for a number of cancers including oral cancer,14 renal cell carcinoma,15 breast cancer,16 hepatocellular carcinoma,17 lung cancer,18 endometrial carcinoma19 and nasopharyngeal carcinoma.20 Quantitative proteomics approaches have been used to study the ESCC proteome.21 However, in these studies, only a limited number of proteins were identified. An analysis of the literature clearly indicates that there is a need for more in-depth proteomics to explore the ESCC proteome further for biomarker discovery.
Here, we describe the use of an iTRAQ labeling strategy for quantitative proteomics using an Accurate Mass QTOF mass spectrometer to compare the protein expression profiles of pooled ESCC samples to pooled matched adjacent normal epithelium. Our study led to the identification of known as well as previously unreported molecules as candidate biomarkers for ESCC proving the utility of multiplexing methods such as iTRAQ for cancer biomarker discovery.
Results and Discussion
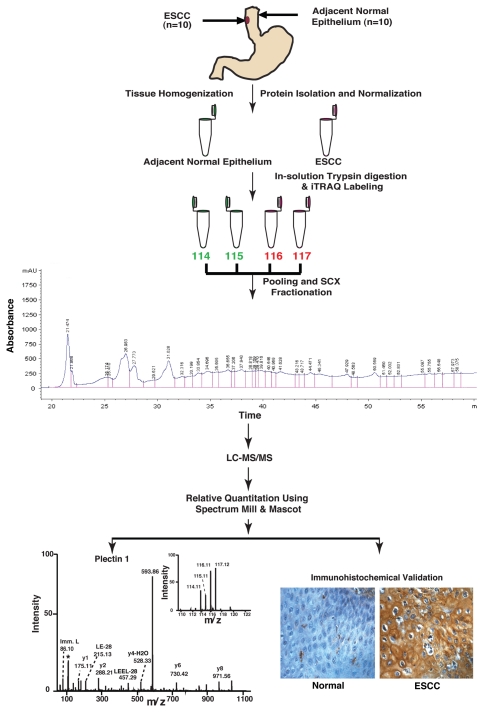
To identify candidate biomarkers for ESCC, a quantitative proteomic analysis was performed using iTRAQ-based in vitro labeling strategy followed by LC-MS/MS analysis. Esophageal squamous cell carcinoma tissue and matched adjacent normal epithelia from 10 patients were used in this study. The workflow employed in this study is shown in Figure 1. The details of the samples used in this study along with tumor grades are provided in Table S1.
Figure 1.
Work flow for quantitative tissue proteomics using iTRAQ labeling and validation of biomarkers for esophageal squamous cell carcinoma. For iTRAQ labeling, proteins were isolated from ten tumor and adjacent normal tissues. Proteins were subjected to trypsin digestion followed by iTRAQ labeling of peptides. Post labeling of peptides the tumor and adjacent normal derived peptide mixture was pooled and fractionated using strong cation exchange (SCX) chromatography, followed by liquid chromatography tandem mass spectrometry (LC-MS/MS) on a QTOF mass spectrometer. The data was searched using Mascot and Spectrum Mill search engines. Some of the overexpressed proteins that were not previously described (e.g., PLEC1) were validated using IHC labeling using tissue microarrays.
iTRAQ labeling and LC-MS/MS analysis.
Lysates were processed by labeling the adjacent normal epithelia-derived peptides with 114 and 115 iTRAQ reagents and ESCC tissue-derived peptides with 116 and 117 iTRAQ reagents thus providing technical replicates within a single run. The data from a total of 41,151 MS/MS spectra generated by LC-MS/MS analysis of 157 SCX fractions were searched against the human RefSeq database using Spectrum Mill and Mascot search engines. An FDR cut off of 1% was applied to eliminate false positive identifications. This led to the identification of 687 proteins. A list of proteins identified in this study is provided in Table S2.
Quantitative analysis of LC-MS/MS data.
Differentially expressed proteins in ESCC were quantified based on the iTRAQ ratios of peptides identified for these proteins. Quantitative analysis using Spectrum Mill and Mascot search engines led to the identification of 687 proteins. Of these, 147 proteins were upregulated >2-fold in tumor as compared with the adjacent normal epithelia, while 91 proteins were downregulated <2-fold in tumor. The list of the proteins with identified peptides using Spectrum Mill and Mascot are mentioned in Tables S3 and S4 respectively.
Bioinformatics analysis of the data.
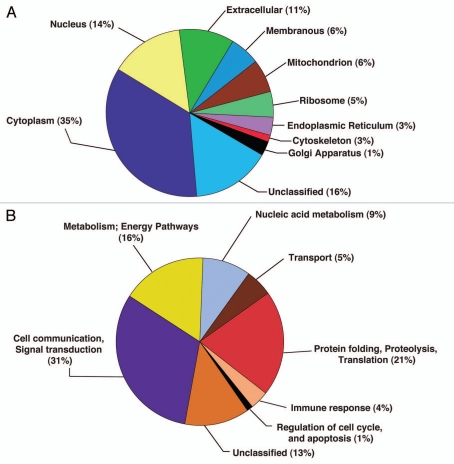
Bioinformatics analysis was performed to classify proteins based on subcellular localization and biological function. We performed classification based on annotations in the Human Protein Reference Database (HPRD; www.hprd.org) in compliance with Gene Ontology (GO) standards. The summary includes fold-changes for differentially expressed proteins in ESCC. It also includes biological domains and motifs obtained from HPRD. The distribution of proteins identified in our study based on subcellular localization is shown in Figure 2A and according to biological processes is shown in Figure 2B. Among the 687 identified proteins, 206 contained signal peptides (SP), 26 contained a transmembrane (TM) domain and 13 contained both a TM domain and a SP motif. The MS and MS/MS spectra of representative known and novel proteins are represented in Figure 3.
Figure 2.
Classification of proteins by gene ontology based on their cellular localization and biological process. (A) Distribution of proteins based on their cellular component using gene ontology classifier. (B) Distribution of proteins based on their biological process using gene ontology classifier.
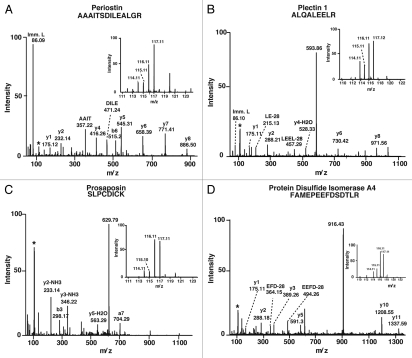
Figure 3.
MS and MS/MS spectra of known and novel upregulated proteins in ESCC tissues as compared with the adjacent normal epithelia. MS and MS/MS spectra of peptide from representative differentially expressed proteins identified in this study. (A) Periostin (POSTN); (B) Plectin 1 (PLEC1); (C) Prosaposin (PSAP); and (D) Protein disulfide isomerase A4 (PDIA4).
Known overexpressed proteins identified in this study. Among the overexpressed proteins in ESCC tissues, we found number of proteins that had been previously described in ESCC, confirming the validity of the quantitative proteomic approach undertaken by us in this study. A partial list of known upregulated proteins is shown in Table 1. Among the proteins previously shown to be upregulated in ESCC were periostin (POSTN),22,23 thrombospondin 1 (THBS1) and fascin 1 (FSCN1). In one of the earlier studies on mRNA profiling of ESCC tissues, periostin was 11-fold upregulated in ESCC.24 In the current quantitative tissue proteomic study periostin was 7-fold upregulated at the protein level in cancer tissue as compared with adjacent normal tissue.
Table 1.
Partial list of overexpressed proteins that were previously reported in esophageal squamous cell carcinoma
| S. no | Gene symbol | Protein | Biological features | Citation | Fold change upregulation in this study | |
| Spectrum Mill | Mascot | |||||
| 1. | THBS1 | Thrombospondin 1 | It is a adhesive glycoprotein that mediates cell-to-cell and cell-to-matrix interactions. | Zhou, et al. (2009)24 | 8.5 | 8.2 |
| 2. | POSTN | Periostin | It is an extracellular matrix (ECM) protein found to be overexpressed in majority of the cases in epithelial and stromal compartments in ESCC. It is involved in EMT and possibly plays an important role in tumor invasion and metastasis. | Kashyap, et al. (2009)22 | 7.6 | - |
| 3. | CTTN | Cortactin | It is reported to be upregulated and overexpressed in early and late stages of ESCC. It is involved in organizing the cell adhesion complex. | Hsu, et al. (2009) | 3.6 | 2.5 |
| 4. | STOML2 | Stomatin like protein 2 | An integral membrane protein involved in tumor progression. It has been shown to be overexpressed in ESCC. | Wang, et al. (2009)26 | 2.9 | - |
| 5. | FSCN1 | Fascin homolog 1 | An actin bundling protein involved in increasing cell motility in various transformed cells. It is overexpressed in ESCC. | Zhang, et al. (2006)25 | 2.5 | 2.8 |
| 6. | FADD | Fas-associated via death domain | Involved in death signaling initiated by TNF receptor. It was found to be overexpressed in ESCC and its corresponding precursor lesions. | Xue, et al. (2006) | 2.7 | - |
| 7. | CALR | Calreticulin | An ER protein. It is a Ca(+2)-binding protein. It's overexpression was correlated with poor prognosis of ESCC patients. | Du, et al. (2009) | 2.6 | 1.3 |
| 8. | HSPA9 | Heat shock 70 kDa protein 9 | HSP70 overexpression has been reported in ESCC and the presence of autoantibodies against HSPA 9. | Fujita, et al. (2008) | 1.77 | 2.5 |
Thrombospondin 1 is an extracellular adhesive glycoprotein that mediates cell-cell contact and is widely expressed. In the current study, thrombospondin 1 was 8-fold upregulated in cancer. Thrombospondin 1 has been previously shown to be overexpressed in ESCC and its overexpression correlated with regional lymph node invasion and poor survival of patients.24 We have identified fascin 1 (FSCN1), an actin bundling protein, which plays diverse roles in cell-cell interaction, cell migration and is involved in increasing cell motility in transformed cells which was 2.5-fold upregulated in ESCC. In one of the previous studies, fascin 1 was shown to be upregulated in ESCC as compared with normal esophageal epithelia.25 Stomatin-like protein 2 (SLP2) is a mitochondrial membrane protein involved in maintaining the mitochondrial membrane potential. Stomatin-like protein 2 affects cell motility and proliferation potentially by effecting the energy cycle within the cell, since it has a control on mitochondrial membrane potential. Stomatin-like protein 2 overexpression has been correlated with tumorigenesis and metastasis. In our study, Stomatin-like protein 2 was 3-fold upregulated in ESCC. Wang et al. showed the correlation of Stomatin-like protein 2 downregulation with inhibition of proliferation and cell motility using wound healing assay and transwell assay in KYSE150 cell line. They also showed by IHC labeling that Stomatin-like protein 2 was overexpressed in ESCC tissues.26
Fas associated via death domain (FADD) is a member of the TNF superfamily which mediates apoptotic signaling via its death domain. It is an adaptor protein that binds to Fas and TNF receptors upon ligand binding, thus mediating downstream apoptotic signaling. In a study by Xue et al. FADD was shown to be overexpressed in ESCC.27 In our study, FADD was found to be 2-fold upregulated in ESCC tissue. Transgelin (TAGLN), also known as SM22, is an actin bundling protein which contains an actin binding region which is made of positively charged amino acids and CLIK domain (C-terminal calponin-like module). It is involved in binding actin filaments and in the process form podosomes. It is known that transgelin dysregulation results in cells becoming more malignant and invasive.28 A proteomic study performed by Qi et al. showed that transgelin was upregulated in ESCC based on a comparative 2DE analysis of ESCC and normal esophageal tissue.29 Our study is in concordance with study performed by Qi et al. as we also observed 2-fold upregulation of transgelin in ESCC.
Novel overexpressed proteins identified in this study. There was a subset of proteins that were observed to be upregulated, which have not been previously described in the context of ESCC. A partial list of these novel and upregulated proteins is shown in Table 2. Among these upregulated proteins, PDIA4, PLEC1 and PSAP, have not been described in the context of ESCC. Cofilin 1 (CFL1) was found to be 3-fold upregulated in ESCC. Among other upregulated proteins in ESCC, ribophorin II (RPN2) was found to be 2-fold upregulated in our study. Ribophorin II is an ER membranous protein involved in N-glycosylation of newly synthesized proteins in ER. Overexpression of RPN2 with respect to lymphovascular invasion is associated with poor survival in gastric cancer.30 Carbonyl reductase 1 (CBR1) and carbonyl reductase 3 (CBR3) are cytoplasmic NADPH-dependent reductase which has been shown to be involved in reduction of a broad range of molecules including quinones, prostaglandins and antitumor agents. Carbonyl reductase 1 was shown to be overexpressed in hepatocellular carcinoma under oxidative stress conditions like hypoxia.31 In the current study, carbonyl reductase 1 and carbonyl reductase 3 were 2-fold upregulated in ESCC. Cofilin 1 is an actin binding protein which is involved in polymerization and depolymerization of actin. It has been shown to be overexpressed in non-small cell lung cancer (NSCLC) and in this study overexpression of cofilin 1 was positively correlated with increased cellular invasiveness and resistance. Hence it was shown to be an effective prognostic and drug resistance marker.32
Table 2.
Partial list of novel proteins identified as overexpressed in esophageal squamous cell carcinoma
| S. no. | Gene symbol | Protein | Features | Fold upregulation | |
| Spectrum Mill | Mascot | ||||
| 1. | ILF3 | Interleukin enhancer binding factor 3 | It is part of heterodimeric transcription factor NFAT (nuclear factor of activated T-cells). It is involved in mRNA binding, stabilization and in export of mRNA to cytoplasm. | 2.17 | 2.36 |
| 2. | CES1 | Carboxylesterase 1 | It is involved in detoxification of xenobiotic and toxic compounds by hydrolysis. It is part of the drug metabolizing machinery in liver and other organs. | 2.37 | 2.3 |
| 3. | CFL1 | Cofilin 1 | It is involved in polymerization and depolymerization of actin. It is shown to be overexpressed in non-small cell lung cancer (NSCLC) and overexpression of cofilin 1 was positively correlated with increased cellular invasiveness. | 2.5 | 3.39 |
| 4. | TAF15 | TBP-associated factor 15 | It is part of the transcription machinery. It is involved in various activities like Promoter recognition and transcription initiation. | 2.33 | 1.95 |
| 5 | PPIB | Peptidylprolyl isomerase B | It is endoplasmic reticulum (E.R.) resident chaperone involved in binding to cyclosporine A. It modulates the immunosuppressive action of cyclosporine. It also protects cells from E.R. stress induced cell death. | 2.13 | 2.18 |
| 6. | NCL | Nucleolin | It is a nucleolus resident phosphoprotein involved in various processes like rRNA transcription, ribosome assembly and transport of ribosome components from nucleus to cytoplasm. | 2.29 | 2.19 |
| 7. | LYN | V-yes-1 Yamaguchi sarcoma viral related oncogene homolog | A protein kinase involved in erythroid differentiation. Reported as potential therapeutic target for breast cancer and involved in epithelial-mesenchymal transition (EMT) in breast cancer. | - | 3.6 |
| 8. | CBR1 | Carbonyl reductase 1 | It catalyzes the reduction of many endogenous and xenobiotic carbonyl compounds. It is shown that during post-treatment of breast cancer cells with chemotherapeutic agents, expression of CBR1 drastically increased possibly for detoxification purpose. | 1.1 | 2.39 |
Downregulated proteins in ESCC. There was a subset of proteins that were downregulated in the context of ESCC. 97 proteins were downregulated ≥2-fold in ESCC tissue as compared with adjacent normal epithelia. Among the downregulated and known proteins, junction plakoglobin (JUP) was found to be 3-fold downregulated in ESCC as compared with normal. We have also identified Cystatin A (CSTA) and was found to be 3-fold downregulated in cancer. Osteoglycin (OGN) is a small protein belonging to the family of proteoglycans, which contains leucine rich repeats (LRR). It is involved in cellular growth, differentiation, cell growth and migration. It has been shown in many studies that downregulation of osteoglycin is related to increased metastasis.33–35 In the current study, osteoglycin was 3-fold downregulated in ESCC tissue as compared with adjacent normal epithelia.
Annexin 1 (ANXA1) is a calcium dependent phosholipid binding protein, phospholipids are precursor for arachidonic acid which are in turn used in biosynthesis of prostaglandins and leukotrienes known proinflammatory molecules. Annexin 1 is localized to the cytoplasmic face of the plasma membrane, where they bind to phospholipids upon stimulation and prevent access to phospholipase A2 (PLA2G4A). Annexin 1 is known to be downregulated in esophageal adenocarcinoma and squamous cell carcinomas,27,36–41 many studies have showed that ANXA1 downregulation correlates with poorly differentiated status of oral squamous cell carcinoma cells. We also found that ANXA1 was 3-fold downregulated in ESCC tissue.
Another molecule found to be downregulated in ESCC was cytokeratin 4 (KRT4), which is a basic cytokeratin usually present as a heterodimer with the acidic cytokeratin 13 (KRT13). It is expressed in differentiated layers of the mucosal and esophageal epithelia and has been shown to be downregulated during progression of ESCC. Xue et al. analyzed cytokeratin 4 in a panel of proteins using tissue microarrays of different stages of tumors varying from precursor lesions to metastasis.27 Downregulation of cytokeratin 4 was observed during progression from dysplasia and carcinoma in situ to metastasis. In our study, cytokeratin 4 was 9-fold downregulated in ESCC. Lastly, our quantitative proteomics study provides validation for several molecules at the protein level that were reported earlier at the mRNA level and in concordance with current study, including, cystatin A (CSTA) and small proline-rich protein 3 (SPRR3).38
Validation of novel candidate biomarkers by immunohistochemical staining.
Some of the candidate proteins identified in our study were further tested by IHC labeling to assess whether they could serve as potential biomarkers for ESCC. The selection of proteins was based on the extent of overexpression, biological importance, involvement in other tumors and availability of commercial antibodies suitable for IHC labeling. We selected plectin 1 (PLEC1), prosaposin (PSAP) and protein disulfide isomerase 4 (PDIA4) for further validation in formalin fixed paraffin embedded tissue sections. All of these proteins were overexpressed at the protein level with ≥2-fold change in ESCC as compared with the adjacent normal epithelium (see Table 2), and were validated using IHC labeling. A statistical analysis was performed to determine the significance of differential expression of PDIA4, PSAP and PLEC1 between ESCC and normal tissues using a Chi-Square test. There was a statistically significant (p < 0.05) difference in the expression of PDIA4, PLEC1 and PSAP between tumor and normal controls. The IHC staining pattern of these molecules in tumor and normal tissues is summarized in Table 3. The IHC scores for all the ESCC patients for plectin 1, prosaposin and protein disulfide isomerase A 4 are provided in Table S5.
Table 3.
Summary of IHC labeling for the validated molecules—PLEC1, PSAP and PDIA4 in tumor and normal tissue
| S. no. | Staining pattern | PLEC1 | PSAP | PDIA4 |
| 1. | Positive | 84 | 94 | 92 |
| 2. | Negative | 16 | 6 | 8 |
| 3. | Strong | 29 | 50 | 23 |
| 4. | Moderate | 30 | 34 | 45 |
| 5. | Weak | 25 | 10 | 24 |
| 6. | Normals stained | 32 | 23 | 19 |
| 7. | p value | 2.74E-013 | 9.59E-024 | 1.25E-024 |
| 8. | Subcellular location of staining | Cytoplasmic and membranous | Cytoplasmic | Cytoplasmic |
Plectin 1 (PLEC1).
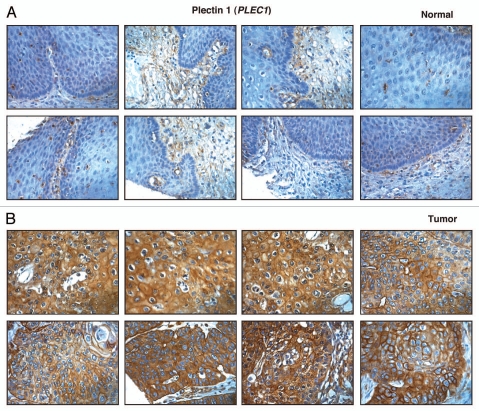
Plectin 1 (PLEC1) is the member of the plakin family. It is a multi-domain protein involved in crosslinking different cytoskeletal proteins and plays an important role in maintaining cell architecture and shape.42 Plectin was originally isolated as an intermediate filament binding protein. It is localized in the basal layer of epidermal cells where it appears to be a component of both hemidesmosomes and desmosomes.43,44 Plectin is also involved in connecting cytokeratins to α6β4 integrin at hemidesmosomes. It has been shown that PLEC1 is involved in microfilament network reorganization during apoptosis since caspase-8 cleaves PLEC1 upon activation during early stages of apoptosis. However this cytoskeletal remodeling is disturbed in cancer cells since most apoptotic pathways are dysregulated in cancer cells leading to accumulation of PLEC1. In one study on pancreatic intraductal papillary mucinous neoplasms (IPMN), it was shown that PLEC1 was upregulated in malignant IPMNs.45 In our study, PLEC1 was 2-fold upregulated in ESCC tissue. Immunohistochemical labeling for PLEC1 showed overexpression of PLEC1 in 84/100 ESCC cases and expression in the majority of cases was cytoplasmic and membranous. The staining pattern of PLEC1 in representative ESCC and normal esophageal tissue is shown in Figure 4.
Figure 4.
Validation of Plectin 1 using immunohistochemical labeling. Representative sections from tissue microarrays stained with anti-Plectin is shown (A) expression of Plectin 1 in representative normal esophageal squamous mucosa and (B) expression of plectin 1 in ESCC.
Prosaposin (PSAP).
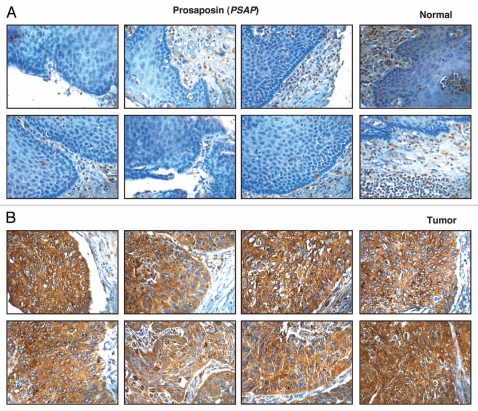
Prosaposin is a lysosomal compartmental protein involved in catabolism of sphingolipids with small sugar chains. It is 526 amino acids long with a molecular weight of 58 kDa and is present as both membrane and secreted forms in the lysosome. It facilitates degradation of sphingolipids by making the lipid substrate more accessible to the soluble degradative enzymes in the lumen of lysosome.46 Prosaposin is a precursor protein which is partially cleaved into structurally related activator proteins saposin A, B, C and D. These saposins are known to play different biological roles. Prosaposin is a known neurotrophic factor47 and prosaposin deficiency or deletion is lethal in human and mice. It has been shown that prosaposin is amplified and overexpressed in number of androgen independent human prostate cancer cell lines (metastatic cell lines-PC-3, DU-145, MDA-PCa 2b, M-12, NCI-H660).48,49 In some of the early studies PSAP has been shown to prevent apoptosis and promote survival in prostate cancer cells50 and PSAP is shown to upregulate androgen receptor (AR), prostate specific antigen (PSA) in prostate cancer cells (LNCaP cells).51 In the process, it acts as androgen-agonist and its growth promoting effect may give a selective growth advantage to these prostate cancer cells, hence in the process it acts as an androgen regulated gene. In our study, PSAP was 4-fold upregulated in ESCC tissue. Immunohistochemical labeling for PSAP showed overexpression of PSAP in 94/100 ESCC cases and expression in the majority of cases was cytoplasmic. The staining pattern of PSAP in representative ESCC and normal esophageal tissue is shown in Figure 5.
Figure 5.
Validation of Prosaposin using immunohistochemical labeling. Representative sections from tissue microarrays stained with anti-prosaposin is shown (A) expression of prosaposin in representative normal esophageal squamous mucosa and (B) expression of prosaposin in ESCC.
Protein disulfide isomerase 4 (PDIA4).
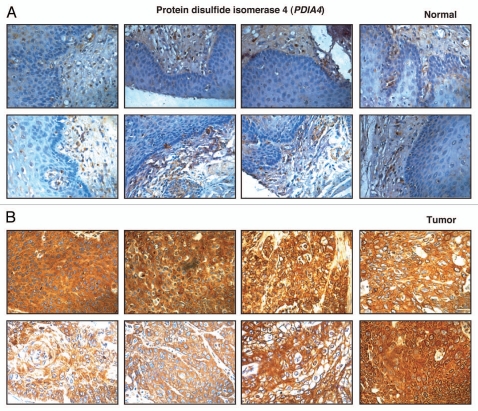
Protein disulfide isomerase 4 is an enzyme resident in the endoplasmic reticulum which is a thiol isomerase involved in facilitating the formation, reduction and rearrangement of disulfide bonds in nascent proteins. The extent of sequence identity between different members of PDI family is relatively low; however they share active-site motifs and domain structure. PDIA4 is 645 amino acids long and has a molecular weight of 72 kDa; thus it is also known as ERP72. Most protein disulfide isomerases have three conserved domains—A, B and C.52,53 Domain A is a catalytically active thioredoxin like domain, domain B is a catalytically inactive thioredoxin like domain and domain C is a highly acidic region which is frequently used to bind to peptides.54 PDIA4 differs from other PDIA family members other in having 3 active thioredoxin domains rather than two with the domain distribution being C-A-A-B-B-A. Like other PDIA family members, PDIA4 is a stress induced protein and hence it is seen in many different cancers especially in tumor induced hypoxic states. In one of our earlier studies where we analyzed the secretome from ESCC and normal esophageal cell lines, PDIA4 along with PDIA3 was shown to be upregulated in the secretome of ESCC cell lines as compared with normal esophageal cell line.55 In this study, PDIA4 was again observed to be 2-fold upregulated in ESCC tissue. Immunohistochemical labeling for PDIA4 showed overexpression of PDIA4 in 92/100 ESCC cases and expression in the majority of cases was cytoplasmic. The staining pattern of PDIA4 in representative ESCC and normal esophageal tissue is shown in Figure 6.
Figure 6.
Validation of protein disulfide isomerase A4 using immunohistochemical labeling. Representative sections from tissue microarrays stained with anti-protein disulfide isomerase A4 as shown (A) expression of protein disulfide isomerase A4 in representative normal esophageal squamous mucosa and (B) expression of protein disulfide isomerase A4 in ESCC.
Public availability and accessibility of proteomic data.
To make our observations publicly available and accessible to other researchers, we have submitted our data on the immunohistochemical analysis and the list of proteins and peptides identified to Human Proteinpedia (HUPA, www.humanproteinpedia. org).56 The raw data described in this study is freely available from ProteomeCommons.org. Online versions of the data may be found at www.proteomecommons.org/dataset.jsp?i=75,354 from the proteomecommons.org main page or alternatively it can be located in the Tranche data repository and can be downloaded through the Tranche server Tranche Hash. Data Set 1-“bxX7cPEuBv9wUkgqq4uCRwWqc415/tGX+X5x 6BNFZRu9tK3ZXhDqZwvOVzWFeChRFIh3O2t7sxptZsNF 1YULs/sEmcoAAAAAAAL3Zg==,” Data Set 2-“KNSTIp1uYntNYp9n9NWDKh5zeU1CcmSapzEL467Tiiys MNyDE2deYGpBJH4Rm8jySKVkFWSBnKwRwOMwH5FRjLNkcusAAAAAAAMjxw==” and Data Set 3 -“cMQvka/5CR93DWSJ/oVeTVNpOo3Wi7UQsnXDanR9 htbnUtbjcDvdajvDCx3BMDX0Ec/vC4TEuQcf5ZqD2g2lgYOOnjoAAAAAAABS0Q==”.
Materials and Methods
Tissue collection and storage.
This study was approved by the institutional review board (IRB) at the Kidwai Memorial Institute of Oncology, Bangalore. The tumor tissues with their adjacent normal epithelium were obtained after surgical resection. All patients were classified as T3N1M0 with the histopathological grade ranging from well to poorly differentiated ESCCs. Informed consent was obtained from all subjects enrolled in the study prior to collection of tissue samples. The normal tissue was at least 5 cm away from the margins of ESCC. All specimens were histologically confirmed by an expert pathologist (RVK).
Protein isolation and iTRAQ labeling.
In this study, we used ESCC tumor tissues samples from ten patients along with the corresponding adjacent normal epithelia. Tissue homogenization and lysate preparation was performed as previously described in reference 17. Briefly, 10 mg of esophageal tissue was homogenized in 0.5% SDS using cell disperser (IKA works, Wilmington, NC) followed by sonication. The cell debris from the tissue homogenates was removed by centrifugation at 14,000 rpm for 30 min at 4°C. The cleared supernatant was transferred into a microfuge tube and the protein concentration determined using the Lowry assay. Equal amounts of lysates (100 µg) from normal and cancer tissue samples were treated with 2 µl of reducing agent [tris(2-carboxyethyl) phosphine (TCEP)] at 60°C for 1 h and alkylated with cysteine blocking reagent, methyl methanethiosulfonate (MMTS) for 10 min at room temperature. The samples were digested overnight with sequencing grade trypsin (Promega, Madison, WI) (1:20) at 37°C and peptides labeled with iTRAQ reagents as follows: pooled adjacent normal tissue derived peptides were labeled with reagents containing reporter ions of m/z 114 and 115; while those from pooled ESCC tumor tissue were labeled with reagents containing reporter ions of m/z 116 and 117. The labeled samples were pooled and mixed with Solvent A (10 mM KH2PO4, 20% acetonitrile, pH 2.8). The diluted samples were manually injected onto a 200 µl bed volume of a strong cation exchange (SCX) column (PolyLC Inc.). Peptides were loaded with a flow rate of 200 µl per min and subsequently washed for 20 min. A gradient of 30 min from 8% Solvent B (350 mM KCl in solvent A) to 50% Solvent B was used for fractionation. Fractions collected were dried and stored at −20°C until LC-MS/MS analysis.
LC-MS/MS analysis.
iTRAQ labeled peptides after SCX fractionation were analyzed using the 6520 Accurate Mass QTOF mass spectrometer (Agilent Technologies, Santa Clara, California, USA) interfaced with an HPLC Chip system (Agilent Technologies, Santa Clara, CA). We used the HPLC Chip system comprising of a 40 nl enrichment column and a 43 mm × 75 µm analytical column, both made up of a reversed-phase material Zorbax 300SB-C18 with a particle size of 5 µm. The samples were loaded on the enrichment column using Agilent 1200 series capillary liquid chromatography system equipped with a micro-well plate autosampler at a flow rate of 3 µl/min using solvent A (0.1% formic acid). An injection flush volume of 4 µl was applied during the enrichment step. The peptides were eluted from the analytical column at the flow rate of 400 nl/min using a gradient of 3–40% solvent B (90% Acetonitrile, 0.1% formic acid) over 30 min. Data dependent acquisition was performed using MassHunter data acquisition software in auto MS/MS mode with precursor survey scans for 1 sec (from 350–1,800 m/z) followed by three MS/MS scans at a rate of 3 spectra/s (50–2,000 m/z). Three most intense ions were chosen for fragmentation with charge state preference in the order of +2, +3 and +4. Dynamic exclusion was set for 2 MS/MS scans of the same precursor ion and released after 2 min.
Data analysis.
The mass spectrometry data was searched using Spectrum Mill (Agilent Technologies, Rev. A.03.03.) and Mascot (Matrix Science Inc., Version 2.2.0). MS/MS data were processed to generate mascot generic format (mgf) files and searched against human RefSeq Build 35 protein sequence database (34,906 sequences) by selecting oxidation of methionine, iTRAQ 4-plex modification at peptide N-terminus and lysine (K) as variable modifications and carbamidomethylation of cysteine as a fixed modification. MS and MS/MS tolerance were set to 100 ppm and 0.1 Da, respectively. Two missed cleavages were allowed. False discovery rate (FDR) was calculated by a decoy database search strategy. Peptide spectrum matches (PSMs) at 1% FDR were used for protein identifications. Relative protein quantitation was performed using Spectrum Mill and Mascot.
Bioinformatics analysis.
Proteins were classified into groups based on their primary subcellular localization, biological process and molecular function. These analyses were performed through the Human Protein Reference Database (HPRD)57 which complies with GO terminologies for functional description of proteins.
Antibodies.
Immunohistochemical staining was performed on formalin fixed paraffin-embedded sections using different antibodies. Of the three antibodies used, one was rabbit monoclonal (anti-PLEC1) and the other two were rabbit polyclonal antibodies. The anti-PDIA4 antibody was used at a dilution of 1:20 (catalog # HPA006140, Human Protein Atlas, Stockholm, Sweden), anti-PSAP antibody at a dilution of 1:250 (catalog # sc-32875, Santa Cruz Biotechnology Inc., Santa Cruz, CA) and anti-PLEC1 antibody at a dilution of 1:100 (catalog # ab32528, Abcam, Cambridge, MA).
Immunohistochemical labeling for validation.
Archived paraffin embedded tissue sections of ESCC samples were obtained from the Department of Pathology, Kidwai Memorial Institute of Oncology, Bangalore. Validation of potential biomarker was performed on a large number of samples using tissue microarrays (TMAs) (n = 100). Tissue microarrays for formalin fixed ESCC from different sources were also used. Commercially available tissue microarrays were obtained from Creative Bioarray (New York) consisting of 60 ESCCs with matched normal esophageal epithelia. The ESCCs varied from tumor grade I to grade III from patients in the age group of 39–79 y. A second group of tissue microarrays obtained from FolioBio (Columbus, OH) consisted of 40 ESCC tissues with matched adjacent normal esophageal squamous epithelium. The ESCC tissues varied from tumor grade I to III from patients in the age group of 43–76 y. The IHC staining was performed as described previously in reference 58. The TMAs were deparaffinized by incubating the slides at 58°C for 2 h. The statistical significance of differential staining was determined using a Chi-square test. Results for expression of PSAP, PLEC1 and PDIA4 were considered statistically significant if the p value was < 0.05. The statistical analysis was performed using R version R-2.13. The scoring of immunohistochemical staining of tissue sections was performed as previously in reference 55.
Conclusions
This is the most extensive quantitative proteomic study in ESCC performed to date. Using an iTRAQ-based quantitative proteomic approach to compare ESCC tissue with its adjacent normal epithelium, we have identified 687 proteins from ESCC tissue which include several proteins reported in previous studies emphasizing their overexpression. We also report many novel proteins which were not previously reported by any other study to be associated with ESCC. The immunohistochemical labeling for PDIA4, PSAP and PLEC1 further support the potential of these molecules to be pursued as biomarkers. Their overexpression in ESCC tissue proteome could lead to overexpression in blood and body fluids of the affected patients. Further testing will be necessary to assess them as biomarkers for early detection of ESCC.
Acknowledgments
We thank the Department of Biotechnology (DBT), Government of India for research support to the Institute of Bioinformatics. T.S.K.P. and R.V.K. are supported by DBT grant (DBT/CSH/GIA/1583/2010–2011). T.S.K.P. is also a recipient of young investigator award from DBT, India. H.G. is a Wellcome Trust/DBT India Alliance Early Career Fellow. H.P., N.A.S. and J.S. are recipients of Senior Research Fellowship from Council of Scientific and Industrial Research (CSIR), Government of India. S.R. is a recipient of Senior Research Fellowship from University Grants Commission (UGC), Government of India. We would also like to thank Drs. S. Krishna Murthy and Vijayalaxmi Deshmane of Department of Surgical Oncology, Kidwai Memorial Institute of Oncology, for providing tissue samples for this study. We would like to thank Drs. S.K. Shankar and Anita Mahadevan of National Institute of Mental Health and Neurological Sciences (NIMHANS), for providing access to the imaging facility. We thank Agilent Technologies for access to instrumentation.
Abbreviations
- ESCC
esophageal squamous cell carcinoma
- PSAP
prosaposin
- PDIA
protein disulfide isomerase
- PLEC1
plectin 1
- POSTN
periostin
- iTRAQ
isobaric tags for relative and absolute quantitation
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Eslick GD. Epidemiology of esophageal cancer. Gastroenterol Clin North Am. 2009;38:17–25. doi: 10.1016/j.gtc.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 3.Holmes RS, Vaughan TL. Epidemiology and pathogenesis of esophageal cancer. Semin Radiat Oncol. 2007;17:2–9. doi: 10.1016/j.semradonc.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Kamangar F, Chow WH, Abnet CC, Dawsey SM. Environmental causes of esophageal cancer. Gastroenterol Clin North Am. 2009;38:27–57. doi: 10.1016/j.gtc.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Islami F, Boffetta P, Ren JS, Pedoeim L, Khatib D, Kamangar F. High-temperature beverages and foods and esophageal cancer risk—a systematic review. Int J Cancer. 2009;125:491–524. doi: 10.1002/ijc.24445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ribeiro U, Jr, Posner MC, Safatle-Ribeiro AV, Reynolds JC. Risk factors for squamous cell carcinoma of the oesophagus. Br J Surg. 1996;83:1174–1185. doi: 10.1002/bjs.1800830905. [DOI] [PubMed] [Google Scholar]
- 7.Zhu X, Ding M, Yu ML, Feng MX, Tan LJ, Zhao FK. Identification of galectin-7 as a potential biomarker for esophageal squamous cell carcinoma by proteomic analysis. BMC Cancer. 2010;10:290. doi: 10.1186/1471-2407-10-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan S, Zhou C, Lou X, Xiao Z, Zhu H, Wang Q, et al. PTTG overexpression promotes lymph node metastasis in human esophageal squamous cell carcinoma. Cancer Res. 2009;69:3283–3290. doi: 10.1158/0008-5472.CAN-08-0367. [DOI] [PubMed] [Google Scholar]
- 9.Uemura N, Nakanishi Y, Kato H, Saito S, Nagino M, Hirohashi S, et al. Transglutaminase 3 as a prognostic biomarker in esophageal cancer revealed by proteomics. Int J Cancer. 2009;124:2106–2115. doi: 10.1002/ijc.24194. [DOI] [PubMed] [Google Scholar]
- 10.Fu L, Qin YR, Xie D, Chow HY, Ngai SM, Kwong DL, et al. Identification of alpha-actinin 4 and 67 kDa laminin receptor as stage-specific markers in esophageal cancer via proteomic approaches. Cancer. 2007;110:2672–2681. doi: 10.1002/cncr.23110. [DOI] [PubMed] [Google Scholar]
- 11.Liu WL, Zhang G, Wang JY, Cao JY, Guo XZ, Xu LH, et al. Proteomics-based identification of autoantibody against CDC25B as a novel serum marker in esophageal squamous cell carcinoma. Biochem Biophys Res Commun. 2008;375:440–445. doi: 10.1016/j.bbrc.2008.08.039. [DOI] [PubMed] [Google Scholar]
- 12.Fujita Y, Nakanishi T, Miyamoto Y, Hiramatsu M, Mabuchi H, Miyamoto A, et al. Proteomics-based identification of autoantibody against heat shock protein 70 as a diagnostic marker in esophageal squamous cell carcinoma. Cancer Lett. 2008;263:280–290. doi: 10.1016/j.canlet.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 13.Chaerkady R, Thuluvath PJ, Kim MS, Nalli A, Vivekanandan P, Simmers J, et al. O labeling for a quantitative proteomic analysis of glycoproteins in hepatocellular carcinoma. Clin Proteomics. 2008;4:137–155. doi: 10.1007/s12014-008-9013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bijian K, Mlynarek AM, Balys RL, Jie S, Xu Y, Hier MP, et al. Serum proteomic approach for the identification of serum biomarkers contributed by oral squamous cell carcinoma and host tissue microenvironment. J Proteome Res. 2009;8:2173–2185. doi: 10.1021/pr800979e. [DOI] [PubMed] [Google Scholar]
- 15.Siu KW, DeSouza LV, Scorilas A, Romaschin AD, Honey RJ, Stewart R, et al. Differential protein expressions in renal cell carcinoma: new biomarker discovery by mass spectrometry. J Proteome Res. 2009;8:3797–3807. doi: 10.1021/pr800389e. [DOI] [PubMed] [Google Scholar]
- 16.Ho J, Kong JW, Choong LY, Loh MC, Toy W, Chong PK, et al. Novel breast cancer metastasis-associated proteins. J Proteome Res. 2009;8:583–594. doi: 10.1021/pr8007368. [DOI] [PubMed] [Google Scholar]
- 17.Chaerkady R, Harsha HC, Nalli A, Gucek M, Vivekanandan P, Akhtar J, et al. A quantitative proteomic approach for identification of potential biomarkers in hepatocellular carcinoma. J Proteome Res. 2008;7:4289–4298. doi: 10.1021/pr800197z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eriksson H, Lengqvist J, Hedlund J, Uhlen K, Orre LM, Bjellqvist B, et al. Quantitative membrane proteomics applying narrow range peptide isoelectric focusing for studies of small cell lung cancer resistance mechanisms. Proteomics. 2008;8:3008–3018. doi: 10.1002/pmic.200800174. [DOI] [PubMed] [Google Scholar]
- 19.DeSouza LV, Grigull J, Ghanny S, Dube V, Romaschin AD, Colgan TJ, et al. Endometrial carcinoma biomarker discovery and verification using differentially tagged clinical samples with multidimensional liquid chromatography and tandem mass spectrometry. Mol Cell Proteomics. 2007;6:1170–1182. doi: 10.1074/mcp.M600378-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Xiao Z, Li G, Chen Y, Li M, Peng F, Li C, et al. Quantitative proteomic analysis of formalin-fixed and paraffin-embedded nasopharyngeal carcinoma using iTRAQ labeling, two-dimensional liquid chromatography and tandem mass spectrometry. J Histochem Cytochem. 2010;58:517–527. doi: 10.1369/jhc.2010.955526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang LY, Ying WT, Mao YS, He HZ, Liu Y, Wang HX, et al. Loss of clusterin both in serum and tissue correlates with the tumorigenesis of esophageal squamous cell carcinoma via proteomics approaches. World J Gastroenterol. 2003;9:650–654. doi: 10.3748/wjg.v9.i4.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kashyap MK, Marimuthu A, Kishore CJ, Peri S, Keerthikumar S, Prasad TS, et al. Genomewide mRNA profiling of esophageal squamous cell carcinoma for identification of cancer biomarkers. Cancer Biol Ther. 2009;8:36–46. doi: 10.4161/cbt.8.1.7090. [DOI] [PubMed] [Google Scholar]
- 23.Michaylira CZ, Wong GS, Miller CG, Gutierrez CM, Nakagawa H, Hammond R, et al. Periostin, a cell adhesion molecule, facilitates invasion in the tumor microenvironment and annotates a novel tumor-invasive signature in esophageal cancer. Cancer Res. 2010;70:5281–5292. doi: 10.1158/0008-5472.CAN-10-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou ZQ, Cao WH, Xie JJ, Lin J, Shen ZY, Zhang QY, et al. Expression and prognostic significance of THBS1, Cyr61 and CTGF in esophageal squamous cell carcinoma. BMC Cancer. 2009;9:291. doi: 10.1186/1471-2407-9-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, Xu L, Xiao D, Xie J, Zeng H, Cai W, et al. Fascin is a potential biomarker for early-stage oesophageal squamous cell carcinoma. J Clin Pathol. 2006;59:958–964. doi: 10.1136/jcp.2005.032730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Cao W, Yu Z, Liu Z. Downregulation of a mitochondria associated protein SLP-2 inhibits tumor cell motility, proliferation and enhances cell sensitivity to chemotherapeutic reagents. Cancer Biol Ther. 2009;8:1651–1658. doi: 10.4161/cbt.8.17.9283. [DOI] [PubMed] [Google Scholar]
- 27.Xue LY, Hu N, Song YM, Zou SM, Shou JZ, Qian LX, et al. Tissue microarray analysis reveals a tight correlation between protein expression pattern and progression of esophageal squamous cell carcinoma. BMC Cancer. 2006;6:296. doi: 10.1186/1471-2407-6-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Assinder SJ, Stanton JA, Prasad PD. Transgelin: an actin-binding protein and tumour suppressor. Int J Biochem Cell Biol. 2009;41:482–486. doi: 10.1016/j.biocel.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 29.Qi Y, Chiu JF, Wang L, Kwong DL, He QY. Comparative proteomic analysis of esophageal squamous cell carcinoma. Proteomics. 2005;5:2960–2971. doi: 10.1002/pmic.200401175. [DOI] [PubMed] [Google Scholar]
- 30.Dicken BJ, Graham K, Hamilton SM, Andrews S, Lai R, Listgarten J, et al. Lymphovascular invasion is associated with poor survival in gastric cancer: an application of gene-expression and tissue array techniques. Ann Surg. 2006;243:64–73. doi: 10.1097/01.sla.0000194087.96582.3e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tak E, Lee S, Lee J, Rashid MA, Kim YW, Park JH, et al. Human carbonyl reductase 1 upregulated by hypoxia renders resistance to apoptosis in hepatocellular carcinoma cells. J Hepatol. 2011;54:328–339. doi: 10.1016/j.jhep.2010.06.045. [DOI] [PubMed] [Google Scholar]
- 32.Castro MA, Dal-Pizzol F, Zdanov S, Soares M, Muller CB, Lopes FM, et al. CFL1 expression levels as a prognostic and drug resistance marker in nonsmall cell lung cancer. Cancer. 2010;116:3645–3655. doi: 10.1002/cncr.25125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui XN, Tang JW, Song B, Wang B, Chen SY, Hou L. High expression of osteoglycin decreases gelatinase activity of murine hepatocarcinoma Hca-F cells. World J Gastroenterol. 2009;15:6117–6122. doi: 10.3748/wjg.15.6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cui X, Song B, Hou L, Wei Z, Tang J. High expression of osteoglycin decreases the metastatic capability of mouse hepatocarcinoma Hca-F cells to lymph nodes. Acta Biochim Biophys Sin (Shanghai) 2008;40:349–355. doi: 10.1111/j.1745-7270.2008.00392.x. [DOI] [PubMed] [Google Scholar]
- 35.Lee JY, Eom EM, Kim DS, Ha-Lee YM, Lee DH. Analysis of gene expression profiles of gastric normal and cancer tissues by SAGE. Genomics. 2003;82:78–85. doi: 10.1016/S0888-7543(03)00098-3. [DOI] [PubMed] [Google Scholar]
- 36.Jazii FR, Najafi Z, Malekzadeh R, Conrads TP, Ziaee AA, Abnet C, et al. Identification of squamous cell carcinoma associated proteins by proteomics and loss of beta tropomyosin expression in esophageal cancer. World J Gastroenterol. 2006;12:7104–7112. doi: 10.3748/wjg.v12.i44.7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu N, Flaig MJ, Su H, Shou JZ, Roth MJ, Li WJ, et al. Comprehensive characterization of annexin I alterations in esophageal squamous cell carcinoma. Clin Cancer Res. 2004;10:6013–6022. doi: 10.1158/1078-0432.CCR-04-0317. [DOI] [PubMed] [Google Scholar]
- 38.Luo A, Kong J, Hu G, Liew CC, Xiong M, Wang X, et al. Discovery of Ca2+-relevant and differentiation-associated genes downregulated in esophageal squamous cell carcinoma using cDNA microarray. Oncogene. 2004;23:1291–1299. doi: 10.1038/sj.onc.1207218. [DOI] [PubMed] [Google Scholar]
- 39.Zhi H, Zhang J, Hu G, Lu J, Wang X, Zhou C, et al. The deregulation of arachidonic acid metabolismrelated genes in human esophageal squamous cell carcinoma. Int J Cancer. 2003;106:327–333. doi: 10.1002/ijc.11225. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Wang HX, Lu N, Mao YS, Liu F, Wang Y, et al. Translocation of annexin I from cellular membrane to the nuclear membrane in human esophageal squamous cell carcinoma. World J Gastroenterol. 2003;9:645–649. doi: 10.3748/wjg.v9.i4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia SH, Hu LP, Hu H, Ying WT, Xu X, Cai Y, et al. Three isoforms of annexin I are preferentially expressed in normal esophageal epithelia but downregulated in esophageal squamous cell carcinomas. Oncogene. 2002;21:6641–6648. doi: 10.1038/sj.onc.1205818. [DOI] [PubMed] [Google Scholar]
- 42.Sonnenberg A, Liem RK. Plakins in development and disease. Exp Cell Res. 2007;313:2189–2203. doi: 10.1016/j.yexcr.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 43.Huber O. Structure and function of desmosomal proteins and their role in development and disease. Cell Mol Life Sci. 2003;60:1872–1890. doi: 10.1007/s00018-003-3050-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfendner E, Rouan F, Uitto J. Progress in epidermolysis bullosa: the phenotypic spectrum of plectin mutations. Exp Dermatol. 2005;14:241–249. doi: 10.1111/j.0906-6705.2005.00324.x. [DOI] [PubMed] [Google Scholar]
- 45.Bausch D, Mino-Kenudson M, Fernandez-Del Castillo C, Warshaw AL, Kelly KA, Thayer SP. Plectin-1 is a biomarker of malignant pancreatic intraductal papillary mucinous neoplasms. J Gastrointest Surg. 2009;13:1948–1954. doi: 10.1007/s11605-009-1001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Brien JS, Kishimoto Y. Saposin proteins: structure, function and role in human lysosomal storage disorders. FASEB J. 1991;5:301–308. doi: 10.1096/fasebj.5.3.2001789. [DOI] [PubMed] [Google Scholar]
- 47.Misasi R, Hozumi I, Inuzuka T, Capozzi A, Mattei V, Kuramoto Y, et al. Biochemistry and neurobiology of prosaposin: a potential therapeutic neuro-effector. Cent Nerv Syst Agents Med Chem. 2009;9:119–131. doi: 10.2174/187152409788452045. [DOI] [PubMed] [Google Scholar]
- 48.Koochekpour S, Zhuang YJ, Beroukhim R, Hsieh CL, Hofer MD, Zhau HE, et al. Amplification and overexpression of prosaposin in prostate cancer. Genes Chromosomes Cancer. 2005;44:351–364. doi: 10.1002/gcc.20249. [DOI] [PubMed] [Google Scholar]
- 49.Lee TJ, Sartor O, Luftig RB, Koochekpour S. Saposin C promotes survival and prevents apoptosis via PI3K/Akt-dependent pathway in prostate cancer cells. Mol Cancer. 2004;3:31. doi: 10.1186/1476-4598-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koochekpour S, Lee TJ, Wang R, Culig Z, Delorme N, Caffey S, et al. Prosaposin upregulates AR and PSA expression and activity in prostate cancer cells (LNCaP) Prostate. 2007;67:178–189. doi: 10.1002/pros.20513. [DOI] [PubMed] [Google Scholar]
- 51.Koochekpour S, Lee TJ, Wang R, Sun Y, Delorme N, Hiraiwa M, et al. Prosaposin is a novel androgen-regulated gene in prostate cancer cell line LNCaP. J Cell Biochem. 2007;101:631–641. doi: 10.1002/jcb.21207. [DOI] [PubMed] [Google Scholar]
- 52.Hogg PJ. Disulfide bonds as switches for protein function. Trends Biochem Sci. 2003;28:210–214. doi: 10.1016/S0968-0004(03)00057-4. [DOI] [PubMed] [Google Scholar]
- 53.Ferrari DM, Soling HD. The protein disulphide-isomerase family: unravelling a string of folds. Biochem J. 1999;339:1–10. doi: 10.1042/0264-6021:3390001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jordan PA, Gibbins JM. Extracellular disulfide exchange and the regulation of cellular function. Antioxid Redox Signal. 2006;8:312–324. doi: 10.1089/ars.2006.8.312. [DOI] [PubMed] [Google Scholar]
- 55.Kashyap MK, Harsha HC, Renuse S, Pawar H, Sahasrabuddhe NA, Kim MS, et al. SILAC-based quantitative proteomic approach to identify potential biomarkers from the esophageal squamous cell carcinoma secretome. Cancer Biol Ther. 2010;10:796–810. doi: 10.4161/cbt.10.8.12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mathivanan S, Ahmed M, Ahn NG, Alexandre H, Amanchy R, Andrews PC, et al. Human Proteinpedia enables sharing of human protein data. Nat Biotechnol. 2008;26:164–167. doi: 10.1038/nbt0208-164. [DOI] [PubMed] [Google Scholar]
- 57.Peri S, Navarro JD, Kristiansen TZ, Amanchy R, Surendranath V, Muthusamy B, et al. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 2004;32:497–501. doi: 10.1093/nar/gkh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kashyap MK, Marimuthu A, Peri S, Kumar GSS, Jacob HK, Prasad TSK, et al. Overexpression of periostin and lumican in esophageal squamous cell carcinoma. Cancers. 2010;2:133–142. doi: 10.3390/cancers2010133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hsu KF, Lin CK, Yu CP, Tzao C, Lee SC, Lee YY, et al. Cortactin, fascin and survivin expression associated with clinicopathological parameters in esophageal squamous cell carcinoma. Dis Esophagus. 2009;22:402–408. doi: 10.1111/j.1442-2050.2008.00921.x. [DOI] [PubMed] [Google Scholar]
- 60.Du XL, Yang H, Liu SG, Luo ML, Hao JJ, Zhang Y, et al. Calreticulin promotes cell motility and enhances resistance to anoikis through STAT3-CTTN-Akt pathway in esophageal squamous cell carcinoma. Oncogene. 2009;28:3714–3722. doi: 10.1038/onc.2009.237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.