Abstract
Filopodia are long, slender, actin-rich cellular protrusions, which recently have become a focus of cell biology research because of their proposed roles as sensory and exploratory organelles that allow for “intelligent” cell behavior. Actin nucleation, elongation and bundling are believed to be essential for filopodia formation and functions. However, the identity of actin filament nucleators responsible for the initiation of filopodia remains controversial. Two alternative models, the convergent elongation and tip nucleation, emphasize two different actin filament nucleators, the Arp2/3 complex or formins, respectively, as key players during filopodia initiation. Although these two models in principle are not mutually exclusive, it is important to understand which of them is actually employed by cells. In this review, we discuss the existing evidence regarding the relative roles of the Arp2/3 complex and formins in filopodia initiation.
Key words: filopodia, Arp2/3 complex, formins, Ena/VASP, actin
Introduction
Filopodia were first documented more than a century ago and defined as long, slender cellular protrusions, as reflected in their name.1,2 Filopodia are found in many different cell types and typically display active protrusive, retractile and sweeping motility, which may be necessary for their proposed functions as cellular sensors. During cell and tissue motility, filopodia are believed to explore the adhesive surfaces and sense soluble cues to determine the direction of cell locomotion. Thus, filopodia in growth cones of neuronal cells recognize attractive and repulsive cues and direct the axon to its destination,3,4 whereas filopodia of endothelial tip cells guide the growth of blood vessels during angiogenesis.5 A critical filopodial protein, fascin, is dramatically overexpressed in certain cancers, which may explain why metastatic cancer cells are better able to find their way to blood vessels compared to non-metastatic cells.6 In cell communication, filopodia find a proper cellular partner and make initial contacts, which subsequently maturate into functional junctions specific for a particular cell-cell interaction. In epithelia, initial filopodial point contacts zipper into continuous cell-cell junctions characteristic for epithelial sheets.7,8 In dendrites of neuronal cells, filopodia serve as precursors of dendritic spines which establish excitatory synapses.9–11 Filopodia are also involved in ability of antigen-presenting dendritic cells to interact with and activate T lymphocytes.12 The many essential roles of filopodia in morphogenesis and disease call for a better understanding of how these structures are organized structurally and molecularly, how they sense the signals, and how they transform signals into actions.
Much progress in understanding the biology of filopodia has been made over the past century, especially in last three decades, and it has been recently discussed in several excellent and comprehensive reviews covering a broad range of questions including potential functions of filopodia, roles of individual molecules and signaling pathways, and physical and mathematical aspects of filopodia protrusion.13–20 Interestingly, the accumulated knowledge fed back onto the original, shape-based definition of filopodia resulting in more detailed, but variable, definitions, which include many structural and dynamic features of filopodia.14,21–24 Although the precise definition has a clear scientific value, it also has limitations, because information obtained in one cell type and/or experimental system may not be applicable to other cells and systems. Initially, when most studies dealt with filopodia formed at the leading edge of various migrating cells, the advantages of exhaustive definitions prevailed over disadvantages. However, it is becoming increasingly clear that filopodia defined purely by shape significantly vary in their structure, behavior and molecular composition. A striking example of this point is dendritic filopodia of neuronal cells, which share very few structural and molecular features with the leading edge filopodia except for their thin elongated shape.25 On the other hand, certain structures that do not have a thread-like shape, such as microspikes, are highly homologous to the leading edge filopodia by structure, kinetics and molecular composition.26 Therefore, detailed definitions of filopodia become impractical. Furthermore, if someone begins to study a totally novel experimental system and observes thin elongated protrusions, it is convenient to use a general term until all physiological details of newly discovered protrusions are understood. To overcome this problem, Higgs and coworkers proposed to use “linear protrusions” as a general term that would encompass all types of filopodia.22 However, since it is virtually literal translation of the term “filopodia”, we propose to continue using “filopodia” as a general term to refer to any thin elongated cell surface protrusions, as defined originally and attach a specific adjective, such as “leading edge filopodia” or “dendritic filopodia” when biological features of these structures become known and they can be further categorized.
For our review, we chose to concentrate on the design of the cytoskeletal machinery responsible for initiation of the best studied type of filopodia, the leading edge filopodia of migrating cells. Much information about this class of filopodia has been obtained by studying various cell types that migrate as a whole cell, but also from analyses of neuronal growth cones that migrate away from a relatively stationary cell body causing cell elongation and generation of the axon. Remarkably, even though growth cone filopodia and dendritic filopodia in hippocampal neurons are generated by the same cells, they are dramatically different from each other, with growth cone filopodia sharing all typical features of leading edge filopodia of other cell types and dendritic filopodia being very divergent.25,27
Leading edge filopodia are characterized by the presence of an actin filament bundle, in which individual filaments span the entire length of the filopodium, are uniformly oriented with barbed ends toward the filopodial tip, and are cross-linked by fascin. During protrusion, actin subunits are incorporated at the filopodial tips, move away from the tip as a part of the filament lattice, and are released at the rear of filopodia in the process termed treadmilling. Actin filament elongation in filopodia is assisted by barbed end-binding proteins, formins and Ena/VASP proteins. Although unrelated, both classes of proteins allow for incorporation of new actin subunits to the barbed end without dissociating from it. Elongation may be even accelerated because both formins and Ena/VASP proteins bind profilin, through which they recruit multiple profilin-actin complexes and thus increase the local concentration of actin subunits near the barbed end. In addition, both formins and Ena/VASP proteins prevent binding of capping proteins to the barbed ends, which would terminate elongation, and keep the elongating ends near the membrane to increase efficiency of pushing.28–32
In contrast to overall consensus regarding mechanisms of actin filament elongation and turnover in leading edge filopodia, the mechanisms of filopodia initiation remain a matter of debate. Two alternative models, the convergent elongation and tip nucleation, emphasize two different actin filament nucleators, the Arp2/3 complex or formins, respectively, as key players during filopodia initiation. The Arp2/3 complex nucleates branched filaments33 and is responsible for generation of the dendritic actin filament network in lamellipodia34,35 and several other actin-based structures.36–38 Intrinsically inactive Arp2/3 complex can be activated in cells by numerous nucleation-promoting factors (NPFs), the best studied of which are the WAVE (or Scar) complex that functions primarily in lamellipodia, and N-WASP that appears to be mainly involved in endocytosis.39–42 Formins are characterized by the presence of conserved formin homology domain 2 (FH2)43 that in biochemical assays can nucleate unbranched actin filaments, protect barbed ends from capping and promote elongation in cooperation with the adjacent FH1 domain; some formins also have actin cross-linking activity.37,44–46
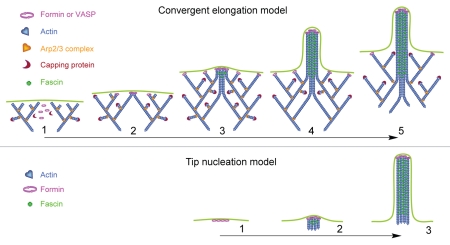
According to the convergent elongation model of filopodia initiation,26,47 filopodia are formed by reorganization of the dendritic actin network, which is assembled through nucleating activity of the Arp2/3 complex (Fig. 1, top). The reorganization is driven primarily by barbed end-associated elongation factors, such as formins and/or Ena/VASP proteins. Because these proteins promote actin filament elongation and can oligomerize, they cause gradual clustering of elongating barbed ends and force filaments to converge and elongate together. Subsequent fascinmediated cross-linking of these parallel filaments completes the formation of filopodial bundles, while elongation factors remain associated with the filopodial tip supporting filament elongation. At later stages, the Arp2/3 complex dissociates from the pointed ends of filopodial filaments and filopodia are maintained by actin filament treadmilling. In contrast, the tip nucleation model of filopodial initiation proposes that filopodial filaments are directly nucleated by a cluster of formins on the plasma membrane, which would instantly produce a bunch of elongating actin filaments that become bundled by fascin (Fig. 1, bottom).
Figure 1.
Mechanistic models of filopodia initiation. Convergent elongation model (top): (1) Branched actin network is formed in lamellipodil by Arp2/3-mediated nucleation. (2) Elongation factors (Ena/VASP or formin) maintain continuous elongation of some barbed end. (3) Interaction between barbed ends, likely also mediated by Ena/VASP or formin, results in synchronized parallel elongation of several converged filaments. (4) Parallel filaments are cross-linked by fascin resulting in formation of an actin bundle. (5) Over time, the Arp2/3 complex that nucleated filopodial filaments dissociates leaving free pointed ends of splayed actin filament. Tip nucleation model (bottom): (1) Activated formin is clustered on the plasma membrane. (2) Formin cluster nucleates a bunch of actin filaments and maintain their elongation. (3) Elongating filaments are cross-linked by fascin to form an actin bundle.
Although these two models in principle are not mutually exclusive, it is important to know which cytoskeletal machinery is actually employed by the cell to generate filopodia and which molecules play key roles in this process. Considering many important roles that filopodia are thought to play in tissue morphogenesis, cell communication and motility, such information will contribute to the development of strategies to control the filopodia-dependent processes and to intervene to correct pathological conditions. In this review, we discuss the existing evidence regarding the relative roles of the Arp2/3 complex and formins in filopodia initiation.
Convergent Elongation Model
A role of the Arp2/3 complex in filopodia seems counterintuitive because of the absence of branched filaments and the Arp2/3 complex in established filopodial bundles.35 However, a growing body of evidence supports an idea that the Arp2/3 complex is important for filopodia initiation, although it is dispensable for subsequent filopodia maintenance. The earliest evidence supporting a role of the Arp2/3 complex in filopodia formation has been provided by Machesky and Insall,39 who found that disrupting the Arp2/3 complex localization by overexpression of the Arp2/3 complex-binding WA (or VCA) domain of Scar inhibited naturally formed filopodia in macrophages. Subsequently, using thorough structural-kinetic analyses of filopodia initiation in mouse melanoma cells, we have shown that filopodia emerge from the lamellipodial dendritic network, so that individual filaments of the filopodial bundle originate at branching points on other filaments. These branch points are usually scattered around the base of the filopodium, so that filaments converge into the bundle after coming from a broad area of the lamellipodium. These findings allowed us to propose the convergent elongation model.26 A similar pathway of filopodia initiation was also observed in growth cones of neuronal cells, where filopodia emerged from small regions of dendritic network within interfilopodial veils,27 and in Drosophila embryos undergoing dorsal closure, where live imaging of GFP-Ena in leading edge cells revealed that filopodia were formed from lamellipodia.48 Immunoelectron microscopy of the Arp2/3 complex localization in platelets also provided data consistent with this view.49
The convergent elongation model has now been supported by functional studies that test contribution of individual molecules to filopodia formation using a range of experimental approaches from in vitro reconstitution systems to cultured cells to whole organisms. The reorganization of dendritic networks (like in lamellipodia) into parallel bundles (like in filopodia) has been reconstituted in cell-free systems using cytoplasmic extracts or purified proteins.50,51 These studies showed that fascin and, importantly, the Arp2/3 complex were necessary for the formation of bundles, whereas capping protein had an inhibitory effect. Moreover, mathematical modeling of bundle formation in this system supported the convergent elongation mechanism, rather than the tip nucleation model.52 A strong support for the convergent elongation model of filopodia initiation has been recently provided by in vitro reconstitution of filopodia-like structures self-assembling on supported PI(4,5)P2-containing lipid bilayers in the presence of cytoplasmic extracts.53 Time-resolved recruitment of individual molecules to filopodia-like structures during their initiation in this system revealed that an F-BAR domain protein Toca-1 is first to be recruited to lipid bilayers through interaction with PI(4,5)P2. Toca-1 recruitment is followed by appearance of its interaction partner, N-WASP, after which the Arp2/3 complex and F-actin appear simultaneously, whereas elongation factors, formin mDia2 and Ena/VASP, are recruited subsequently, and fascin is recruited last. Moreover, Lee et al. have found that Arp2/3 complex-dependent nucleation is vital for the formation of filopodia-like structures in this reconstitution system. These findings are consistent with the convergent elongation model, but contradict the model of tip nucleation.
Various loss-of-function and gain-of-function approaches in cultured cells and in animals, also provided supportive evidence for the convergent elongation model. Thus, knockdown of the Arp2/3 complex in cultured neurons significantly inhibited the rate of filopodia initiation, which led to a decreased number of filopodia in growth cones.27 Similarly, Arp2/3 complex loss-offunction mutations caused significant defects of filopodia initiation in developing neuronal growth cones of C. elegans in vivo54 and in cultured Drosophila neurons.55 Furthermore, delocalization of the Arp2/3 complex by Scar-WA in macrophages39 or by N-WASP-VCA in HeLa cells56 resulted in inhibition of endogenous39 or syndapin-induced56 filopodia. Filopodia inhibition was also observed after interference with NPFs. Thus, in cultured Drosophila cells, RNAi-mediated depletion of SCAR, the only member of the WAVE subfamily of Arp2/3 complex activators, strongly inhibited both lamellipodia and filopodia.57 Inhibition of filopodia was also observed after expression of dominant negative Rac1 in cultured fibroblasts,58 and in developing Drosophila pupae.59 Since the most prominent effect of Rac1 on the actin cytoskeleton is induction of lamellipodia through activation of the WAVE complex, which activates the Arp2/3 complex, these data can be best explained from a point of view of the convergent elongation model. Involvement of another potent NPF, N-WASP, in filopodia formation has been repeatedly observed using gainof-function approaches,60–63 although loss-of-function data indicated that N-WASP is not essential for filopodia formation.64,65 A potential explanation of these results is that overactive N-WASP increases a pool of branched filaments in cells, which can be then used by elongating and cross-linking proteins for remodeling into filopodial bundles regardless of the originally intended destiny of these filaments. On the other hand, inhibition of N-WASP may have no effect on filopodia because of the presence of other NPFs. It should be pointed out, however, that although activation of the Arp2/3 complex appears to be the main biochemical activity of N-WASP, N-WASP may also function as a barbed end-associated elongation factor66 or an Arp2/3 complex-independent nucleator67 leaving the exact mechanism of potential contribution of N-WASP to filopodia formation uncertain.
Functional analyses of capping protein and elongation factors, Ena/VASP proteins and formins, also produced results consistent with the convergent elongation model. Thus, inhibition of capping protein in mouse melanoma cells resulted in enhanced formation of filopodia at the expense of lamellipodia.68 Similar results have been reported for cultured Drosophila neurons.55 These results indicate that lamellipodia and filopodia are dynamically interrelated and their filaments may be generated by common nucleator(s), as proposed by the convergent elongation model. However, it is hard to explain this phenomenon based on the tip nucleation model. Experiments modulating functions of Ena/VASP proteins revealed that their activity positively correlates with the expression of filopodia in mammalian69,70 or Drosophila55 neurons, in Dictyostelium,71 and in Drosophila embryos.72 Similar correlation exists for formins.17,47,73–75 Although the filopodia-promoting role of formins could be attributed to their nucleating activity, some of filopodia-inducing formins are poor nucleators,75,76 whereas Ena/VASP proteins do not have appreciable nucleating activity in physiological conditions,77 suggesting that other activities of these proteins, such as promotion of filament elongation31,32 and clustering of barbed ends,75,78 are more essential for filopodia induction. The dynamic behavior of Ena/VASP proteins and formins at the leading edge is also consistent with this possibility. Indeed, during filopodia initiation, both VASP and mDia2 are initially evenly distributed along the leading edge of lamellipodia, but gradually converge into a tight cluster at the tip of a newly formed filopodium.26,47 This behavior indicates that clustering of formins is a relatively late event during filopodia initiation, thus contrasting the postulate of the tip nucleation model, which assumes that pre-clustered formins begin the process of filopodia formation. Existence of two distinct families of elongation factors, formins and Ena/VASP, raises a question of whether they have redundant or specialized functions during filopodia formation. At present, it appears that much redundancy exist among them,79 but some specific functions also begin to emerge.55,72
Tip Nucleation Model
The combination of biochemical activities of formins, which are able to nucleate, elongate, protect from capping and cross-link actin filaments, makes them perfect candidates to both initiate and maintain filopodia, thus setting a stage for the tip nucleation model. Formin mDia2 (or its non-mammalian homologs) is the primary candidate to play a role in filopodia,47,73,74,80 but other formins are also able to perform this function.24,75,81 However, despite attractive biochemical properties and beautiful simplicity of the tip nucleation model, direct evidence supporting the use of this mechanism by cells is missing. In fact, our analysis of mDia2 functions in cells47 was motivated exactly by this model and we were aiming to prove it experimentally. By performing correlative light and electron microscopy of filopodia that were induced in cells by constitutively active mDia2, we hoped to observe a birth of a filopodium from a spot-like cluster of fluorescently labeled mDia2, as predicted by the tip nucleation model. However, we failed to observe such events and instead found that GFP-mDia2 was initially evenly distributed along the lamellipodial leading edge and then gradually condensed to the tip of a newly formed filopodium concomitantly with the convergence of actin filaments into a bundle from their initially broad distribution in lamellipodia.47 This observation indicated that the filopodial actin filaments were born all over the lamellipodium, not at a focal point as predicted by the tip nucleation model, and then gradually converged during elongation with their barbed ends congregating into the filopodial tip. These data, however, did not discriminate whether lamellipodial filaments that that ended up in the filopodial bundle were directly nucleated by mDia2 or they were nucleated by the Arp2/3 complex and their barbed ends were subsequently captured by mDia2 to assist the processive elongation.
The existing arguments in favor of the tip nucleation model and direct nucleation of filopodial actin filament by formins are based on two sets of data: (1) Induction of filopodia by upregulated formins or inhibition of filopodia after their downregulation,80,82 and (2) persistence of filopodia upon downregulation of the Arp2/3 complex or its activators.83 In the former set of arguments, the role of formins in filopodia generation can be equally well explained by the ability of formins to function as nucleators and as elongation factors, so these data are consistent with both models. However, when constitutively active mDia2 is overexpressed in cells, it indeed appears to nucleate actin filaments, as can be inferred from our observation of multiple unattached pointed ends of actin filaments appearing at the rear of induced filopodia and lamellipodia,47 as well as from the unusual clublike shape of induced filopodia having a thicker tip and a thinner base.47,82 However, we never observed such features in control cells, suggesting that properly regulated mDia2 may not nucleate actin filaments at the cell leading edge in normal physiological conditions, but instead primarily use its ability to elongate actin filaments.
The second set of data, preservation of filopodia after inhibition of the Arp2/3 complex or NPFs is frequently used as an argument against the convergent elongation model and a role of the Arp2/3 complex in filopodia initiation, but in favor of the tip nucleation model and a role of formins in nucleation of filopodial filaments. In the most frequently cited paper of this kind,83 the authors inhibited components of the Arp2/3 complex or WAVE complex in mouse melanoma cells co-transfected with constitutively active Cdc42 and dominant negative Rac1 to exaggerate the formation of filopodia. They reported that although lamellipodia were abrogated in these cells, filopodia remained normal. To support this observation, they quantified a total fraction of filopodia-containing cells in the population in different conditions, and also fractions of cells containing <5, 6–10 or >10 filopodia, and found no differences with either approach. However, a closer look at their quantitative data reveals that although the first approach indeed shows no differences between control and siRNA-treated samples, as expected for cells expressing this cocktail of GTPases, the quantification after binning showed a clear decrease of the “>10” category with a concomitant increase of the “<5” category in Arp3-depleted cells, which is also consistent with visual impression from images provided in the paper.83 A similar, albeit less clear, trend can also be seen in WAVE-WA-expressing cells, suggesting that either depletion or delocalization of the Arp2/3 complex might decrease the overall number of filopodia in cells, supporting a role of this nucleator in filopodia generation contrary to the claim in the paper. We propose that quantification of the actual number of filopodia per cell, or even better, quantification of the rate of filopodia initiation, is a more suitable way to evaluate the phenotype in such studies.
Even when the number of filopodia decreases after Arp2/3 complex knockdown, many of them do persist, raising a question of how the remaining filopodia are initiated. Surprisingly, a common way of reasoning in this situation is that if any filopodia remain, they should be nucleated by formins. However, an alternative possibility is that they are nucleated by surviving Arp2/3 complex, because knockdowns are not absolute. In cases, when NPFs are inhibited instead of the Arp2/3 complex, the situation is even more uncertain because of the presence of other NPFs. Therefore, additional experiments are needed to address this point. In our study,27 we performed electron microscopy of newly formed filopodia in Arp2/3 complex-depleted cells to distinguish these possibilities. We found that individual filaments in remaining filopodia originated as branches on other filaments at the base of the filopodium, as expected for Arp2/3 complexmediated nucleation. These data allowed us to conclude that remaining filopodia were initiated by the remaining Arp2/3 complex. Notably, it is essential to investigate nascent filopodia in these experiments, because actin filaments debranch very rapidly and filopodial filaments are subsequently maintained by treadmilling, although they may continue to recruit additional Arp2/3-nucleated filaments. As a result, old filopodia do not display branched filaments at their roots, whereas nascent filopodia do. Although Steffen et al. provided electron microscopic images of filopodia in p16 (Arp2/3 complex component) knockdown cells, they did not determine whether these filopodia were newly formed. Also, they did not show the pointed ends of filopodial filaments at sufficient resolution to determine whether they originate as branches on other filaments or not. However, their images showed that filopodial filaments did not start at the same focal spot, as predicted by the tip nucleation model, but gathered into a bundle from a broad area, as predicted by the convergent elongation model. In another study, the origin of filopodia was evaluated by light microscopy with a conclusion that filopodia arise from regions not containing lamellipodia.24 However, light microscopy does not have enough resolution to detect small patches of branched network that might be used for filopodia initiation, as we observed in neuronal growth cones.27 Therefore, arguments in favor of the tip nucleation model that are built on the “negative” argument that filopodia remain after Arp2/3 complex inhibition are not solid enough to claim that this mechanism is actually used by cells.
At first glance, it seems surprising that after dramatic depletion, sometimes up to 95%, the Arp2/3 complex still can function for filopodia initiation. However, numeric estimates of the remaining amount of the Arp2/3 complex in such cases makes things less puzzling. In normal conditions, fibroblast-like cells have large excess of the Arp2/3 complex, which can be used only in challenging situations. As a result, even significant knockdown of the Arp2/3 complex leaves enough activity to maintain limited lamellipodia production.22 As compared to lamellipodia, filopodia need much less Arp2/3 complex and only during initiation. As estimated by Higgs and colleagues,22 after ∼95% depletion, the cytoplasmic concentration of the Arp2/3 complex in 3T3 cells is expected to be ∼76 nM. For the cytoplasmic (excluding nucleus) volume of ∼2,000–3,000 µm3 for these cells, this concentration translates into tens of thousands Arp2/3 complex molecules per cell. Considering that a typical filopodium has 10–30 filaments,18,84 the Arp2/3 complex-depleted cell can produce up to a thousand filopodia, if all Arp2/3 complexes are activated. Since the life time of a filopodium is significantly longer than the life time of the Arp2/3 complex in its root, the number of filopodia existing in the cell any given moment may increase several fold. Of course, this estimate gives an upper limit for the amount of filopodia per cell, which unlikely can be reached in reality, but it shows that a knockdown cell has much more Arp2/3 complex than it needs to nucleate the observed number of filopodia in such cells. Therefore, the presence of filopodia even after deep depletion of the Arp2/3 complex does not rule out its role in filopodia initiation.
A likely cause of confusion regarding the role of the Arp2/3 complex in filopodia initiation is that a level of filopodia inhibition is frequently disproportionally lower than the degree of Arp2/3 complex depletion. One potential explanation for this phenomenon is that many cells form microspikes, or “hidden” filopodia, that are completely embedded into the lamellipodial network and protrude at the same speed as lamellipodia. Inhibition of the Arp2/3 complex has a more dramatic effect on the protrusion rate of lamellipodia, which require constant Arp2/3 complex-dependent nucleation of actin filaments, as compared to microspikes, which can continue to protrude through filament elongation. As a result, microspikes “win the race” and begin to protrude faster than lamellipodia, thus transforming into typical finger-like filopodia, which may give an impression that even more filopodia are formed after inhibition the Arp2/3 complex.85,86
Another possibility is that the Arp2/3 complex knock-down cells switch to a higher rate of lamellipodia-to-filopodia conversion in conditions of insufficient supply of the Arp2/3 complex. A potential underlying mechanism is that the molecular machinery converting the branched network into filopodial bundles, such as elongation factors and cross-linkers, keeps working with the same efficiency and overwhelms the weakened nucleation machinery. This idea is consisted with results obtained in reconstitution studies that showed that more and longer actin bundles were formed in vitro, when the concentration of the Arp2/3 complex was decreased, while the concentrations of other components, such as WASP-VCA, actin and fascin, kept constant.87 A naturally high rate of lamellipodia-to-filopodia transition is observed in neuronal growth cones, which do not form extensive lamellipodia, but rapidly reorganize their small lamellipodial regions into filopodia.27 Another factor that may contribute to this phenomenon in Arp2/3 complex knock-down cells is that when actin monomers are less consumed by Arp2/3 complex, a higher concentration of them becomes available for actin filament elongation, thus shifting the actin dynamics at the leading edge toward filopodia.
Concluding Remarks
Two proposed models of filopodia initiation, the convergent elongation and tip nucleation, are both theoretically possible. Moreover, they are not mutually exclusive and can easily co-exist, especially considering the diversity and variability of broadly defined filopodial protrusions. However, the challenge is to determine which of these theoretical possibilities is actually realized in cells. Evaluation of experimental evidence provided in this review indicates that some data fit both models, whereas other data are only consistent with the convergent elongation model. Moreover, direct evidence that the convergent elongation mechanism is used by at least some cells is available, whereas this is not the case for the tip nucleation model, although the existing data do not exclude a possibility that the tip nucleation mechanism functions in yet to be discovered physiological situations. The major problem in interpretation of formin gain-of-function and loss-of-function phenotypes is that all effects can be explained by actin nucleation, but equally well by actin elongation. Since the actin-nucleating and actin-elongating activities of formins reside in the same FH2 domain and require the same amino acid residues, it is hard, if not impossible, to design mutations that would separate these two activities in formins and therefore allow for obtaining a definitive proof that formins nucleate actin filaments for filopodia.
Acknowledgments
This work was supported by NIH grant GM70898 to T.S.
Abbreviations
- F-BAR
a subfamily of BAR (Bin-Amphiphysin-Rvs) domain
- FH1
formin-homology 1
- FH2
formin-homology 2
- N-WASP
neural Wiskott-Aldrich syndrome protein
- NPF
nucleation-promoting factor
- PI(4,5)P2
phosphatidylinositol-4,5-bisphosphate
- WA
WASP-homology region and acidic region
- WAVE
WASP-family verprolin-homologous protein
- VCA
verprolin homology, central and acidic region
References
- 1.Ellermann V. On the detection of rhizopods in two cases of acute anterior poliomyelitis. In: Bruce A, Bramwell E, Campbell M, editors. Review of neurology and psychiatry. Edinburgh: Otto Schulze Co.; 1906. p. 353. [Google Scholar]
- 2.Harrison RG. Observations on the living developing nerve fiber. Anat Rec. 1907;1:116–118. [Google Scholar]
- 3.Gallo G, Letourneau PC. Regulation of growth cone actin filaments by guidance cues. J Neurobiol. 2004;58:92–102. doi: 10.1002/neu.10282. [DOI] [PubMed] [Google Scholar]
- 4.Drees F, Gertler FB. Ena/VASP: proteins at the tip of the nervous system. Curr Opin Neurobiol. 2008;18:53–59. doi: 10.1016/j.conb.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Smet F, Segura I, De Bock K, Hohensinner PJ, Carmeliet P. Mechanisms of vessel branching: filopodia on endothelial tip cells lead the way. Arterioscler Thromb Vasc Biol. 2009;29:639–649. doi: 10.1161/ATVBAHA.109.185165. [DOI] [PubMed] [Google Scholar]
- 6.Machesky LM, Li A. Fascin: Invasive filopodia promoting metastasis. Commun Integr Biol. 2010;3:263–270. doi: 10.4161/cib.3.3.11556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasioukhin V, Bauer C, Yin M, Fuchs E. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 2000;100:209–219. doi: 10.1016/s0092-8674(00)81559-7. [DOI] [PubMed] [Google Scholar]
- 8.Wood W, Martin P. Structures in focus-filopodia. Int J Biochem Cell Biol. 2002;34:726–730. doi: 10.1016/s1357-2725(01)00172-8. [DOI] [PubMed] [Google Scholar]
- 9.Bourne JN, Harris KM. Balancing structure and function at hippocampal dendritic spines. Annu Rev Neurosci. 2008;31:47–67. doi: 10.1146/annurev.neuro.31.060407.125646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hotulainen P, Hoogenraad CC. Actin in dendritic spines: connecting dynamics to function. J Cell Biol. 2010;189:619–629. doi: 10.1083/jcb.201003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heiman MG, Shaham S. Twigs into branches: how a filopodium becomes a dendrite. Curr Opin Neurobiol. 2010;20:86–91. doi: 10.1016/j.conb.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Alwan MM, Rowden G, Lee TD, West KA. Fascin is involved in the antigen presentation activity of mature dendritic cells. J Immunol. 2001;166:338–345. doi: 10.4049/jimmunol.166.1.338. [DOI] [PubMed] [Google Scholar]
- 13.Faix J, Rottner K. The making of filopodia. Curr Opin Cell Biol. 2006;18:18–25. doi: 10.1016/j.ceb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Gupton SL, Gertler FB. Filopodia: the fingers that do the walking. Sci STKE. 2007;2007:5. doi: 10.1126/stke.4002007re5. [DOI] [PubMed] [Google Scholar]
- 15.Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- 16.Faix J, Breitsprecher D, Stradal TE, Rottner K. Filopodia: Complex models for simple rods. Int J Biochem Cell Biol. 2009;41:1656–1664. doi: 10.1016/j.biocel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 17.Mellor H. The role of formins in filopodia formation. Biochim Biophys Acta. 2010;1803:191–200. doi: 10.1016/j.bbamcr.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 18.Mogilner A, Rubinstein B. The physics of filopodial protrusion. Biophys J. 2005;89:782–795. doi: 10.1529/biophysj.104.056515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mogilner A. Mathematics of cell motility: have we got its number? J Math Biol. 2009;58:105–134. doi: 10.1007/s00285-008-0182-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlsson AE, Sept D. Mathematical modeling of cell migration. Methods Cell Biol. 2008;84:911–937. doi: 10.1016/S0091-679X(07)84029-5. [DOI] [PubMed] [Google Scholar]
- 21.Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007;9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- 22.Nicholson-Dykstra SM, Higgs HN. Arp2 depletion inhibits sheet-like protrusions but not linear protrusions of fibroblasts and lymphocytes. Cell Motil Cytoskeleton. 2008;65:904–922. doi: 10.1002/cm.20312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmed S, Goh WI, Bu W. I-BAR domains, IRSp53 and filopodium formation. Cemin Cell Dev Biol. 2010;21:350–356. doi: 10.1016/j.semcdb.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 24.Goh WI, Sudhaharan T, Lim KB, Sem KP, Lau CL, Ahmed S. Rif-mDia1 interaction is Involved in filopodium formation independent of Cdc42 and Rac effectors. J Biol Chem. 2011;286:13681–13694. doi: 10.1074/jbc.M110.182683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korobova F, Svitkina T. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol Biol Cell. 2010;21:165–176. doi: 10.1091/mbc.E09-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, et al. Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korobova F, Svitkina T. Arp2/3 complex is important for filopodia formation, growth cone motility and neuritogenesis in neuronal cells. Mol Biol Cell. 2008;19:1561–1574. doi: 10.1091/mbc.E07-09-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, et al. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002;297:612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- 29.Romero S, Le Clainche C, Didry D, Egile C, Pantaloni D, Carlier MF. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 30.Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci USA. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breitsprecher D, Kiesewetter AK, Linkner J, Urbanke C, Resch GP, Small JV, et al. Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. EMBO J. 2008;27:2943–2954. doi: 10.1038/emboj.2008.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansen SD, Mullins RD. VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J Cell Biol. 2010;191:571–584. doi: 10.1083/jcb.201003014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping and formation of branching networks of filaments. Proc Natl Acad Sci USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Svitkina TM, Verkhovsky AB, McQuade KM, Borisy GG. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 37.Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 38.Le Clainche C, Carlier MF. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev. 2008;88:489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- 39.Machesky LM, Insall RH. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- 40.Innocenti M, Gerboth S, Rottner K, Lai FP, Hertzog M, Stradal TE, et al. Abi1 regulates the activity of N-WASP and WAVE in distinct actin-based processes. Nat Cell Biol. 2005;7:969–976. doi: 10.1038/ncb1304. [DOI] [PubMed] [Google Scholar]
- 41.Derivery E, Gautreau A. Generation of branched actin networks: assembly and regulation of the N-WASP and WAVE molecular machines. Bioessays. 2010;32:119–131. doi: 10.1002/bies.200900123. [DOI] [PubMed] [Google Scholar]
- 42.Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Higgs HN, Peterson KJ. Phylogenetic analysis of the formin homology 2 domain. Mol Biol Cell. 2005;16:1–13. doi: 10.1091/mbc.E04-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Higgs HN. Formin proteins: a domain-based approach. Trends Biochem Sci. 2005;30:342–353. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 45.Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- 46.Paul AS, Pollard TD. Review of the mechanism of processive actin filament elongation by formins. Cell Motil Cytoskeleton. 2009;66:606–617. doi: 10.1002/cm.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang C, Czech L, Gerboth S, Kojima S, Scita G, Svitkina T. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 2007;5:317. doi: 10.1371/journal.pbio.0050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gates J, Mahaffey JP, Rogers SL, Emerson M, Rogers EM, Sottile SL, et al. Enabled plays key roles in embryonic epithelial morphogenesis in Drosophila. Development. 2007;134:2027–2039. doi: 10.1242/dev.02849. [DOI] [PubMed] [Google Scholar]
- 49.Falet H, Hoffmeister KM, Neujahr R, Hartwig JH. Normal Arp2/3 complex activation in platelets lacking WASp. Blood. 2002;100:2113–2122. [PubMed] [Google Scholar]
- 50.Vignjevic D, Yarar D, Welch MD, Peloquin J, Svitkina T, Borisy GG. Formation of filopodia-like bundles in vitro from a dendritic network. J Cell Biol. 2003;160:951–962. doi: 10.1083/jcb.200208059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haviv L, Brill-Karniely Y, Mahaffy R, Backouche F, Ben-Shaul A, Pollard TD, et al. Reconstitution of the transition from lamellipodium to filopodium in a membrane-free system. Proc Natl Acad Sci USA. 2006;103:4906–4911. doi: 10.1073/pnas.0508269103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brill-Karniely Y, Ideses Y, Bernheim-Groswasser A, Ben-Shaul A. From branched networks of actin filaments to bundles. Chemphyschem. 2009;10:2818–2827. doi: 10.1002/cphc.200900615. [DOI] [PubMed] [Google Scholar]
- 53.Lee K, Gallop JL, Rambani K, Kirschner MW. Self-assembly of filopodia-like structures on supported lipid bilayers. Science. 2010;329:1341–1345. doi: 10.1126/science.1191710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Norris AD, Dyer JO, Lundquist EA. The Arp2/3 complex, UNC-115/abLIM and UNC-34/Enabled regulate axon guidance and growth cone filopodia formation in Caenorhabditis elegans. Neural Dev. 2009;4:38. doi: 10.1186/1749-8104-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goncalves-Pimentel C, Gombos R, Mihaly J, Sanchez-Soriano N, Prokop A. Dissecting regulatory networks of filopodia formation in a Drosophila growth cone model. PLoS One. 2011;6:18340. doi: 10.1371/journal.pone.0018340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qualmann B, Kelly RB. Syndapin isoforms participate in receptor-mediated endocytosis and actin organization. J Cell Biol. 2000;148:1047–1062. doi: 10.1083/jcb.148.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Biyasheva A, Svitkina T, Kunda P, Baum B, Borisy G. Cascade pathway of filopodia formation downstream of SCAR. J Cell Sci. 2004;117:837–848. doi: 10.1242/jcs.00921. [DOI] [PubMed] [Google Scholar]
- 58.Johnston SA, Bramble JP, Yeung CL, Mendes PM, Machesky LM. Arp2/3 complex activity on filopodia of spreading cells. BMC Cell Biol. 2008;9:65. doi: 10.1186/1471-2121-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cohen M, Georgiou M, Stevenson NL, Miodownik M, Baum B. Dynamic filopodia transmit intermittent Delta-Notch signaling to drive pattern refinement during lateral inhibition. Dev Cell. 2010;19:78–89. doi: 10.1016/j.devcel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 60.Miki H, Sasaki T, Takai Y, Takenawa T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature. 1998;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- 61.Cory GO, Garg R, Cramer R, Ridley AJ. Phosphorylation of tyrosine 291 enhances the ability of WASp to stimulate actin polymerization and filopodium formation. J Biol Chem. 2002;277:45115–45121. doi: 10.1074/jbc.M203346200. [DOI] [PubMed] [Google Scholar]
- 62.Bu W, Chou AM, Lim KB, Sudhaharan T, Ahmed S. The Toca-1-N-WASP Complex Links Filopodial Formation to Endocytosis. J Biol Chem. 2009;284:11622–11636. doi: 10.1074/jbc.M805940200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sheldon H, Andre M, Legg JA, Heal P, Herbert JM, Sainson R, et al. Active involvement of Robo1 and Robo4 in filopodia formation and endothelial cell motility mediated via WASP and other actin nucleation-promoting factors. FASEB J. 2009;23:513–522. doi: 10.1096/fj.07-098269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lommel S, Benesch S, Rottner K, Franz T, Wehland J, Kuhn R. Actin pedestal formation by enteropathogenic Escherichia coli and intracellular motility of Shigella flexneri are abolished in N-WASP-defective cells. EMBO Rep. 2001;2:850–857. doi: 10.1093/embo-reports/kve197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Snapper SB, Takeshima F, Anton I, Liu CH, Thomas SM, Nguyen D, et al. N-WASP deficiency reveals distinct pathways for cell surface projections and microbial actin-based motility. Nat Cell Biol. 2001;3:897–904. doi: 10.1038/ncb1001-897. [DOI] [PubMed] [Google Scholar]
- 66.Co C, Wong DT, Gierke S, Chang V, Taunton J. Mechanism of actin network attachment to moving membranes: barbed end capture by N-WASP WH2 domains. Cell. 2007;128:901–913. doi: 10.1016/j.cell.2006.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takano K, Watanabe-Takano H, Suetsugu S, Kurita S, Tsujita K, Kimura S, et al. Nebulin and N-WASP cooperate to cause IGF-1-induced sarcomeric actin filament formation. Science. 2010;330:1536–1540. doi: 10.1126/science.1197767. [DOI] [PubMed] [Google Scholar]
- 68.Mejillano MR, Kojima S, Applewhite DA, Gertler FB, Svitkina TM, Borisy GG. Lamellipodial versus filopodial mode of the actin nanomachinery; pivotal role of the filament barbed end. Cell. 2004;118:363–373. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 69.Lebrand C, Dent EW, Strasser GA, Lanier LM, Krause M, Svitkina TM, et al. Critical role of Ena/VASP proteins for filopodia formation in neurons and in function downstream of netrin-1. Neuron. 2004;42:37–49. doi: 10.1016/s0896-6273(04)00108-4. [DOI] [PubMed] [Google Scholar]
- 70.Kwiatkowski AV, Rubinson DA, Dent EW, Edward van Veen J, Leslie JD, Zhang J, et al. Ena/VASP is required for neuritogenesis in the developing cortex. Neuron. 2007;56:441–455. doi: 10.1016/j.neuron.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 71.Han YH, Chung CY, Wessels D, Stephens S, Titus MA, Soll DR, et al. Requirement of a vasodilator-stimulated phosphoprotein (VASP) family member for cell adhesion, the formation of filopodia and chemotaxis in Dictyostelium. J Biol Chem. 2002;17:17. doi: 10.1074/jbc.M209107200. [DOI] [PubMed] [Google Scholar]
- 72.Homem CC, Peifer M. Exploring the roles of diaphanous and enabled activity in shaping the balance between filopodia and lamellipodia. Mol Biol Cell. 2009;20:5138–5155. doi: 10.1091/mbc.E09-02-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peng J, Wallar BJ, Flanders A, Swiatek PJ, Alberts AS. Disruption of the Diaphanous-related formin Drf1 gene encoding mDia1 reveals a role for Drf3 as an effector for Cdc42. Curr Biol. 2003;13:534–545. doi: 10.1016/s0960-9822(03)00170-2. [DOI] [PubMed] [Google Scholar]
- 74.Pellegrin S, Mellor H. The Rho family GTPase Rif induces filopodia through mDia2. Curr Biol. 2005;15:129–133. doi: 10.1016/j.cub.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 75.Harris ES, Gauvin TJ, Heimsath EG, Higgs HN. Assembly of filopodia by the formin FRL2 (FMNL3) Cytoskeleton (Hoboken) 2010;67:755–772. doi: 10.1002/cm.20485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harris ES, Rouiller I, Hanein D, Higgs HN. Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J Biol Chem. 2006;281:14383–14392. doi: 10.1074/jbc.M510923200. [DOI] [PubMed] [Google Scholar]
- 77.Lambrechts A, Kwiatkowski AV, Lanier LM, Bear JE, Vandekerckhove J, Ampe C, et al. cAMP-dependent protein kinase phosphorylation of EVL, a Mena/VASP relative, regulates its interaction with actin and SH3 domains. J Biol Chem. 2000;275:36143–36151. doi: 10.1074/jbc.M006274200. [DOI] [PubMed] [Google Scholar]
- 78.Applewhite DA, Barzik M, Kojima S, Svitkina TM, Gertler FB, Borisy GG. Ena/VASP proteins have an anti-capping independent function in filopodia formation. Mol Biol Cell. 2007;18:2579–2591. doi: 10.1091/mbc.E06-11-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dent EW, Kwiatkowski AV, Mebane LM, Philippar U, Barzik M, Rubinson DA, et al. Filopodia are required for cortical neurite initiation. Nat Cell Biol. 2007;9:1347–1359. doi: 10.1038/ncb1654. [DOI] [PubMed] [Google Scholar]
- 80.Schirenbeck A, Bretschneider T, Arasada R, Schleicher M, Faix J. The Diaphanous-related formin dDia2 is required for the formation and maintenance of filopodia. Nat Cell Biol. 2005;7:619–625. doi: 10.1038/ncb1266. [DOI] [PubMed] [Google Scholar]
- 81.Matusek T, Gombos R, Szecsenyi A, Sanchez-Soriano N, Czibula A, Pataki C, et al. Formin proteins of the DAAM subfamily play a role during axon growth. J Neurosci. 2008;28:13310–13319. doi: 10.1523/JNEUROSCI.2727-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Block J, Stradal TE, Hanisch J, Geffers R, Kostler SA, Urban E, et al. Filopodia formation induced by active mDia2/Drf3. J Microsc. 2008;231:506–517. doi: 10.1111/j.1365-2818.2008.02063.x. [DOI] [PubMed] [Google Scholar]
- 83.Steffen A, Faix J, Resch GP, Linkner J, Wehland J, Small JV, et al. Filopodia formation in the absence of functional WAVE- and Arp2/3-complexes. Mol Biol Cell. 2006;17:2581–2591. doi: 10.1091/mbc.E05-11-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pronk S, Geissler PL, Fletcher DA. Limits of filopodium stability. Phys Rev Lett. 2008;100:258102. doi: 10.1103/PhysRevLett.100.258102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Beli P, Mascheroni D, Xu D, Innocenti M. WAVE and Arp2/3 jointly inhibit filopodium formation by entering into a complex with mDia2. Nat Cell Biol. 2008;10:849–857. doi: 10.1038/ncb1745. [DOI] [PubMed] [Google Scholar]
- 86.Sarmiento C, Wang W, Dovas A, Yamaguchi H, Sidani M, El-Sibai M, et al. WASP family members and formin proteins coordinate regulation of cell protrusions in carcinoma cells. J Cell Biol. 2008;180:1245–1260. doi: 10.1083/jcb.200708123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ideses Y, Brill-Karniely Y, Haviv L, Ben-Shaul A, Bernheim-Groswasser A. Arp2/3 branched actin network mediates filopodia-like bundles formation in vitro. PLoS ONE. 2008;3:3297. doi: 10.1371/journal.pone.0003297. [DOI] [PMC free article] [PubMed] [Google Scholar]