Abstract
Epithelial-mesenchymal transition (EMT) is an essential developmental program that becomes reactivated in adult tissues to promote the progression of cancer. EMT has been largely studied by examining the beginning epithelial state or the ending mesenchymal state without studying the intermediate stages. Recent studies using trophoblast stem (TS) cells paused in EMT have defined the molecular and epigenetic mechanisms responsible for modulating the intermediate “metastable” stages of EMT. Targeted inactivation of MAP3K4, knockdown of CBP or overexpression of SNAI1 in TS cells induced similar metastable phenotypes. These TS cells exhibited epigenetic changes in the histone acetylation landscape that cause loss of epithelial maintenance while preserving self-renewal and multipotency. A similar phenotype was found in claudin-low breast cancer cells with properties of EMT and stemness. This intersection between EMT and stemness in TS cells and claudin-low metastatic breast cancer demonstrates the usefulness of developmental EMT systems to understand EMT in cancer.
Key words: EMT, metastable EMT, TS cells, claudin-low breast cancer, EMT and stemness, epigenetics, MAP3K4, CBP, histone acetylation
Introduction to Epithelial-Mesenchymal Transition
Epithelial-mesenchymal transition (EMT) is a morphogenic cellular program, whereby stationary epithelial cells convert to a motile mesenchymal morphology. The initiation and subsequent completion of EMT occurs through the precise coordination of numerous molecular events, including activation of EMTinducing transcription factors, altered expression of cell-surface proteins, reorganization of the actin cytoskeleton and enhanced invasive properties.1 Epithelial cells exhibit an organized apical-basal polarity maintained by the precise arrangement of actin filaments and adhesive structures such as tight junctions, adherens junctions and desmosomes. Specialized adhesive molecules, such as cadherins, integrins and other cell-surface proteins, are essential for maintenance of the epithelial phenotype by stabilizing cell-cell contacts.2 Conversely, mesenchymal cells are characterized by a unique spindle morphology defined by a front-back-end polarity and enhanced invasive potential.3,4 In addition to promoting cellular migration and invasion, the transient phenotypic changes associated with formation of the mesenchymal state during EMT have been associated with the acquisition of stem-like properties.5 This intermediate stage of EMT, coined the metastable phenotype, describes the simultaneous existence of both epithelial and mesenchymal characteristics and is of great importance for understanding the cellular changes associated with progression of the EMT program.6 Due to the tremendous difficulty in capturing cells in the intermediate states of EMT, most studies have focused on the initiation or completion of EMT. Our recent study describes the development of new models using trophoblast stem (TS) cells to examine the intermediate stages of EMT.7 One of these TS cell models was isolated from conceptuses with a point mutation in MAP3K4, a kinase regulating JNK and p38 MAPK pathways, rendering the kinase inactive (TSKI4 cells) and resulting in cells in a metastable state with properties of both stemness and EMT.8 Studies of TSKI4 cells allowed the discovery of epigenetic mechanisms regulating EMT in trophoblasts and highlighted an EMT signature overlapping with the control of EMT in metastatic breast cancer.7
Classifications of EMT
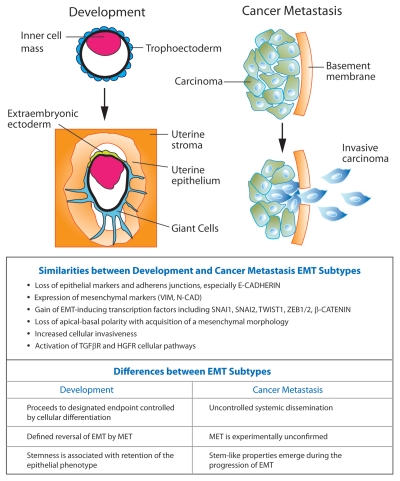
EMT is essential for the proper formation of the body plan and differentiation of many tissues and organs during embryonic development. During organ development, epithelia convert between epithelia and mesenchyme through multiple rounds of EMT and the reversible process mesenchymal-epithelial transition (MET).9 In adult tissue, the EMT program is reactivated during wound healing, organ fibrosis and tumor progression. EMTs are classified into three main categories based upon involvement in different biological processes (Fig. 1).4,10 Developmental EMTs constitute the first EMT subtype and include implantation, gastrulation and neural crest formation. Developmental EMT events occur in a temporally controlled setting and contribute to embryonic morphogenesis and tissue remodeling during development. The second EMT subtype is associated with wound healing and tissue regeneration in adult tissue and occurs in response to inflammation. Persistent inflammatory signals produce continuous activation of EMT resulting in organ fibrosis. The third EMT subtype occurs during cancer progression, whereby EMT in epithelial cancer cells produces cells with increased invasive and metastatic capacity. Although the three subtypes of EMT have seemingly diverse outcomes, a common set of molecular features underlies each EMT event. Critical developmental signaling pathways (i.e., Wnt, Notch, Hedgehog and TGFβ) with demonstrated importance in promoting developmental EMT are reactivated in the generation of EMT-associated pathologies, including organ fibrosis and the metastatic spread of cancer (Fig. 1).2,11–15 Using trophoblast stem (TS) cells as a model system, we will discuss the molecular and epigenetic mechanisms responsible for the first developmental EMT event, implantation and the initiation of placenta formation. Furthermore, we will examine the connections between EMT and stemness and discuss the considerable overlap between signaling pathways that regulate developmental EMT and EMT-associated pathologies.
Figure 1.
Comparison of EMT subtypes in development and cancer metastasis. The first developmental EMT that occurs during implantation of the blastocyst into the uterine epithelium (left part) and EMT associated with the metastatic progression of cancer (right part) are shown. MET indicates mesenchymal-epithelial transition.
The First Developmental EMT Event
Implantation is the first developmental EMT event and is essential for proper placental development necessary to support a developing fetus. Failure of implantation prevents all successive EMT processes, such as gastrulation and neural crest formation. 16 During implantation, the epithelial trophoectoderm of the blastocyst undergoes EMT with changes in cell polarity, motility and adhesiveness (Fig. 1). These changes in cell polarity are accompanied by extension of protrusive structures and loss of expression of the master-regulator of the epithelial phenotype E-cadherin, which are necessary for trophoblast invasion of the uterine epithelium. After implantation, the epithelial polar trophoectoderm overlying the inner cell mass of the blastocyst differentiates into specialized trophoblast lineages to form a functional placenta. Invading trophoblasts generated through differentiation penetrate and remodel the maternal blood vessels, forming the chorionic villi. Considered the functional unit of the placenta, the chorionic villi provide a large surface area for nutrient exchange between mother and developing fetus. Formation of the chorionic villi, requiring extensive blood vessel remodeling, constitutes the second developmental EMT event.17–20 Precise regulation of trophoblast EMT is essential for successful implantation and placentation. Under normal conditions, the trophoblast invasive potential is restricted both temporally and spatially according to trophoblast differentiation signals. Several disease states are resultant from dysregulated trophoblast EMT. Two notable pathologies, preeclampsia and accreta, manifest as clinical outcomes directly linked to aberrant trophoblast invasion.21 Preeclampsia is the result of decreased or absent trophoblast invasion and remodeling of the maternal arteries.22,23 Conversely, placenta accreta is defined as abnormal adherence of the placenta to the uterine epithelium resulting from excessive invasion of hyperinvasive trophoblasts into the uterine wall. Trophoblasts at the maternal-placental interface have demonstrated increased invasive capacity. Rather than being more differentiated and syncytiotrophoblastic in nature, these trophoblasts resemble self-renewing and highly proliferative cytotrophoblasts (i.e., trophoblast stem cells) in the early stages of implantation and placenta development.24,25 Due to the similarity between invasive trophoblasts and malignant cancer cells, it is likely that similar regulatory paradigms persist between these EMT subtypes.26
Molecular Mechanisms of Trophoblast EMT
Trophoblast stem (TS) cells can be isolated from the trophoectoderm of the developing blastocyst and the extraembryonic ectoderm of the E6.5 conceptus.16,27 Self-renewal in TS cells, defined as cell division with the maintenance of multipotency, is maintained in vitro culture conditions by FGF4.28,29 E-cadherin and cortical actin are localized around the periphery of TS cell epithelial colonies and maintain the integrity of the epithelial colonies. Removal of FGF4 induces TS cell differentiation into different trophoblast lineages. Differentiating trophoblasts undergo an EMT involving downregulation of E-cadherin expression, loss of cortical actin with concomitant gain of filamentous actin, and the acquisition of invasiveness. After four days of TS cell differentiation following FGF4 withdrawal, the trophoblast invasive capacity peaks, as demonstrated by Matrigel invasion assays.7,8,30–32 Since TS cell differentiation pathways are essential to EMT processes of implantation and placentation, the in vitro TS cell differentiation model induced by removal of FGF4 is highly useful for interrogating the connections between EMT and TS cell differentiation.
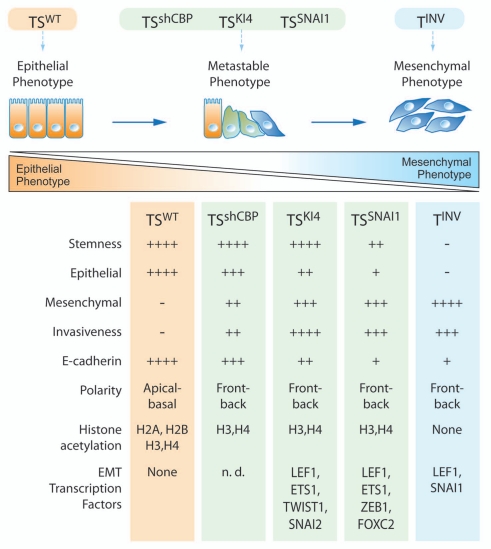
The genetically engineered mouse model with targeted inactivation of MAP3K4 (KI4) exhibits defective decidualization, fetal growth restriction and implantation defects attributable to trophoblast hyperinvasion and dysregulated EMT.33 When cultured in non-differentiating conditions in the presence of FGF4, TSKI4 cells demonstrate a 20-fold increase in invasion compared with wild-type trophoblast stem cells (TSWT cells). In addition, TSKI4 cells cultured in the presence of FGF4 exhibit a more robust invasive capacity than wild-type trophoblasts differentiated for four days (TINV cells) by removal of FGF4. Furthermore, the acquisition of the mesenchymal phenotype in TSKI4 cells is demonstrated by several phenotypic and molecular changes, including loss of epithelial apical-basal polarity and a gain of mesenchymal front-back polarity, reduced levels of peripheral E-cadherin and cortical actin, and acquired expression of mesenchymal markers such as N-cadherin and vimentin. Importantly, TSKI4 cells maintain self-renewal properties as demonstrated by expression of the stem cell markers CDX2 and ESRRβ at levels similar to TSWT cells.7,8,34,35 Maintenance of multipotency is also exhibited by the ability of TSKI4 cells to differentiate into each of the trophoblast lineages. These findings in TSKI4 cells directly contrast with that of terminally differentiated TINV cells that completely lack stemness markers CDX2 and ESRRB and no longer divide. Stemness properties of TSKI4 cells were definitively demonstrated by blastocyst chimera experiments, whereby TSKI4 cells injected into donor blastocysts reconstituted the extraembryonic stem cell compartment and differentiated in vivo into giant cells and cells of the ectoplacental cone.7,8 The simultaneous existence of stemness and EMT properties indicates that TSKI4 cells represent a metastable EMT state (Fig. 2). Metastable EMT is defined as an intermediate stage in the progression of the EMT program, whereby cells express attributes of both epithelial and mesenchymal phenotypes while simultaneously expressing stem cell markers. Due to the transient nature of the metastable phenotype, it is difficult to observe this intermediate stage in the progression of EMT.4,6 From studies with TSKI4 cells, we have shown that the simultaneous maintenance of stemness and the acquisition of EMT properties promote an unrestricted trophoblast invasive potential. During both development and cancer progression, hyperinvasion resulting from uncontrolled EMT signaling events can be detrimental to survival. TSKI4 cells in metastable EMT have proven to be a unique tool to define novel transcriptional and epigenetic reprogramming events and signaling pathways important for induction of the EMT program.
Figure 2.
Characteristics of intermediate stages of EMT in trophoblast stem cells. TSWT and TSKI4 cells were isolated from wild-type or MAP3K4 kinase-inactive conceptuses, respectively. TSshCBP cells are TSWT cells expressing CBP shRNA. TSSNAI1 cells are TSWT cells overexpressing SNAI1. TSWT, TSKI4, TSshCBP and TSSNAI1 cells are cultured under non-differentiating conditions in FGF4. TINV cells are invasive trophoblasts cultured for four days under differentiating conditions and isolated from Matrigel-coated transwell invasion chambers. The + symbol indicates presence of the trait; the − symbol indicates absence of the trait. The quantity of + symbols indicates the relative degree of presence of the trait.
Transcriptional Regulation of Trophoblast EMT
Expression of the adhesive glycoprotein and master-regulator of the epithelial phenotype, E-cadherin, is transcriptionally repressed by several transcription factors of the SNAIL and ZEB superfamilies. In multiple EMT subtypes, SNAI1, SNAI2, ZEB1 and ZEB2 are coordinately upregulated during the initiation and progression of EMT. SNAI1 and SNAI2 are well-characterized transcriptional repressors of E-cadherin and function by binding a canonical E-box consensus sequence within the E-cadherin gene promoter.36–40 Ectopic expression of SNAI1 can induce EMT via direct repression of E-cadherin in several epithelial cell models, including human mammary epithelial cells.5,41,42 As TS cells are differentiated in vitro, via the withdrawal of FGF4, SNAI1 expression gradually increases starting at 4 d of differentiation. The gradual increase in SNAI1 expression coincides with the peak of trophoblast invasion, thereby functionally linking the trophoblast invasive potential to the levels of SNAI1 expression.7 The expression of SNAI1 has also been demonstrated in the invading extravillous trophoblasts in human placenta development, further signifying the importance of SNAI1 to the induction of trophoblast EMT.20,25 Concurrent with the increases in SNAI1 expression upon TS cell differentiation, expression levels of the TGFβ-responsive transcription factor LEF1 exhibit an almost identical pattern of induction. The SNAI1 promoter contains LEF1 binding elements, suggesting that LEF1 and SNAI1 function cooperatively during TGFβ-mediated EMT programs.43 TSKI4 cells with properties of stemness and EMT express elevated levels of the EMT-inducing transcription factors SNAI2, TWIST1 and LEF1, but not SNAI1. This expression pattern contrasts with EMT induced by differentiation where SNAI1 and LEF1 are induced, but not SNAI2 or TWIST1, suggesting that different sets of transcription factors may be used to induce EMT in the context of stemness.
As mentioned above, ectopic expression of SNAI1 has been shown to induce EMT in several cell types. Similarly, in TS cells, expression of SNAI1 (TSSNAI1 cells) robustly induces EMT.5 Characteristic of EMT, TSSNAI1 cells have a loss of E-cadherin expression and exhibit a mesenchymal spindle-like morphology, increased expression of mesenchymal markers N-cadherin and vimentin, and a functional increase in trophoblast invasiveness. Interestingly, TSSNAI1 cells are 66% less invasive than TSKI4 cells. In addition, TSSNAI1 cells are less stem-like than TSKI4 cells, showing a 20–50% decrease in the expression of stemness genes and a reduced growth rate.7 These findings suggest that maintenance of stemness is important for regulating the trophoblast invasive potential during the induction of EMT. From a full panel of seven well-characterized EMT-regulatory transcription factors (Fig. 2), ETS1, LEF1, ZEB1 and FOXC2 were the only EMTregulatory transcription factors significantly induced during the SNAI1-mediated EMT program in TS cells. Expression levels of LEF1 and ZEB1 were induced 50 to 100-fold, indicating that these transcription factors function simultaneously with SNAI1 to execute trophoblast EMT.7 It is possible that members of the EMT-regulatory transcription factor repertoire of SNAI1, LEF1, ZEB1 and ETS1 have different roles in the initiation vs. execution of trophoblast EMT.
Epigenetic Regulation of Trophoblast EMT
Recent studies have demonstrated an increasingly significant role of epigenetic mechanisms for regulation of critical developmental programs important for establishing and maintaining cell lineage fates.7,44,45 The first cell fate decision during development generates two primary lineages of the blastocyst, the trophoectoderm that differentiates into the trophoblast subtypes of the placenta and the inner cell mass that forms the embryo.19,27 The main mechanisms of trophoblast epigenetic regulation include DNA methylation, histone modifications and X-chromosome inactivation.46,47
In contrast to pluripotent embryonic stem cells responsible for the formation of the embryo, DNA of the trophoblast epigenome is hypomethylated. This hypomethylation is critical for implantation and trophoblast differentiation. As trophoblasts differentiate into a more invasive subtype, occurring simultaneously with the progression of EMT, the trophoblast epigenome is continually demethylated. Reduced DNA methylation promotes activation of genetic information critical to trophoblast lineage commitment.47–51 Despite global hypomethylation of the trophoblast lineage, DNA methylation is indispensable for extraembryonic development. For example, the DNA methyltransferase regulatory factor DNMT3L is highly expressed in the epithelial trophoectoderm and DNMT3L-deficient mice exhibit multiple trophoblast defects, including failure to form syncytiotrophoblasts of the placenta.52–54 Furthermore, recent genome-wide sequencing studies have demonstrated that DNA hypomethylation is located primarily in intergenic regions, which raises the question of the importance of DNA methylation for the direct control of genetic information.
Histone modifications comprise the second layer of trophoblast epigenetic regulation. Gene expression levels are controlled by the degree of chromatin compaction, which is mediated by wrapping DNA around an octamer of four core histones H2A, H2B, H3 and H4. The charged tails of histones are often modified by acetylation and methylation marks, which generally contribute to the respective activation or repression of genetic information. The repressive histone modification H3K27me3 conferred by the multi-subunit Polycomb complex has been extensively studied for control of gene expression during differentiation of embryonic lineages. Immunohistochemistry of the mouse blastocyst reveals reduced levels of H3K27me3 in the trophectoderm compared with the inner cell mass.47,55–58 Furthermore, genome-wide sequencing studies demonstrate that few promoters in TS cells are marked by this modification.59 These findings contrast with post-implantation stage extraembryonic tissues, where high gene-specific levels of H3K27me3 are detected. These results suggest that an alternative repressive modification is dominant in TS cells and that the repressive H3K27me3 modification becomes important only after implantation, likely driving later lineage commitment decisions critical to the formation of the placenta. This could explain why mutants of Polycomb complex members, such as EZH2, SUZ12 and EED, demonstrate defects later during development of extraembryonic tissues while not directly affecting TS cells.55,56 Specifically, EED mutants exhibit trophoblast differentiation defects demonstrated by failure to produce secondary invasive trophoblast giant cells, suggesting that H3K27me3 modifications might be important during activation of a specific transcriptional program.57,60 For example, invading trophoblast giant cells play important roles during the EMT events of placentation. Furthermore, the activating H3K4me3 and the repressive H3K9me3 modifications demonstrate importance in TS cell maintenance, but the biological connections to EMT are unclear.47 Further studies with these histone methylation marks could provide insight into the importance of these modifications in specific transcriptional and EMT programs.
Histone acetylation is associated with active gene transcription due to its ability to decondense and open chromatin.61,62 Recent work from our lab has demonstrated that global loss of histone acetylation is a dominant mechanism for the activation of TS cell differentiation programs. As TS cells are differentiated by FGF4 withdrawal, acetylation of all four core histones H2A, H2B, H3 and H4 is globally reduced over a 5-day time-course of differentiation. Occurring in parallel with trophoblast differentiation programs, the induction of trophoblast EMT is also epigenetically regulated by histone acetylation patterns.7 The discovery that trophoblast EMT programs are regulated by epigenetic mechanisms of histone acetylation stemmed from the observation of overlapping phenotypes between KI4 mice and mice deficient for the histone acetyltransferase CBP.33,63,64 KI4 and CBP knockout mice both display skeletal, neural tube and craniofacial defects combined with growth retardation and embryonic lethality. Biochemical assays, including kinase and histone acetylation assays, demonstrate that regulation of CBP acetyltransferase activity is controlled by MAP3K4/JNK-dependent phosphorylation of CBP. Loss of MAP3K4 activity results in diminished CBP histone acetyltransferase activity. Furthermore, loss of CBP activity through either loss of MAP3K4 activity in TSKI4 cells or by shRNA knockdown of CBP in TSshCBP cells results in the selective loss of H2A and H2B acetylation and the induction of a mesenchymal phenotype. Similar to TSKI4 cells, TSshCBP cells exhibit loss of apical-basal polarity and increased expression of mesenchymal markers and invasiveness, while maintaining the expression of stemness genes (Fig. 2). Examination of self-renewing TSKI4 and TSSNAI1 cells with properties of stemness and EMT reveals selective loss of acetylation from histones H2A and H2B, suggesting a specific role for H2A and H2B acetylation in regulation of TS cell EMT (Fig. 2). In support of this conclusion, three separate models of TS cell EMT (TSKI4, TSSNAI1 and TSshCBP cells) exhibit selective inhibition of H2A and H2B acetylation (Fig. 2). For TSKI4 cells, this selective loss of H2A and H2B acetylation occurs independently of changes in H3K4 and H3K9 histone methylation patterns, indicating that changes in histone acetylation but not methylation are responsible for the induction of EMT in TS cells. In addition to the examination of histone acetylation patterns, genome-wide H2BK5Ac ChIP-seq studies coupled with gene expression analysis revealed that genes both significantly downregulated and hypoacetylated in TSKI4 cells clustered into pathways critical for maintenance of the actin cytoskeleton, focal adhesions and the extracellular matrix.7 These results highlight the importance of H2B acetylation for maintenance of the trophoblast epithelial phenotype. To our knowledge, this is the first analysis directly connecting selective histone acetylation marks to the transcriptional activation of the trophoblast EMT program. We hypothesize that loss of H2A and H2B acetylation is an epigenetic signature critical to the induction of EMT metastability. Although this intermediate EMT phenotype is usually transient in nature, TSKI4 and TSshCBP cells are uniquely paused in the metastable EMT state due to perturbation of the upstream modifier CBP and subsequent incomplete modulation of the epigenetic landscape.
Trophoblast EMT and Stemness
Several studies have reported the interrelationship between EMT and stemness. The transcription factor SNAI1 is well characterized for its ability to induce EMT in multiple cell-based systems. Interestingly in mammary epithelial cells, overexpression of SNAI1 or TWIST1 not only induces EMT, but also results in the acquisition of stem cell properties.5,41 Furthermore, TWIST1 promotes EMT while bypassing cellular senescence, connecting EMT to an unrestricted proliferative capacity.65–67 More recently, mesenchymal-epithelial transition (MET), the reverse process of EMT, was shown to occur in parallel with the reprogramming of fibroblasts to induced pluripotent stem cells (iPS).68,69 Similarly in TS cells, the coexistence of EMT and stemness was demonstrated by the comparison of properties between TSKI4 and TSSNAI1 cells. For example, TSKI4 cells are more invasive than TSSNAI1 cells even though TSSNAI1 cells exhibit more profound mesenchymal properties (Fig. 2). TSKI4 cells also have more robust stem cell features, including greater expression of the trophoblast stem cell marker CDX2.7,70 Thus, TSKI4 cells, characterized as self-renewing, multipotent stem cells in a highly invasive intermediate EMT state, highlight the association between stemness properties and metastable EMT. Obviously, such a phenotype has the potential to cause severe disease pathologies. During the pathophysiological progression of placenta accreta, trophoblasts at the maternal-placental interface with increased invasive capacity resemble selfrenewing and highly proliferative TS cells in the early stages of implantation and placenta development. Although the genetic basis for accreta is unknown, the phenotype highlights the ability of stem cells to display EMT properties.25 In addition to TSKI4 cells, EMT metastability has been reported in colorectal cancer cells and progenitor cells of various organs undergoing EMT. During colorectal tumor remodeling and metastasis, immunohistochemistry of colon adenocarcinomas demonstrated the simultaneous retention of branched epithelial structures stabilized by E-cadherin and invading mesenchymal protrusions marked by high levels of β-catenin, SNAI1 and fibronectin expression.71,72 Furthermore, the claudin-low subtype of breast cancer, which is characterized by its EMT features, also exhibits stem-like properties, as demonstrated by overlapping gene expression profiles with breast cancer stem cells identified by the CD44+/CD24-/low antigenic phenotype. Collectively, these studies reveal a connection between the cellular plasticity associated with the induction of EMT and the maintenance of stem-like characteristics.73–78 In the context of EMT and stemness, the importance of identifying TSKI4 cells is that they were the first example of a point mutation targeting the activity of a kinase (MAP3K4) that causes loss of epithelial cell maintenance, while allowing a self-renewing, multipotent stem cell to be in a permanent metastable EMT. The TSKI4 cell phenotype clearly shows that dysregulation of kinase signaling networks can induce a metastable EMT with properties arguably similar to those proposed for the controversial tumor initiating or cancer stem cell.79 These results suggest that characterization of these signaling networks might provide new therapeutic strategies to reverse these EMT-associated pathologies.
Connections between Trophoblast EMT and Tumor Progression
Cancer metastasis is responsible for the majority of cancer related deaths, and EMT is believed to induce the cellular traits associated with the metastatic progression of cancer. In recent years, evidence for the role of EMT in the metastatic progression of cancer has continued to build.80,81 Multiple reports demonstrate that the very same transcription factors (i.e., SNAI1, SNAI2, TWIST1), adhesive structures (i.e., aν-β-integrin family) and signaling pathways (i.e., Tgfβ, Notch, Hedgehog) controlling defined EMT programs are also responsible for regulating tumor progression.15,82 For instance, loss of E-cadherin expression is a hallmark of metastatic carcinoma and expression of the transcriptional repressor of E-cadherin SNAI1 correlates with poor survival rates. Additionally, SNAI1 expression levels are elevated at the invading front of colorectal tumors.36,38,40,83–92 The controversy surrounding the role of EMT in cancer metastasis stems from the difficulty of directly following the progression of EMT during tumor formation. In support of the connection between EMT and cancer metastasis, both proteomic and multiphoton microscopy analysis demonstrate the conversion of circulating mammary tumor cells to the mesenchymal phenotype.93–95 Furthermore, the phenotypic similarity between the primary mammary tumor and the secondary tumor metastasis suggests a role for both EMT and MET in the metastatic progression of cancer.
In 1902 John Beard proposed that many cancers are trophoblastic-like, in part, because their tissue invasive nature is similar to the invasiveness of the trophoblast during placentation.26,96 Today, the origin of cancer stem cells is controversial. It remains unclear if different cancer stem cells arise from multipotent tissue stem cells (similar to TSKI4 or TSSNAI1 cells) or from reprogramming of differentiated cells that revert to a stem cell-like phenotype (similar to primary mammary epithelial cells expressing SNAI1).97–100 To begin addressing this question, we identified an intersecting EMT gene signature from analysis of microarray gene expression data shared by TSKI4 cells developmentally entering EMT and claudin-low breast cancer. Similar to human claudin-low breast cancer cell lines and tumors, TSKI4 cells exhibit an increase in the mesenchymal markers VIM, CDH2, SNAI2 and TWIST1 with loss of the epithelial differentiation and cell adhesion markers CD24, KRT7/8/19 and CLDN4.7 In concordance with the stem cell-like CD44+/CD24-/low and EpCAM-/CD49f+ antigenic phenotypes of breast tumor initiating cells and mammary stem cells, gene expression profiling demonstrated that claudin-low tumors have the lowest expression of epithelial differentiation markers, while exhibiting the highest expression of mesenchymal markers.76 With a few notable exceptions such as SNAI2, TWIST1, VIM and CDH2 (N-cadherin), many of the genes in this EMT signature are uncharacterized with respect to EMT. It is hypothesized that this gene signature defines a novel set of genes with importance in EMT and potentially cancer metastasis. Many of the intersecting genes have a correlative loss of expression and loss of H2BK5Ac in both TSKI4 and claudin-low breast cancer cells.7 Thus, even though claudin-low breast cancer cells do not have the selective loss of histone H2A and H2B acetylation seen in TSKI4 cells, specific genes within this intersecting EMT signature have loss of H2BK5 acetylation. These findings suggest that epigenetic changes in the acetylation landscape of tumor cells in EMT may be important for regulating the metastatic progression of cancer.
Conclusions and Outlook
Unlike cancer metastasis, trophoblast invasive potential is temporally and spatially restricted during normal development. We propose that studies aimed at understanding the regulatory mechanisms restricting EMT and invasiveness of primary epithelial stem cells can provide insight into signaling networks responsible for management of metastatic cancer. Due to the intersecting features of EMT subtypes, a complete picture of this complex cellular program, which is fundamental to development and reactivated in adult tissues during disease progression, can now be studied from multiple biological perspectives. The change in histone acetylation observed with TSKI4, TSshCBP, TSSNAI1 and a subset of intersecting EMT signature genes in claudin-low breast cancer suggests that an “EMT acetylome” may control signaling networks regulating properties of epithelial maintenance and EMT. Defining the acetylome network could potentially reveal a strategy to reverse metastable EMT by stabilizing the non-invasive epithelial phenotype. Significant therapeutic interest in the reversal of EMT is highlighted by the exploration of synthetic molecular compounds to restore E-cadherin expression.101–103 The controlled induction of MET could have benefit for the reversal of several pathophysiological states where there is a dysregulated EMT.
Acknowledgments
G.L.J. is supported by NIH grants GM30324 and DK37871 and the University Cancer Research Fund for support of the deep Sequencing Genomics Facility. N.V.J. is supported by NIH training grant GM007040. We thank Betsy Clarke for graphical work.
Abbreviations
- EMT
epithelial-mesenchymal transition
- MET
mesenchymal-epithelial transition
- TS cells
trophoblast stem cells
- KI4
MAP3K4 kinase-inactive mice
- TSKI4 cells
MAP3K4 kinase-inactive TS cells
- TSWT cells
wild-type TS cells
- TSSNAI1 cells
TS cells overexpressing SNAI1
- TSshCBP cells
TS cells expressing CBP shRNA
- TINV cells
invasive trophoblasts differentiated for four days
- H3K27me3
H3 lysine 27 tri-methylation
- H3K4me3
H3 lysine 4 tri-methylation
- H3K9me3
H3 lysine 9 tri-methylation
- H2BK5Ac
H2B lysine 5 acetylation
References
- 1.Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 3.Hay ED. The mesenchymal cell, its role in the embryo, and the remarkable signaling mechanisms that create it. Dev Dyn. 2005;233:706–720. doi: 10.1002/dvdy.20345. [DOI] [PubMed] [Google Scholar]
- 4.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: new insights in signaling, development and disease. J Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abell AN, Jordan NV, Huang W, Prat A, Midland AA, Johnson NL, et al. MAP3K4/CBP-Regulated H2B Acetylation Controls Epithelial-Mesenchymal Transition in Trophoblast Stem Cells. Cell Stem Cell. 2011;8:525–537. doi: 10.1016/j.stem.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abell AN, Granger DA, Johnson NL, Vincent-Jordan N, Dibble CF, Johnson GL. Trophoblast Stem Cell Maintenance by Fibroblast Growth Factor 4 Requires MEKK4 Activation of Jun N-Terminal Kinase. Mol Cell Biol. 2009;29:2748–2761. doi: 10.1128/MCB.01391-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest. 2009;119:1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. 2009;119:1429–1437. doi: 10.1172/JCI36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalluri R. EMT: when epithelial cells decide to become mesenchymal-like cells. J Clin Invest. 2009;119:1417–1419. doi: 10.1172/JCI39675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Timmerman LA, Grego-Bessa J, Raya A, Bertrán E, Pérez-Pomares JM, Díez J, et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18:99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zavadil J, Böttinger EP. TGFβ and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 14.Zavadil J, Cermak L, Soto-Nieves N, Böttinger EP. Integration of TGFbeta/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. EMBO J. 2004;23:1155–1165. doi: 10.1038/sj.emboj.7600069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuxe J, Vincent T, Garcia de Herreros A. Transcriptional crosstalk between TGFβ and stem cell pathways in tumor cell invasion: role of EMT promoting Smad complexes. Cell Cycle. 2010;9:2363–2374. doi: 10.4161/cc.9.12.12050. [DOI] [PubMed] [Google Scholar]
- 16.Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat Rev Genet. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- 17.Gonzales DS, Jones JM, Pinyopummintr T, Carnevale EM, Ginther OJ, Shapiro SS, et al. Trophectoderm projections: a potential means for locomotion, attachment and implantation of bovine, equine and human blastocysts. Hum Reprod. 1996;11:2739–2745. doi: 10.1093/oxfordjournals.humrep.a019201. [DOI] [PubMed] [Google Scholar]
- 18.Pafilis J, Batistatou A, Iliopoulou A, Tsanou E, Bakogiannis A, Dassopoulos G, et al. Expression of adhesion molecules during normal pregnancy. Cell Tissue Res. 2007;329:1–11. doi: 10.1007/s00441-007-0406-6. [DOI] [PubMed] [Google Scholar]
- 19.Sutherland A. Mechanisms of implantation in the mouse: differentiation and functional importance of trophoblast giant cell behavior. Dev Biol. 2003;258:241–251. doi: 10.1016/S0012-1606(03)00130-1. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Y, Fisher SJ, Janatpour M, Genbacev O, Dejana E, Wheelock M, et al. Human cytotrophoblasts adopt a vascular phenotype as they differentiate. A strategy for successful endovascular invasion? J Clin Invest. 1997;99:2139–2151. doi: 10.1172/JCI119387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldman-Wohl D, Yagel S. Regulation of trophoblast invasion: from normal implantation to preeclampsia. Mol Cell Endocrinol. 2002;187:233–238. doi: 10.1016/S0303-7207(01)00687-6. [DOI] [PubMed] [Google Scholar]
- 22.McMaster MT, Zhou Y, Fisher SJ. Abnormal placentation and the syndrome of preeclampsia. Semin Nephrol. 2004;24:540–547. doi: 10.1016/S0270-9295(04)00124-X. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Y, Damsky CH, Fisher SJ. Preeclampsia is associated with failure of human cytotrophoblasts to mimic a vascular adhesion phenotype. One cause of defective endovascular invasion in this syndrome? J Clin Invest. 1997;99:2152–2164. doi: 10.1172/JCI119388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khong TY. The pathology of placenta accreta, a worldwide epidemic. J Clin Pathol. 2008;61:1243–1246. doi: 10.1136/jcp.2008.055202. [DOI] [PubMed] [Google Scholar]
- 25.Perry JK, Lins RJ, Lobie PE, Mitchell MD. Regulation of invasive growth: similar epigenetic mechanisms underpin tumour progression and implantation in human pregnancy. Clin Sci. 2009;118:451–457. doi: 10.1042/CS20090503. [DOI] [PubMed] [Google Scholar]
- 26.Ferretti C, Bruni L, Dangles-Marie V, Pecking AP, Bellet D. Molecular circuits shared by placental and cancer cells, and their implications in the proliferative, invasive and migratory capacities of trophoblasts. Hum Reprod Update. 2007;13:121–141. doi: 10.1093/humupd/dml048. [DOI] [PubMed] [Google Scholar]
- 27.Rossant J. Stem cells and lineage development in the mammalian blastocyst. Reprod Fertil Dev. 2007;19:111–118. doi: 10.1071/RD06125. [DOI] [PubMed] [Google Scholar]
- 28.Erlebacher A, Price KA, Glimcher LH. Maintenance of mouse trophoblast stem cell proliferation by TGFbeta/activin. Dev Biol. 2004;275:158–169. doi: 10.1016/j.ydbio.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of Trophoblast Stem Cell Proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 30.Kokkinos MI, Murthi P, Wafai R, Thompson EW, Newgreen DF. Cadherins in the human placenta—epithelial-mesenchymal transition (EMT) and placental development. Placenta. 2010;31:747–755. doi: 10.1016/j.placenta.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 31.Larue L, Ohsugi M, Hirchenhain J, Kemler R. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc Natl Acad Sci USA. 1994;91:8263–8267. doi: 10.1073/pnas.91.17.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shih IM, Hsu MY, Oldt RJ, Herlyn M, Gearhart JD, Kurman RJ. The Role of E-cadherin in the Motility and Invasion of Implantation Site Intermediate Trophoblast. Placenta. 2002;23:706–715. doi: 10.1016/s0143-4004(02)90864-7. [DOI] [PubMed] [Google Scholar]
- 33.Abell AN, Rivera-Perez JA, Cuevas BD, Uhlik MT, Sather S, Johnson NL, et al. Ablation of MEKK4 kinase activity causes neurulation and skeletal patterning defects in the mouse embryo. Mol Cell Biol. 2005;25:8948–8959. doi: 10.1128/MCB.25.20.8948-59.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, et al. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 35.Ralston A, Rossant J. Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev Biol. 2008;313:614–629. doi: 10.1016/j.ydbio.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 36.Hajra KM, Chen DYS, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62:1613–1618. [PubMed] [Google Scholar]
- 37.Nieto MA. THE SNAIL SUPERFAMILY OF ZINC-FINGER TRANSCRIPTION FACTORS. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 38.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 39.Vandewalle C, van Roy F, Berx G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci. 2009;66:773–787. doi: 10.1007/s00018-008-8465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vandewalle C, Comijn J, De Craene B, Vermassen P, Bruyneel E, Andersen H, et al. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cellcell junctions. Nucleic Acids Res. 2005;33:6566–6578. doi: 10.1093/nar/gki965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Battula VL, Evans KW, Hollier BG, Shi Y, Marini FC, Ayyanan A, et al. Epithelial-Mesenchymal Transition-Derived Cells Exhibit Multilineage Differentiation Potential Similar to Mesenchymal Stem Cells. Stem Cells. 2010;28:1435–1445. doi: 10.1002/stem.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci USA. 2010;107:15449–15454. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Medici D, Hay ED, Goodenough DA. Cooperation between snail and LEF-1 transcription factors is essential for TGFbeta1-induced epithelial-mesenchymal transition. Mol Biol Cell. 2006;17:1871–1879. doi: 10.1091/mbc.E05-08-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 45.Heredia F, Nieto MA. An epigenetic mark that protects the epithelial phenotype in health and disease. Cell Stem Cell. 2011;8:462–463. doi: 10.1016/j.stem.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 46.Hemberger M. Genetic-epigenetic intersection in trophoblast differentiation: implications for extraembryonic tissue function. Epigenetic. 2010;5:24–29. doi: 10.4161/epi.5.1.10589. [DOI] [PubMed] [Google Scholar]
- 47.Hemberger M. Epigenetic landscape required for placental development. Cell Mol Life Sci. 2007;64:2422–2436. doi: 10.1007/s00018-007-7113-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chapman V, Forrester L, Sanford J, Hastie N, Rossant J. Cell lineage-specific undermethylation of mouse repetitive DNA. Nature. 1984;307:284–286. doi: 10.1038/307284a0. [DOI] [PubMed] [Google Scholar]
- 49.Ng RK, Dean W, Dawson C, Lucifero D, Madeja Z, Reik W, et al. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat Cell Biol. 2008;10:1280–1290. doi: 10.1038/ncb1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rossant J, Sanford JP, Chapman VM, Andrews GK. Undermethylation of structural gene sequences in extraembryonic lineages of the mouse. Dev Biol. 1986;117:567–573. doi: 10.1016/0012-1606(86)90325-8. [DOI] [PubMed] [Google Scholar]
- 51.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 52.Arima T, Hata K, Tanaka S, Kusumi M, Li E, Kato K, et al. Loss of the maternal imprint in Dnmt3Lmat-/- mice leads to a differentiation defect in the extraembryonic tissue. Dev Biol. 2006;297:361–373. doi: 10.1016/j.ydbio.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 53.Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 54.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 55.O'Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-6.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang J, Mager J, Schnedier E, Magnuson T. The mouse PcG gene eed is required for Hox gene repression and extraembryonic development. Mamm Genome. 2002;13:493–503. doi: 10.1007/s00335-002-2182-7. [DOI] [PubMed] [Google Scholar]
- 58.Wang J, Mager J, Chen Y, Schneider E, Cross JC, Nagy A, et al. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat Genet. 2001;28:37137–37135. doi: 10.1038/ng574. [DOI] [PubMed] [Google Scholar]
- 59.Rugg-Gunn PJ, Cox BJ, Ralston A, Rossant J. Inaugural Article: Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc Natl Acad Sci USA. 2010;107:10783–10790. doi: 10.1073/pnas.0914507107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kalantry S, Mills KC, Yee D, Otte AP, Panning B, Magnuson T. The Polycomb group protein Eed protects the inactive X-chromosome from differentiationinduced reactivation. Nat Cell Biol. 2006;8:195–202. doi: 10.1038/ncb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hebbes TR, Thorne AW, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-Q. [DOI] [PubMed] [Google Scholar]
- 63.Giles RH, Peters DJ, Breuning MH. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 1998;14:178–183. doi: 10.1016/S0168-9525(98)01438-3. [DOI] [PubMed] [Google Scholar]
- 64.Tanaka Y, Naruse I, Maekawa T, Masuya H, Shiroishi T, Ishii S. Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein-Taybi syndrome. Proc Natl Acad Sci USA. 1997;94:10215–10220. doi: 10.1073/pnas.94.19.10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ansieau S, Bastid J, Doreau A, Morel AP, Bouchet BP, Thomas C, et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14:79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 66.Smit MA, Peeper DS. Epithelial-mesenchymal transition and senescence: two cancer-related processes are crossing paths. Aging (Albany NY) 2010;2:735–741. doi: 10.18632/aging.100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smit MA, Peeper DS. Deregulating EMT and senescence: double impact by a single twist. Cancer Cell. 2008;14:5–7. doi: 10.1016/j.ccr.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 68.Li R, Liang J, Ni S, Zhou T, Qing X, Li H, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 69.Samavarchi-Tehrani P, Golipour A, David L, Sung Hk, Beyer TA, Datti A, et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 70.Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, et al. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- 71.Brabletz T, Jung A, Reu S, Porzner M, Hlubek F, Kunz-Schughart LA, et al. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc Natl Acad Sci USA. 2001;98:10356–10361. doi: 10.1073/pnas.171610498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zlobec I, Lugli A. Epithelial mesenchymal transition and tumor budding in aggressive colorectal cancer: tumor budding as oncotarget. Oncotarget. 2010;1:651–661. doi: 10.18632/oncotarget.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, et al. Characterization of a Naturally Occurring Breast Cancer Subset Enriched in Epithelial-to-Mesenchymal Transition and Stem Cell Characteristics. Cancer Res. 2009;69:4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pang R, Law WL, Chu ACY, Poon JT, Lam CSC, Chow AKM, et al. A subpopulation of CD26+ cancer stem cells with metastatic capacity in human colorectal cancer. Cell Stem Cell. 2010;6:603–615. doi: 10.1016/j.stem.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 76.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 78.Sheridan C, Kishimoto H, Fuchs RK, Mehrotra S, Bhat-Nakshatri P, Turner CH, et al. CD44+/CD24-breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res. 2006;8:59. doi: 10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morel AP, Lièvre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS ONE. 2008;3:2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 81.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 82.Bianchi A, Gervasi ME, Bakin A. Role of β5-integrin in epithelial-mesenchymal transition in response to TGFβ. Cell Cycle. 2010;9:1647–1659. doi: 10.4161/cc.9.8.11517. [DOI] [PubMed] [Google Scholar]
- 83.Côme C, Magnino F, Bibeau F, De Santa Barbara P, Becker KF, Theillet C, et al. Snail and slug play distinct roles during breast carcinoma progression. Clin Cancer Res. 2006;12:5395–5402. doi: 10.1158/1078-0432.CCR-06-0478. [DOI] [PubMed] [Google Scholar]
- 84.De Craene B, Gilbert B, Stove C, Bruyneel E, Van Roy F, Berx G. The transcription factor snail induces tumor cell invasion through modulation of the epithelial cell differentiation program. Cancer Res. 2005;65:6237–6244. doi: 10.1158/0008-5472.CAN-04-3545. [DOI] [PubMed] [Google Scholar]
- 85.Moll R, Mitze M, Frixen UH, Birchmeier W. Differential loss of E-cadherin expression in infiltrating ductal and lobular breast carcinomas. Am J Pathol. 1993;143:1731–1742. [PMC free article] [PubMed] [Google Scholar]
- 86.Moody SE, Perez D, Pan Tc, Sarkisian CJ, Portocarrero CP, Sterner CJ, et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 87.Novak A, Hsu SC, Leung-Hagesteijn C, Radeva G, Papkoff J, Montesano R, et al. Cell adhesion and the integrin-linked kinase regulate the LEF-1 and betacatenin signaling pathways. Proc Natl Acad Sci USA. 1998;95:4374–4379. doi: 10.1073/pnas.95.8.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Peinado H, Quintanilla M, Cano A. Transforming growth factor beta-1 induces snail transcription factor in epithelial cell lines: mechanisms for epithelial mesenchymal transitions. J Biol Chem. 2003;278:21113–21123. doi: 10.1074/jbc.M211304200. [DOI] [PubMed] [Google Scholar]
- 89.Prall F. Tumour budding in colorectal carcinoma. Histopathology. 2007;50:151–162. doi: 10.1111/j.1365-2559.2006.02551.x. [DOI] [PubMed] [Google Scholar]
- 90.Turner FE. Slug Regulates Integrin Expression and Cell Proliferation in Human Epidermal Keratinocytes. J Biol Chem. 2006;281:21321–21331. doi: 10.1074/jbc.M509731200. [DOI] [PubMed] [Google Scholar]
- 91.Vuoriluoto K, Haugen H, Kiviluoto S, Mpindi JP, Nevo J, Gjerdrum C, et al. Vimentin regulates EMT induction by Slug and oncogenic H-Ras and migration by governing Axl expression in breast cancer. Oncogene. 2011;30:1436–1448. doi: 10.1038/onc.2010.509. [DOI] [PubMed] [Google Scholar]
- 92.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 93.Bonnomet A, Brysse A, Tachsidis A, Waltham M, Thompson EW, Polette M, et al. Epithelial-to-Mesenchymal Transitions and Circulating Tumor Cells. J Mammary Gland Biol Neoplasia. 2010;15:261–273. doi: 10.1007/s10911-010-9174-0. [DOI] [PubMed] [Google Scholar]
- 94.Willipinski-Stapelfeldt B, Riethdorf S, Assmann V, Woelfle U, Rau T, Sauter G, et al. Changes in cytoskeletal protein composition indicative of an epithelialmesenchymal transition in human micrometastatic and primary breast carcinoma cells. Clin Cancer Res. 2005;11:8006–8014. doi: 10.1158/1078-0432.CCR-05-0632. [DOI] [PubMed] [Google Scholar]
- 95.Wyckoff JB, Wang Y, Lin EY, Li Jf, Goswami S, Stanley ER, et al. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 2007;67:2649–2656. doi: 10.1158/0008-5472.CAN-06-1823. [DOI] [PubMed] [Google Scholar]
- 96.Burleigh AR. Of germ cells, trophoblasts and cancer stem cells. Integr Cancer Ther. 2008;7:276–281. doi: 10.1177/1534735408326454. [DOI] [PubMed] [Google Scholar]
- 97.Campbell LL, Polyak K. Breast tumor heterogeneity: cancer stem cells or clonal evolution? Cell Cycle. 2007;6:2332–2338. doi: 10.161/cc.6.19.4914. [DOI] [PubMed] [Google Scholar]
- 98.Floor S, van Staveren WC, Larsimont D, Dumont JE, Maenhaut C. Cancer cells in epithelial-to-mesenchymal transition and tumor-propagating-cancer stem cells: distinct, overlapping or same populations. Oncogene. 2011 doi: 10.1038/onc.2011.184. In Press. [DOI] [PubMed] [Google Scholar]
- 99.Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23:2563–2577. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 101.Baritaki S, Huerta-Yepez S, Sahakyan A, Karagiannides I, Bakirtzi K, Jazirehi A, et al. Mechanisms of nitric oxide-mediated inhibition of EMT in cancer: Inhibition of the metastasis-inducer Snail and induction of the metastasis-suppressor RKIP. Cell Cycle. 2010;9:4931–4940. doi: 10.4161/cc.9.24.14229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Halder SK, Beauchamp RD, Datta PK. A specific inhibitor of TGFβ receptor kinase, SB-431542, as a potent antitumor agent for human cancers. Neoplasia. 2005;7:509–521. doi: 10.1593/neo.04640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stoops SL, Pearson AS, Weaver C, Waterson AG, Days E, Farmer C, et al. Identification and optimization of small molecules that restore e-cadherin expression and reduce invasion in colorectal carcinoma cells. ACS Chem Biol. 2011;6:452–465. doi: 10.1021/cb100305h. [DOI] [PMC free article] [PubMed] [Google Scholar]