Building a new cell is a substantial metabolic commitment that requires not only energy, but also building blocks for new proteins, nucleic acids and membranes. How does a cell assess nutrient availability before it makes a growth decision? Studies of nutrient sensing and signaling pathways have revealed important roles for the PKA, TOR and AMPK pathways in conveying metabolic status to steer cells toward growth or quiescence.1 Recently, we discovered that the metabolite acetyl-CoA can induce histone acetylation, which consequently activates the transcriptional growth program in budding yeast.2 Thus, a key output of the activation of growth control pathways could be the upregulated production of acetyl-CoA.
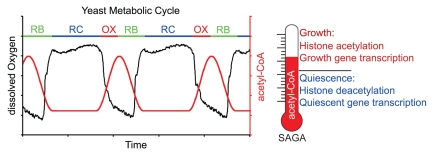
Our study made use of a continuous culture system termed the yeast metabolic cycle (YMC), where a highly synchronized cell population continuously alternates between growth and quiescent phases in each cycle of oxygen consumption (Fig. 1).3,4 We found that the addition of carbon sources, including fermentation products such as ethanol and acetate, could induce cycling cells in the quiescent phase of the YMC to enter growth.2 This is accompanied by a surge in acetyl CoA levels. In an uninterrupted YMC or in batch culture, acetyl-CoA levels also peak during growth and decrease when cells enter stationary phase or quiescence (Fig. 1).2,4
Figure 1.
Metabolic regulation of gene expression as revealed through the yeast metabolic cycle (YMC). Intracellular acetyl-CoA levels increase during growth and induce the acetylation of histones to upregulate those genes specifically important for growth. Upon entry into stationary phase or quiescence, acetyl-CoA levels decrease, histones become deacetylated, and a group of genes associated with stress, starvation, and survival responses are upregulated. These genes are much less dependent on histone acetylation for their activation.
Acetyl-CoA is a key metabolite for numerous anabolic pathways and fuels energy production from the TCA cycle. As it is also the acetyl donor for protein acetylation, we conducted a proteomic screen in search of newly acetylated proteins in response to increasing acetyl-CoA levels during the transition to growth. We hypothesized that such acetylation events might be sensitive to acetyl-CoA levels and could be involved in regulating cell growth and proliferation. We found that Spt7p, Ada3p and Sgf73p, three components of the multisubunit transcriptional coactivator SAGA are auto-acetylated in the complex in tune with acetyl-CoA levels.2 Since SAGA can function as a histone acetyltransferase,5 we examined histone acetylation over the YMC and found that acetylation of K9, K14, K18, K23 and K27 on H3 as well as K5, K8, K12 on H4 are also very dynamic. The acetylation of these sites begins to increase as soon as cells enter growth.2
Histone acetylation has been linked to transcription activation and replication-coupled nucleosome assembly.6 Given that H3K56ac, a marker for nucleosome assembly, increased at later time points of the YMC corresponding to the time of cell division, the earlier increase in the other marks might be associated with transcription activation. We conducted ChIP PCR and ChIP-Seq analysis to detect SAGA and acetylated histone occupancy on different classes of genes when cells were present in different metabolic states. Strikingly, we observed abundant histone acetylation specifically on genes important for growth (e.g., ribosomal, translation, amino acid biosynthesis genes) precisely during growth (Fig. 1).2 In contrast, acetylated histone occupancy was much lower or completely absent on other classes of genes upon activation.
How might a yeast cell “measure” its acetyl-CoA levels? Gcn5p, the catalytic acetyltransferase within SAGA, has been reported to have a slightly lower affinity for acetyl-CoA,7,8 which is in the physiological range of cellular acetyl-CoA fluctuations.2 Therefore, acetylation catalyzed by Gcn5p could conceivably be regulated by acetyl-CoA substrate availability in vivo. In support of this idea, all of the non-histone proteins that exhibited dynamic acetylation over the YMC that we have found so far are exclusively SAGA targets. Furthermore, we also observed that the acetylation of sites on histone H4 that are catalyzed by the NuA4 complex may also be dependent on Gcn5p/SAGA, as they fail to increase in response to acetyl-CoA in the absence of Gcn5p.2
Moreover, the dynamic acetylation of a protein could also be regulated by deacetylation. In particular, the NAD+-dependent sirtuin class of deacetylases also helps the cell relay its metabolic status through important deacetylation events.9 Several proteomic studies have now shown that acetylation is a much more common post-translational modification than previously anticipated.10,11 Our data reveal that certain protein acetylation events which are driven by acetyl-CoA fluctuations might be particularly important for regulating cell growth and proliferation. It will be of great interest and importance to identify additional such proteins from the expanding acetylated proteome as well as the consequences of their acetylation. Thus, a complex interplay between dynamic acetylation and deacetylation might represent the basis for the regulation of numerous cellular and metabolic processes.
Comment on: Cai L, et al. Mol Cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004.
References
- 1.Zaman S, et al. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
- 2.Cai L, et al. Mol Cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tu BP, et al. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 4.Tu BP, et al. Proc Natl Acad Sci USA. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grant PA, et al. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 6.Brownell JE, et al. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/S0959-437X(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 7.Langer MR, et al. J Biol Chem. 2002;277:27337–27344. doi: 10.1074/jbc.M203251200. [DOI] [PubMed] [Google Scholar]
- 8.Berndsen CE, et al. Curr Opin Struct Biol. 2008;18:682–689. doi: 10.1016/j.sbi.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blander G, et al. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 10.Choudhary C, et al. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 11.Zhao S, et al. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]