Abstract
G protein-mediated signal transduction can transduce signals from a large variety of extracellular stimuli into cells and is the most widely used mechanism for cell communication at the membrane. The RhoGTPase family has been well established as key regulators of cell growth, differentiation and cell shape changes. Among G protein-mediated signal transduction, G12/13-mediated signalling is one mechanism to regulate RhoGTPase activity in response to extracellular stimuli. The alpha subunits of G12 or G13 have been shown to interact with members of the RH domain containing guanine nucleotide exchange factors for Rho (RH–RhoGEF) family of proteins to directly connect G protein-mediated signalling and RhoGTPase signalling. The G12/13–RH–RhoGEF signalling mechanism is well conserved over species and is involved in critical steps for cell physiology and disease conditions, including embryonic development, oncogenesis and cancer metastasis. In this review, we will summarize current progress on this important signalling mechanism.
Keywords: G protein, Galpha12, 13, Rho, RhoGEF
G protein-mediated signal transduction
Heterotrimeric guanine nucleotide-binding proteins (G proteins), composed of α, β and γ subunits, act as molecular switches which cycle between an inactive, GDP-bound state and an active, GTP-bound state (1, 2). These G proteins couple to G-protein coupled receptors (GPCRs) that have a common structure of seven transmembrane alpha helices. About 800 GPCRs form the largest gene family in the human genome to sense and transduce huge arrays of extracellular stimuli, ranging from ions, amines, lipids and proteins. GPCR-G protein signalling mechanisms are involved in the regulation of many cellular processes. A ligand-bound GPCR activates its coupled G protein to facilitate a GDP–GTP exchange reaction on the alpha subunit. The GTP-bound Gα subunit dissociates from the Gβγ dimer, both of which regulate the activity of multiple intracellular effectors to elicit cellular responses. The strength and duration of the signal is determined by the amount and lifetime of Gα–GTP in the cell. All Gα subunits have intrinsic GTPase activity that hydrolyses GTP to GDP, and this rate of hydrolysis is enhanced by GAPs (GTPase-activating proteins) such as RGS (regulator of G protein signalling) proteins, which bind the switch regions of activated Gα subunits, and thus accelerate the hydrolysis of GTP to GDP (3). Once in the GDP-bound state, Gα subunits re-associate with Gβγ and are capable of initiating another round of signalling.
Gα subunits are classified into four subfamilies, Gs, Gi, Gq and G12 (4). Gαs and Gαi are mainly involved in the stimulation and inhibition of adenylyl cyclases, respectively, to regulate the intracellular concentration of cAMP (cyclic adenosine 3′–5′ monophosphate). PLCβ (phospholipase C-β) isozymes are well-established effectors for all members of the Gq subfamily and generate IP3 (inositol 1,4,5 triphosphate) and DG (diacylglycerol) from PIP2 (phosphatidylinositol 4,5 bisphosphate) at the plasma membrane. IP3 increases intracellular Ca2+ levels and DG is involved in the activation of PKC (protein kinase C). In addition to the regulation of soluble intracellular second messengers, it has been clearly demonstrated that Gα12/13 and Gαq are directly involved in the activation of RhoGTPases (5). The RhoGTPases are members of the RAS superfamily of monomeric GTP-binding proteins. It has been demonstrated that the activation of RhoGTPase results in a variety of cellular responses including gene transcription, embryogenesis and rearrangement of the actin cytoskeleton (6). Similar to heterotrimeric G proteins, RhoGTPase also cycles between a GDP-bound and GTP-bound state. RhoGEFs (guanine nucleotide exchange factors for Rho) serve a similar role as GPCRs to facilitate GDP–GTP exchange on Rho (7). Most RhoGEFs contain a common domain structure consisting of tandem DH (Dbl homology) and PH (pleckstrin homology) domains. The DH domain is directly involved in the catalytic activity of GDP–GTP exchange. The Dbl family of RhoGEFs consists of about 80 members and is the largest family of oncogenes (8). This strongly reinforces the critical importance of the regulation of RhoGTPases in cell physiology as well as in cancer. In addition, the critical importance of regulation of Rho activity in cardiovascular diseases, neurological disorders and immunological responses has also been reported. In this review, we will focus on the recent progress on the molecular mechanisms for the regulation of RhoGTPase activity through GPCR-heterotrimeric G12/13-signalling pathways.
Gα12 and Gα13 regulates Rho activity
The G12 subfamily consists of two alpha subunits, Gα12 and Gα13, which are ∼70% identical in amino acid sequence and have similar biochemical properties, such as slow rates of intrinsic GTP hydrolysis and a slow rate of GDP–GTP exchange (9, 10). Both Gα12 and Gα13 are expressed ubiquitously. GPCRs for various ligands such as angiotensin II, endothelin, thrombin, bombesin, thromboxane A2 (TXA2), sphingosine-1-phosphate (S1P) and lysophosphatidic acid (LPA) have been shown to couple to G12/13 (11). The coupling specificity between G12 and G13 is usually not strict and most of these ligands can activate both G12 and G13. It is notable that the most GPCRs which couple to G12/13 also couple to Gq. The physiological significance of this co-coupling is not yet clearly understood, but strongly indicates the importance of simultaneous regulation of both signalling pathways.
Although Gα12 and Gα13 are the most homologous among Gα subunits and share similar biochemical properties, their signalling functions are clearly different. For example, in in vitro reconstitution experiments, Gα13 stimulates the RhoGEF activity of p115RhoGEF and LARG (leukaemia associated RhoGEF) (12–14). On the other hand, Gα12 does not activate the RhoGEF activity of these proteins although it directly interacts with these targets with an affinity similar to Gα13 and Gα12 competes with Gα13 to inhibit Gα13-stimulated RhoGEF activity of p115RhoGEF. In gene knockout experiments, Gα13−/− mice die at day E9.5 due to defects in vascular structure formation (15). On the other hand, Gα12−/− mice are viable and do not show any apparent phenotype. Gα12−/−/Gα13−/− double knockout mice die about 1 day earlier than Gα13−/− single knockout mice (16). These results clearly demonstrate that Gα12 and Gα13 have distinct physiological functions.
Phosphorylation of Gα subunits is an important modification that regulates their function. Gα12 is a substrate for phosphorylation by PKC (17). It was demonstrated that Gα12 over-expressed in NIH3T3 cells is phosphorylated following treatment of the cells with PMA (17), and endogenous Gα12 in human platelets is phosphorylated in response to PMA, thrombin and the TXA2 receptor agonist U46619 (18). Although the phosphorylated residue(s) on Gα12 has not been identified, it is located within the first 50 N-terminal amino acid residues (17). It has been demonstrated that phosphorylated Gα12 loses its affinity for Gβγ, and that the association with Gβγ reciprocally inhibits the phosphorylation of Gα12 by PKC (17). This appears to be consistent with the fact that Gβγ interacts with the N-terminal alpha helix of Gα through one of the seven bladed propellers of the Gβ subunit. As to phosphorylation of Gα13, there is a discrepancy between in vitro and cell-based experiments. In vitro experiments demonstrate purified Gα13 is not a substrate for PKCα (17). However, it was reported that endogenous Gα13 in platelets is phosphorylated in response to PMA, and that Gα13 over-expressed in COS cells is effectively phosphorylated by PKC (18). Thus, some additional factors may be required for PKC-mediated phosphorylation of Gα13 in cells. Interestingly, in Caenorhabditis elegans, genetic experiments suggested that the novel calcium-independent PKCθ/δ is a potential downstream target of Gα12 (19). Further investigation may reveal the important unknown molecular link between Gα12 or Gα13 and PKC.
Gα subunits have been known to be subject to lipid modifications that affect their subcellular localization as well as their interactions with other proteins. Gα subunits are subjected to myristoylation and/or palmitoylation. Gα12 and Gα13 are not myristoylated because they lack a glycine residue as the second amino acid residue. However, both Gα12 and 13 are subjected to palmitoylation at cysteine residues near their N-terminus. Cys12 of Gα12 and Cys14 and Cys18 of Gα13 are candidate residues (20, 21). It was demonstrated that palmitoylation of Gα13 is required for its association with the plasma membrane and its ability to activate RhoA through p115RhoGEF (22). It has also been reported that palmitoylated Gα12 but not Gα13 localizes in lipid rafts (23), and that the NIH3T3 transforming activity of constitutively active mutant of Gα12 can be prevented by blocking its palmitoylation (20).
RH domain containing guanine nucleotide exchange factors for Rho serve as direct effectors for Gα12 and Gα13
One of the cellular effects first identified for Gα12 and Gα13 subunits was their potent cell transforming activities compared with other Gα subunits (24). In addition, wild-type Gα12 was the only Gα subunit that was isolated as an oncogene using a focus formation assay in NIH3T3 cells (25). Subsequently, it was demonstrated that over-expression of constitutively active Gα12 or 13 induces several cellular effects which suggest stimulation of Rho activity in cells, such as formation of actin stress fibres or neurite retraction in neuronal cells (26, 27). However, the exact biochemical mechanism connecting the activation of Gα12/13 to Rho activation was not clearly understood.
A direct link between Gα13 and RhoA was established with the discovery of p115RhoGEF, a RhoA-specific GEF (13) (Fig. 1). In mammals, three members of the RH domain containing guanine nucleotide exchange factors for Rho (RH–RhoGEF) family, that contain an RH (RGS homology) domain in their N-terminus, have been identified: p115RhoGEF, PDZ–RhoGEF and LARG (28–30) (Fig. 2). This G12/13-RH–RhoGEF pathway is extremely well conserved over animal species. Homologous signalling mechanisms were identified both in Drosophila and C. elegans (31–33) (Fig. 2). In Drosophila, genetic experiments demonstrated that the deletion of any component of the Gα13–RhoGEF–RhoA-signalling pathway results in a similar phenotype consisting of embryonic lethality at the stage of gastrulation. These results clearly indicate the fundamental biological importance of this signalling pathway in cell growth, differentiation and development. Since the discovery of RH–RhoGEF as their effectors, several other molecules have also been identified as effectors for Gα12 or Gα13. However, the RH–RhoGEFs have been the most extensively studied and characterized. In the following sections, we will summarize our current understanding of the three mammalian RH–RhoGEFs, p115RhoGEF, LARG and PDZ–RhoGEF.
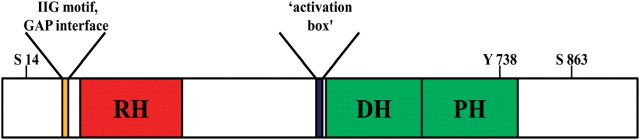
Fig. 3.
Schematic representation of p115RhoGEF highlighting known and potential regulatory elements. The core domains of p115RhoGEF are coloured red (RH) and green (DH/PH module). The region N-terminal to the RH domain containing the IIG motif and residues required for GAP activity is indicated by an orange box (41, 75). The N-terminal extension of the DH domain thought to be important for regulation of basal activity, the ‘activation box,’ is indicated with a blue box (43). Serine (S) and tyrosine (Y) residues reported to be modified by phosphorylation are also indicated. Note that the cartoon is not drawn to scale (53, 54).
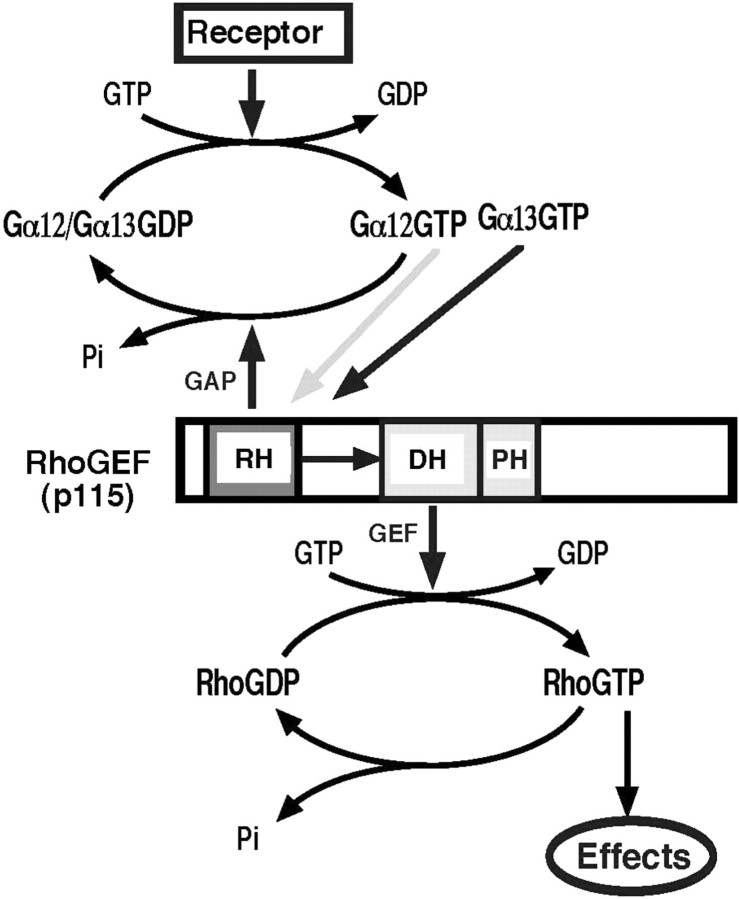
Fig. 1.
Gα12/13-p115RhoGEF-Rho signalling pathway. G12/13-coupled receptor stimulation generates GTP-bound Gα12 or Gα13. Both Gα subunits interact with the RH domain of p115RhoGEF. The interaction with Gα13, but not Gα12, stimulates RhoGEF activity through the DH/PH domains, possibly through a conformational change of the linker region between the RH and DH/PH domains. The RH domain of p115RhoGEF has GAP activity for both Gα12 and Gα13, which terminates the signal.
Fig. 2.

RH–RhoGEF protein family. Upper panel: schematic representation of mammalian RH–RhoGEF members, p115RhoGEF, LARG and PDZ–RhoGEF. Blue:PDZ domain, Red:RH domain, Green: DH-PH domain. Amino acid residue numbers for the domains are shown above. Lower panel: primary sequence alignment of the RH domains from the human RH–RhoGEFs LARG, PDZ–RhoGEF and p115RhoGEF. Identical and similar residues are indicated with a star and a colon, respectively. Semi-conserved residues are indicated with a period. Secondary structural elements were assigned based on the X-ray crystal structure of the RH domain of p115RhoGEF (42), and are indicated below the p115RhoGEF sequence. The IIG motif is highlighted in green while residues required for the GAP activity of p115RhoGEF are highlighted in red. Amino acids are numbered according to the sequences of the full-length proteins. The NCBI reference sequences used to generate the alignment are as follows: LARG (NP_056128.1), PDZ–RhoGEF (NP_055599.1) and p115RhoGEF (NP_004697.2).
p115RhoGEF
Function
p115RhoGEF is a Ras homology (Rho) guanine nucleotide exchange factor that was first discovered through its specificity for binding nucleotide-free RhoA (34). p115RhoGEF binds to the alpha subunits of the G12 family of heterotrimeric G proteins through its N-terminal RH domain. The RH domain allows p115RhoGEF to serve as a GAP, accelerating the intrinsic rate of hydrolysis of GTP to GDP by Gα12 and Gα13 and terminating signalling through Gα12 and Gα13 (12, 34). In addition to the canonical RGS box, the RH domain of p115RhoGEF requires an extra 60 amino acids for full stability of the isolated domain (35). p115RhoGEF activates Rho by catalysing the exchange of GDP for GTP on Rho through its DH domain. Additionally, p115RhoGEF contains a PH domain immediately C-terminal to the DH domain. It is thought that the PH domain works with the RH domain to regulate plasma membrane localization of p115RhoGEF (36).
Activated Gα12 and Gα13 bind p115RhoGEF through the RH domain, which is crucial for its GAP function. However, only Gα13 is able to stimulate the RhoGEF activity of p115RhoGEF in vitro (12, 13, 35). Amino acids 22–31, N-terminal to the RH domain, are necessary for the interaction between p115RhoGEF and activated Gα13 (35, 44) The deletion of 41 amino acids from the N-terminus of the RH domain of p115RhoGEF leads to a loss in the GAP activity of p115RhoGEF, though it can still bind Gα13. Thus, the RH domain itself does not contain the catalytic residues necessary for GAP activity (35). A second, activation-dependent binding site for Gα12/13 was identified between amino acids 288–760 of p115RhoGEF (38). However, Gα13 has no capacity to stimulate the GEF activity of this fragment of p115RhoGEF. Thus, the role of this putative interface in the regulation of p115RhoGEF activity is unclear. Deletion of amino acids 1–288 of p115RhoGEF eliminates its sensitivity to activation by Gα13 (35). Therefore, binding of Gα13 to the RH domain is a critical step in the activation process.
In addition to the interaction with Gα12/13, p115RhoGEF has been shown to interact with a variety of other proteins. For example, p115RhoGEF interacts with the cytoplasmic domain of the human immunodeficiency virus type 1 (HIV-1) transmembrane glycoprotein 41 (gp41) through its DH/PH domains and this interaction inhibits HIV-1 replication (37). p115RhoGEF has also been shown to inhibit thrombin-induced activation of extracellular signal-regulated kinases 1 and 2 (ERK1/2) and p90 ribosomal S6 kinase (p90RSK) through its RH domain in adult rat ventricular myocytes (38). p115RhoGEF interacts with the scaffolding protein connector enhancer of kinase suppressor of Ras 1 (CNKSR1) through its RH and DH/PH domains. CNKSR1 can block stress fibre formation and SRE activation induced by p115RhoGEF, and CNKSR1 acts synergistically with p115RhoGEF to enhance phosphorylation and activation of c-Jun. This enhancement in c-Jun phosphorylation and increase in total c-Jun protein levels indicate increased signalling via the JNK pathway (39). p115RhoGEF functionally couples to CD44 variant 3 (CD44v3), a hyaluronan (HA) receptor found in human metastatic breast tumour cells (MDA-MB-231 cells) (40).
Structure
Although the RH domain of p115RhoGEF exhibits very low primary sequence identity to other RGS proteins, crystallographic analysis indicates that its overall tertiary structure is similar to that of other RGS proteins (12, 41). Additional crystallographic studies have revealed a unique bipartite interaction between the RH domain of p115RhoGEF and Gα13 (41). The interaction between the GAP interface of p115RhoGEF and the switch regions of Gα13 resembles that of RGS4 and RGS9 with their respective Gα proteins (41). Additionally, an effector-like interface between is formed between the RH domain of p115RhoGEF and Gα13. Mutation of residues in Gα13 that contribute to this interface reduce Rho activation in cells, and impair p115RhoGEF activity in vitro (42). Recent mutational analysis of p115RhoGEF revealed the importance of the linker region between RH domain and DH domain for the regulation of GEF activity. In particular, the deletion of amino acids 395–412, which are at the N-terminal extension of the DH domain, significantly decreases the GEF activity of p115RhoGEF in vitro (43). This region of p115 is homologous to the region of LARG that has been shown to be critical in achieving maximum GEF activity (44). This N-terminal extension is well folded in the crystal structure of the DH/PH domains alone, but is disordered in the presence of the whole linker region (43). It is possible that interaction between p115RhoGEF with Gα13 changes the structure of this N-terminal extension region to activate the GEF activity of p115. However, the molecular mechanism by which this conformational change occurs is still unknown.
Mutagenesis and chimeric studies of Gα13 suggest that the binding and activation of RH–RhoGEFs such as p115RhoGEF require most of the GTPase domain of Gα13 in addition to the switch regions (45). The differential GAP response of Gα12 and Gα13 to the RH domain of p115RhoGEF is dependent on the N-terminal α-helical and switch regions, while the C-terminal region (residues 264–377) of Gα13 is responsible for its RhoGEF activity (46). Recently, it has also been shown that the double mutation of two residues in Gα13 that lie in the effector interface with the RH domain of p115RhoGEF, T274E and N278A, eliminates the interaction between p115RhoGEF and Gα13 and abrogates both the GEF and GAP activities of p115RhoGEF in response to Gα13 (42).
p115RhoGEF exhibits synergistic activation of transcription via the serum response element (SRE) in NIH3T3 and HeLa cells when it is co-expressed with a constitutively active mutant of Gα13, but not with constitutively active Gα12 or Gαq (14, 47). Deletion of its C-terminus (amino acids 761–912) allows p115RhoGEF to stimulate SRE activity in NIH 3T3 cells more potently and promotes greater accumulation of GTP-bound endogenous RhoA as compared to full-length p115RhoGEF (48). The C-terminus is also responsible for homo-oligomerization of p115RhoGEF and its murine orthologue, Lsc (Lbc's second cousin) (49, 50), and deletion of the C-terminus (amino acids 801–912) significantly increases the transforming ability of p115RhoGEF in NIH 3T3 cells. However, deletion of the putative dimerization interface fails to enhance Lsc-mediated SRE activation, demonstrating that dimerization and negative regulation are distinct properties of the C-terminus. Deletion of the DH domain or truncation of the PH domain abolishes the transforming activity of p115RhoGEF (49).
Phosphorylation
There is evidence that serine/threonine phosphorylation also plays a role in the regulation of p115RhoGEF activity. For example, thrombin stimulates the phosphorylation of p115RhoGEF in a PKCα-dependent manner (51). PKCα, but not PKCζ, associates with p115RhoGEF, and inhibition of PKCα activity prevents Rho activation and activates endothelial barrier dysfunction (51). PKCα has also recently been reported to phosphorylate p115RhoGEF in response to TNFα stimulation in BEND.3 mouse brain endothelial cells, leading to barrier dysfunction (52). Specific residues that are phosphorylated by PKCα have yet to be identified. Additionally, p115RhoGEF was identified as being phosphorylated in HeLa cells during mitosis, specifically on serine residues 14 and 863 (53). However, the role and mechanism of serine/threonine phosphorylation in regulating p115RhoGEF activity still remains unknown. It has recently been reported that p115RhoGEF is tyrosine phosphorylated by Jak2 on tyrosine residue 738 in a Gαq-dependent manner after stimulation of arterial smooth muscle cells with angiotensin II (54).
Physiology
In mice, Lsc (Lbc's second cousin), the murine orthologue of p115RhoGEF, has primarily been characterized in hematopoietic cells and is implicated in immune system function. Targeted disruption of Lsc in 120/Sv background mice showed that it is required for normal B- and T-lymphocyte function. These Lsc−/− mice have fewer T-cells in the spleen and lymph nodes, a decrease in B-cells in the marginal zone, and impaired thymus-dependent and type 2 thymus-independent immune responses. Lsc−/− marginal zone B (MZB) cells showed enhanced chemotaxis, and proliferation of splenic B-cells is impaired. Lsc−/− T-cells have defects in actin polymerization induced by TXA2 or LPA (55). Lsc−/− mice on the BALB/c background show thymic hyperplasia and an age-dependent increase in thymocytes (56). These Lsc−/− mice are unable to generate a T-dependent (TD) antibody response (IgM). Additionally, their naïve and activated MZB cells exhibit enhanced migration towards S1P, but their integrin-mediated release during migration is impaired (57). In a 50:50 129 Sv/Jae:C57BL/6 background, mice lacking Lsc have peripheral leukocytosis, splenomegaly and extramedullary haematopoiesis, though they show no evidence of local or systemic infections. Cellular recruitment to chemical dermatitis and bacterial peritonitis in these mice is normal. Lsc−/− neutrophils from these mice are unable to generate and sustain dominant pseudopods when stimulated by formyl peptide, and consequently have significantly enhanced chemokinesis and abnormal directional migration. These Lsc−/− neutrophils also have impaired β2 and β1 integrin function (58). Lsc−/− germinal centre B-cells also proliferate faster than wild-type cells (59). Recently, it was also described that mice in the 129/Sv background lacking Lsc exhibit progressive dilatation of the oesophagus. These mice have a thinner muscle layer in the oesophagus, hypertonic lower oesophageal sphincter, and lack regular propulsive peristalsis, traits similar to achalasia in humans. These Lsc−/− mice also have a lower neurite density, reduced NGF (nerve growth factor) expression and fewer GFAP (glial fibrillary acidic protein)-positive glia cells in the oesophagus, which may partially account for the achalasia-like phenotype in these mice (60).
PDZ–RhoGEF
Function
A full-length transcript of PDZ–RhoGEF (known as KIAA380) was first identified in a human brain cDNA library screen for transcripts that could potentially encode large proteins (61). While the function of KIAA380 was not immediately clear, it was recognized to share sequence similarity with Lsc, a recently identified oncogene and putative Dbl family Rho guanine nucleotide exchange factor that was subsequently shown to be a RhoA-specific GEF (62, 63). Kozasa et al. (12) were the first to appreciate the sequence similarity between the N-terminus of KIAA380, canonical GAPs for heterotrimeric G proteins such as RGS2 and RGS4, and other RhoA-specific GEFs. Shortly thereafter, the putative DH/PH domains of KIAA380 were demonstrated to stimulate release of (3H)GDP from RhoA in vitro, but not from Rac1 or Cdc42, directly demonstrating that KIAA380 is a RhoA-specific GEF (64). The murine orthologue of PDZ–RhoGEF, GTRAP48, was identified in a yeast two-hybrid screen for proteins which could interact with the glutamate transporter EAAT4 (65). This study also demonstrated that GTRAP48 could stimulate binding of GTPγS to RhoA but not Rac or CDC42 in vitro, further confirming this protein's function as a RhoA-specific GEF.
Subsequent biochemical analysis of PDZ–RhoGEF confirmed that the isolated DH and PH domains are sufficient to stimulate specific nucleotide exchange on Rho in vitro (66, 67). Furthermore, Togashi et al. demonstrated that over-expression of full-length PDZ–RhoGEF in COS7 cells increased GTP binding to RhoA, but not Rac1 or CDC42, suggesting that it functions as a Rho-specific GEF in a cellular context as well (66). Consistent with this notion, PDZ–RhoGEF has been linked to several Rho-dependent cellular responses such as SRE-mediated gene transcription and neurite retraction (29, 66).
Both GTRAP48 and PDZ–RhoGEF are able to co-immunoprecipitate with activated forms of Gα13, suggesting these proteins act as direct links between heterotrimeric G proteins and RhoA (29, 65). In support of this model, deletion mutants of PDZ–RhoGEF lacking its DH/PH domains abrogate Rho-dependent SRE activation by constitutively active mutants of Gα12/13 (29), or LPA-induced neurite retraction (66), demonstrating indirectly that PDZ–RhoGEF is regulated by G12 family members in a cellular context. In vitro however, neither PDZ–RhoGEF nor GTRAP48 have detectable GAP activity towards Gα12 or Gα13 and stimulation of the guanine nucleotide exchange activity of PDZ–RhoGEF/GTRAP48 by these heterotrimeric G proteins has not been detected under the conditions tested (35). Thus, the precise mechanism by which G12 family members regulate the RhoGEF activity of PDZ–RhoGEF and GTRAP48 remains unknown. PDZ–RhoGEF may regulate Rho activity independently of the interaction with G12 family members by virtue of its PDZ domain, which has been shown to interact with a variety of cell surface receptors including GPCRs and plexins (68–71).
Structure
The structure of full-length PDZ–RhoGEF has not been solved; however the structures of its core domains have been determined individually, in some cases both in the presence and absence of accessory proteins. The primary sequence of the RH domain of PDZ–RhoGEF is distantly related to that of ‘classical’ RGS proteins such as RGS2 and RGS4, with sequence identities of ∼20% (12). Despite this fact, X-ray crystallography revealed that the RH domain of PDZ– RhoGEF adopts a tertiary structure very similar to that of other RGS proteins (72). Like the RGS domain of RGS4 (73), the core RGS box of PDZ–RhoGEF is formed by 9 α helices, with helices 4–7 forming a classical anti-parallel bundle. In contrast to other RGS proteins, the RH domain of PDZ–RhoGEF also contains three additional helices C-terminal to the RGS box which are tightly associated with helices in the core RGS domain via hydrophobic interactions. These additional helices are conserved in both p115RhoGEF and LARG, and are a key feature that distinguishes these RH domains from canonical RGS proteins.
The RH domain of PDZ–RhoGEF has also been crystallized with the inactive (GDP-bound), active (GTPγS-bound) and transition state (AlF4–-bound) forms of Gα13 (74). The interaction between the RH domain and GDP-bound Gα13 was particularly surprising especially in light of the fact that PDZ–RhoGEF was able to maintain the switch regions of Gα13 in an activated conformation. This suggests that GTP hydrolysis may not be sufficient to terminate the Gα13–PDZ–RhoGEF interaction, and perhaps Gα13 must be sequestered by Gβγ to fully conclude the signalling cycle.
The RH domain of PDZ–RhoGEF interacts with Gα13 through multiple intermolecular interfaces. One point of contact is centred on an IIG (isoleucine–isoleucine–glycine) motif found N-terminal to the RGS box. This motif forms multiple contacts with the α helical domain of Gα13, and is conserved in other RH–RhoGEFs. As the primary sequence of Gα subunits is most divergent in the α helical domain, this interface may represent one mechanism by which RH–RhoGEFs specifically recognize Gα13. Interaction with the IIG motif may also facilitate activation of Gα13 by restraining its α helical and Ras-like domains in their active conformation.
Additionally, Gα13 interacts with an acidic stretch of residues N-terminal to the core RGS box of PDZ–RhoGEF. While the analogous residues in p115RhoGEF confer GAP activity for Gα13 (75), PDZ–RhoGEF has no detectable effect on GTP hydrolysis by Gα13 in vitro (35). This lack of GAP activity on the part of PDZ–RhoGEF is readily explained by the structure, as key residues in the acidic patch of PDZ–RhoGEF fail to make productive interactions with catalytic residues in the switch regions of Gα13. In addition to the contacts formed with Gα13 through elements N-terminal to the RGS box, the RH domain of PDZ–RhoGEF also utilizes residues in the unique C-terminal extension of the RGS box to dock into the cleft formed by the α2 and α3 helices of Gα13, which a highly conserved effector binding site on other G protein α subunits (reviewed in (76)). This suggests that this interface may play a role in regulation of PDZ–RhoGEF activity by Gα13. Although Gα13 has been implicated in regulation of PDZ–RhoGEF activity in cell-based experiments (29), direct activation of this GEF by Gα13 in vitro has not been detectable under the conditions tested. Thus, additional regulatory mechanisms may be required to render PDZ–RhoGEF susceptible to regulation by Gα13.
A crystal structure of the DH/PH domains of PDZ–RhoGEF bound to nucleotide-free RhoA has been determined by X-ray crystallography (67). The DH domain is a helical bundle which assumes the classical ‘chaise lounge’ configuration characteristic of Dbl family proteins (reviewed in (8)). As with other RhoGEFs, conserved regions 1 and 3 of the DH domain contact switch I in RhoA, while switch II is engaged by conserved region 3 and the α6 helix. The latter set of interactions is believed to dictate the specificity for the GTPase. Subsequent mutational analysis of RhoA has defined the precise residues dictating its interaction with PDZ–RhoGEF (77).
The PH domain of PDZ–RhoGEF also assumes a fold common to this domain family in the form of an anti-parallel β sandwich capped with an α helix. However, the PH domain of PDZ–RhoGEF has several features that distinguish it from classical PH domains. The β4 strand contains an 18 amino acid residue insertion which includes an uncharacteristic proline. This results in a β-strand which protrudes into the putative phosphoinositide-binding pocket. Additionally, this pocket is lined with fewer positively charged residues than are found in the PH domains of other RhoGEFs like Dbs, suggesting that binding to phosphoinositides may not be a major function of this particular PH domain. Interestingly, the PH domain also makes direct contact with RhoA, though the interaction surface is small, <10% of the total surface area buried in the complex. It remains unclear to what extent the PH domain influences nucleotide exchange under physiological conditions, although, in vitro, the DH/PH tandem is a more efficient GEF than the isolated DH domain (67, 77), suggesting that the PH domain likely influences GEF activity in a positive manner. Studies of the DH/PH domains of PDZ–RhoGEF in the presence and absence of nucleotide-free RhoA in solution are consistent with models determined by X-ray crystallography (78).
Another unusual feature of the PH domain of PDZ–RhoGEF is its capacity to bind directly to the GTPγS-bound form of RhoA (79). Although the precise regulatory implications of this interaction are currently unknown, association with activated RhoA may be one mechanism that recruits PDZ–RhoGEF to the plasma membrane, where its target, geranylgeranylated RhoA, is presumably enriched. This putative feed-forward mechanism is consistent with the observations that while RhoA–GTPγS does not have a detectable effect on the GEF activity of PDZ–RhoGEF in vitro, the DH/PH module of PDZ–RhoGEF can be isolated in a stable ternary complex with both nucleotide-free RhoA and RhoA–GTPγS.
In vitro biochemical studies analysing the basal GEF activity of PDZ–RhoGEF truncation mutants have identified an acidic stretch of residues N-terminal to the DH domain that may play a role in autoinhibition of its exchange activity (80). NMR spectroscopy suggests that residues in this negatively charged sequence motif may suppress basal GEF activity by interacting with positively charged residues in the DH domain of PDZ–RhoGEF. While mutation of amino acids in the acidic cluster could enhance the GEF activity of the isolated DH/PH domains in vitro, in the presence of the RH domain these mutations had little effect, suggesting that the RH domain may work in concert with the acidic motif to regulate nucleotide exchange.
A structure of the PDZ domain of PDZ–RhoGEF has been solved by NMR (Protein Data Bank ID 2dls).
Phosphorylation
Early studies of cytoskeletal reorganization mediated by G12 family members suggested that tyrosine kinases may play a role in regulating Rho activation. Tyrosine kinase inhibitors have been reported to block various Rho-dependent processes such as Gα13-induced neurite retraction and cell rounding, the assembly of stress fibres and focal adhesions in response to LPA stimulation, and Rho activation itself (27, 81–83). More specifically, subsequent work has demonstrated that the non-receptor tyrosine kinase FAK (focal adhesion kinase) may play a role in the regulation of PDZ–RhoGEF activity (83). Tyrosine phosphorylation of over-expressed PDZ–RhoGEF can be enhanced by co-expression with an activated form of FAK, and challenging cells co-expressing PDZ–RhoGEF and wild-type FAK with thrombin also results in an increase in PDZ–RhoGEF phosphorylation. This tyrosine phosphorylation occurs in the region C-terminal to the PH domain of PDZ–RhoGEF, and enhances its exchange activity in cell-based assays. However, the precise mechanism(s) by which FAK-mediated tyrosine phosphorylation of PDZ–RhoGEF regulates its activity has yet to be determined.
PDZ–RhoGEF has also been reported to be tyrosine phosphorylated in rat vascular smooth cells stimulated with angiotensin II, and co-immunoprecipitation studies suggested that PDZ–RhoGEF may associate with the non-receptor tyrosine kinase PYK2 (84, 85). PDZ–RhoGEF immunoprecipitated from these cells can be tyrosine phosphorylated by a constitutively active mutant of PYK2 in vitro, and this modification appears to enhance basal GEF activity in vitro as well (84).
Yeast two-hybrid screening identified a serine/threonine kinase, p21-activated kinase 4 (PAK4), as a putative binding partner of PDZ–RhoGEF (86). PAK4 is able to phosphorylate a peptide consisting of the last 100 amino acid residues of PDZ–RhoGEF in vitro and over-expression of full-length PDZ–RhoGEF and a constitutively active mutant of PAK4 in mammalian cells results in increased serine/threonine phosphorylation of PDZ–RhoGEF, suggesting that PDZ–RhoGEF is a substrate for this kinase. One functional consequence of enhanced phosphorylation of PDZ–RhoGEF appears to be a decrease in LPA-induced Rho activation and stress fibre formation (86). Additionally, the interaction between PAK4 and PDZ–RhoGEF provides a potential mechanism for cross-talk between RhoA and other monomeric GTPases acting upstream of PAK4, such as Cdc42.
Physiology
The phenotype(s) of the PDZ–RhoGEF knockout mouse have not been reported.
LARG
Function
RH–RhoGEFs combine GAP activity and effector activity into a single molecule. In vitro, leukaemia-associated RhoGEF (LARG) and p115RhoGEF act as specific GAPs for Gα12 and Gα13, while the RH domain of PDZ–RhoGEF lacks detectable GAP activity for these Gα subunits (12–14). Biochemical evidence using reconstitution systems with purified proteins has demonstrated that the GEF activity of LARG and p115RhoGEF can be directly stimulated by Gα13, but not Gα12 (13, 14). LARG activation requires not only binding to activated Gα12 but also phosphorylation by a non-receptor tyrosine kinase (13, 14). While the molecular mechanism of RH–RhoGEF activation upon Gα13 binding remains unclear, some studies have recently provided information about the interface between Gα13 and RH–RhoGEFs required for RH–RhoGEF activation. A study utilizing chimeras of Gα12 and Gα13 identified that the C-terminal 100 amino acid residues of Gα13 are required for activation of the GEF activity of p115RhoGEF and LARG, whereas the N-terminal an alpha helical and switch regions of Gα12 and Gα13 are responsible for their differential GAP responses to the RH domain (46). Kinetic and thermodynamic analysis of the interaction between Gα13 and LARG using surface plasmon resonance has demonstrated that the simultaneous binding of the RH domain and DH/PH domains with Gα13 facilitates formation of the high affinity active Gα13–LARG complex (87). Furthermore, Suzuki et al. have demonstrated that the interaction of Gα13 with LARG through the RH domain (a GAP interface) and the DH/PH domains (an effector interface) could coordinate together to stimulate the RhoGEF activity of LARG. This result demonstrated that interaction through multiple interfaces, including the RH domains and DH/PH domains of RH–RhoGEFs, and Gα13 might play an important role in stimulating GEF activity. It is noteworthy that the GEF activity of LARG and PDZ–RhoGEF is activated by binding of their PDZ domains with plexin B1 or non-phosphorylated insulin-like growth factor-1 (IGF-1) receptor, independently of G12/13 activation (69–71, 88, 89). Interestingly, the association was constitutive, and ligand binding induces Rho activation.
It has been shown that deletion of the C-termini of RH–RhoGEFs potentiates their abilities to stimulate Rho activation as compared to the full length protein in cells, while deletion of the C-terminus of PDZ–RhoGEF or p115RhoGEF does not affect or even reduces the RhoGEF activity at the basal state in vitro (48, 49). These results suggest that the RH–RhoGEF's activity may be negatively regulated in vivo through the C-terminus. The discrepancy between in vitro and cell-based studies suggests that ancillary factors in the cellular milieu might release the inhibition of the RH–RhoGEF's activity through the C-terminal region. One possibility is post-translational modification at the C-terminal region. PDZ–RhoGEF is reported to be tyrosine phosphorylated by FAK at its C-terminus (49). Another possibility is the interaction of proteins with their C-termini. Several proteins have been reported to interact with the C-termini of RH–RhoGEFs. The active form of Gα13 binds to the C-terminus of LARG (87). The Rho effector Dia1 also binds to the C-terminus of LARG to potentiate its GEF activity, and its activation constitutes a positive feedback loop between LARG, RhoA and Dia1 (90). Several studies have reported that RH–RhoGEFs oligomerize via their C-terminal regions (49, 50, 91). A co-immunoprecipitation experiment using cells over-expressing RH–RhoGEFs showed that p115RhoGEF, LARG and PDZ–RhoGEF homo-oligomerize via their C-terminal regions and LARG and PDZ–RhoGEF possibly form hetero-oligomers with each other, but not with p115RhoGEF (49). A recent study has suggested that oligomerization of LARG may regulate its intracellular localization. Oligomerization functions to prevent nucleocytoplasmic shuttling and to retain LARG in the cytoplasm, while the mechanism for regulation of LARG oligomerization and the function of LARG localized in the nucleus are unknown (91).
Structure
Structures have been determined for the LARG DH/PH domains, both alone and in complex with RhoA (44). Although the conformation of the DH/PH domains of LARG is comparable to other solved structures of Dbl family members, LARG's DH domain contains a flexible extension at its N-terminus, which binds directly to the Switch I region of RhoA. Deletion or mutation of this region decreases the GEF activity of the DH/PH domains in vitro compared to the wild-type domains. The PH domain directly contacts with RhoA, which is allowed by considerable conformational change between DH and PH domains through the relatively long helical linker upon binding to RhoA. Although this interface contributes to <10% of the total buried surface area in the LARG DH/PH–RhoA complex, mutation of residues in this interface diminished GEF activity to the level of the DH domain alone. On the other hand, mutation of residues in the PDZRhoGEF interface did not lead to significant differences in vitro (77). A conserved hydrophobic patch in the PH domain of LARG is supposed to be in position to bind peripheral membrane proteins or domains at the cell membrane. Mutation of residues in the hydrophobic patch of LARG had no effect on nucleotide exchange activity in vitro but abrogated the ability of LARG to induce RhoA activation and stress fibre formation in cells (92). The hydrophobic patch might contribute to the proper localization of LARG by interacting with unknown targets at the cell membrane.
Phosphorylation
One mechanism regulating RH–RhoGEF activity is post-translational modification in the form of phosphorylation. Several studies have reported that tyrosine kinase modulates the RhoGEF activity of LARG. Chikumi et al. have demonstrated that over-expression of the non-receptor tyrosine kinase FAK induces phosphorylation of both LARG and PDZ–RhoGEF, but not p115RhoGEF in cells, and have concluded that FAK enhances RhoGEF activity even in the absence of Gα12 or Gα13 (83). They also observed that FAK can be activated by thrombin, Gα12, Gα13 and Gαq through both Rho-dependent and -independent mechanisms, and proposed the existence of positive feedback regulation between Rho and FAK (83). Another group showed that LARG is activated through the Src family tyrosine kinase Fyn in response to stimulation of integrins triggered with tensional force (93). Another study using reconstitution and cell-based assays has demonstrated that Gα12 can stimulate the RhoGEF activity of tyrosine-phosphorylated LARG, but not non-phosphorylated LARG (14). The direct phosphorylation of LARG by a non-receptor tyrosine kinase, Tec, in vitro greatly enhances the RhoGEF activity of LARG in response to Gα12, while it does not affect its basal RhoGEF activity. Although binding of Tec to Gα12 in cells was also demonstrated, the mechanism by which Tec is phosphorylated downstream of Gα12 remains unclear. It is interesting to note that thrombin, which can activate the Gα12/13 pathway, can also activate Tec (94), and that Gα12 promotes the kinase activity of Bruton's tyrosine kinase (BTK), another member of the Tec family (95). Furthermore, over-expression of Gα12 and Gα13 stimulates autophosphorylation and transphosphorylation of Tec in NIH 3T3 cells (96). However, it is likely that activated Gα12 may recruit Tec to LARG and facilitate its phosphorylation. In contrast to the study by Suzuki et al., early studies of Gα12/13-mediated cytoskeletal reorganization suggested that tyrosine kinases might play a role in regulating Rho activation downstream of Gα13, but not Gα12 in PC12 cells or SWISS 3T3 cells (81, 82).
Physiology
It is noteworthy that unlike Ras, activating mutations in Rho have not been found in human cancers (97–99). On the other hand, the regulators of Rho activation, RhoGEFs have been isolated in screens for transforming genes. LARG (leukaemia-associated RhoGEF) was one of the very few RhoGEFs that have been found mutated in human cancers. LARG was originally identified as a novel protein fused to the MLL (mixed lineage leukaemia) gene in a patient with acute myeloid leukaemia (30). The MLL–LARG rearrangement is expressed as an in-frame fusion from the MLL–promoter. MLL–LARG fusion protein contains not only the DH/PH domains responsible for Rho activation but also the RH domain and a nuclear localization signal. The mechanism how this molecule causes the aberrant signalling to promote leukaemogenesis remains unknown. It is also reported that LARG expression is dramatically increased in bone marrow of patients with the pre-leukaemic disorder Shwachman–Diamond syndrome (100). A recent study demonstrated that LARG is frequently under-expressed in many breast and colorectal carcinomas, which was significantly associated with genomic loss (101). LARG was then proposed for a candidate tumour suppressor gene, since over-expression of LARG by stable transfection in colorectal cancer cells resulted in reduced cell proliferation and a markedly slower cell migration rate.
It is believed that upon binding with vasoconstrictors, receptors coupling to both Gq/11 and G12/13 stimulate phosphorylation of myosin light chain (MLC) via Ca2+/MLC kinase- and Rho/Rho kinase-mediated signalling pathways, respectively, and regulate vascular smooth muscle tone (26). Recently, mice with conditional Gα12/13 double deficiencies in smooth muscle cells have been developed (102). Aortic segments from Gα12/13 deficient mice show impaired contractile responses to the vasoconstrictors angiotensin II, TXA2 and endothelin I. Furthermore, these mice are almost completely protected from salt-induced hypertension, while their basal blood pressure is unaffected. Similar phenotypes were observed in mice lacking LARG in smooth muscle cells (SMC). These findings suggest that the Gα12/13–LARG pathway is a key regulator of vascular smooth muscle tone in the context of hypertension. On the other hand, a recent study demonstrated that through LARG, S1P receptor 2 signals promote SMC differentiation, but not SMC proliferation or migration (103).
G12/13-RH–RhoGEF pathways in cancer
As stated above it is clear that this signalling pathway is its essentially involvement in the biology of cancer such as, cell transformation, cancer growth, cancer metastasis, cell migration, or angiogenesis. Indeed, many G12/13 coupled receptors, such as LPA, S1P, GRH, endothelin, angiotensin II and CXCR4 have been reported to be involved in many cancer phenotypes (104). Further detailed structural and dynamic studies are critically required to develop novel anti-cancer drugs in the future.
Funding
National Institutes of Health (Grants GM61454 and GM074001 to T.K.); grant for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to N.S.); Funding Program for World-Leading Innovative R&D on Science and Technology (to T.K.); post-doctoral fellowship from the Japan Society for the Promotion of Science (to N.H.).
Conflict of interest
None declared.
References
- 1.Hepler JR, Gilman AG. G proteins. Trends Biochem. Sci. 1992;17:383–387. doi: 10.1016/0968-0004(92)90005-t. [DOI] [PubMed] [Google Scholar]
- 2.Kaziro Y, Itoh H, Kozasa T, Nakafuku M, Satoh T. Structure and function of signal-transducing GTP-binding proteins. Annu. Rev. Biochem. 1991;60:349–400. doi: 10.1146/annurev.bi.60.070191.002025. [DOI] [PubMed] [Google Scholar]
- 3.Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu. Rev. Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- 4.Simon MI, Strathmann MP, Gautam N. Diversity of G proteins in signal transduction. Science. 1991;252:802–808. doi: 10.1126/science.1902986. [DOI] [PubMed] [Google Scholar]
- 5.Aittaleb M, Boguth CA, Tesmer JJ. Structure and function of heterotrimeric G protein-regulated Rho guanine nucleotide exchange factors. Mol. Pharmacol. 2010;77:111–125. doi: 10.1124/mol.109.061234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt A, Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- 8.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell. Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 9.Kozasa T, Gilman AG. Purification of recombinant G proteins from Sf9 cells by hexahistidine tagging of associated subunits. Characterization of alpha 12 and inhibition of adenylyl cyclase by alpha. J. Biol. Chem. 1995;270:1734–1741. doi: 10.1074/jbc.270.4.1734. [DOI] [PubMed] [Google Scholar]
- 10.Singer WD, Miller RT, Sternweis PC. Purification and characterization of the alpha subunit of G13. J. Biol. Chem. 1994;269:19796–19802. [PubMed] [Google Scholar]
- 11.Riobo NA, Manning DR. Receptors coupled to heterotrimeric G proteins of the G12 family. Trends Pharmacol. Sci. 2005;26:146–154. doi: 10.1016/j.tips.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 12.Kozasa T, Jiang X, Hart MJ, Sternweis PM, Singer WD, Gilman AG, Bollag G, Sternweis PC. p115 RhoGEF, a GTPase activating protein for Galpha12 and Galpha13. Science. 1998;280:2109–2111. doi: 10.1126/science.280.5372.2109. [DOI] [PubMed] [Google Scholar]
- 13.Hart MJ, Jiang X, Kozasa T, Roscoe W, Singer WD, Gilman AG, Sternweis PC, Bollag G. Direct stimulation of the guanine nucleotide exchange activity of p115 RhoGEF by Galpha13. Science. 1998;280:2112–2114. doi: 10.1126/science.280.5372.2112. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki N, Nakamura S, Mano H, Kozasa T. Galpha 12 activates Rho GTPase through tyrosine-phosphorylated leukemia-associated RhoGEF. Proc. Natl Acad. Sci. USA. 2003;100:733–738. doi: 10.1073/pnas.0234057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Offermanns S, Mancino V, Revel JP, Simon MI. Vascular system defects and impaired cell chemokinesis as a result of Galpha13 deficiency. Science. 1997;275:533–536. doi: 10.1126/science.275.5299.533. [DOI] [PubMed] [Google Scholar]
- 16.Offermanns S. In vivo functions of heterotrimeric G-proteins: studies in Galpha-deficient mice. Oncogene. 2001;20:1635–1642. doi: 10.1038/sj.onc.1204189. [DOI] [PubMed] [Google Scholar]
- 17.Kozasa T, Gilman AG. Protein kinase C phosphorylates G12 alpha and inhibits its interaction with G beta gamma. J. Biol. Chem. 1996;271:12562–12567. doi: 10.1074/jbc.271.21.12562. [DOI] [PubMed] [Google Scholar]
- 18.Offermanns S, Hu YH, Simon MI. Galpha12 and galpha13 are phosphorylated during platelet activation. J. Biol. Chem. 1996;271:26044–26048. doi: 10.1074/jbc.271.42.26044. [DOI] [PubMed] [Google Scholar]
- 19.van der Linden AM, Moorman C, Cuppen E, Korswagen HC, Plasterk RH. Hyperactivation of the G12-mediated signaling pathway in Caenorhabditis elegans induces a developmental growth arrest via protein kinase C. Curr. Biol. 2003;13:516–521. doi: 10.1016/s0960-9822(03)00164-7. [DOI] [PubMed] [Google Scholar]
- 20.Jones TL, Gutkind JS. Galpha12 requires acylation for its transforming activity. Biochemistry. 1998;37:3196–3202. doi: 10.1021/bi972253j. [DOI] [PubMed] [Google Scholar]
- 21.Ponimaskin E, Behn H, Adarichev V, Voyno-Yasenetskaya TA, Offermanns S, Schmidt MF. Acylation of Galpha(13) is important for its interaction with thrombin receptor, transforming activity and actin stress fiber formation. FEBS Lett. 2000;478:173–177. doi: 10.1016/s0014-5793(00)01845-7. [DOI] [PubMed] [Google Scholar]
- 22.Bhattacharyya R, Wedegaertner PB. Galpha 13 requires palmitoylation for plasma membrane localization, Rho-dependent signaling, and promotion of p115-RhoGEF membrane binding. J. Biol. Chem. 2000;275:14992–14999. doi: 10.1074/jbc.M000415200. [DOI] [PubMed] [Google Scholar]
- 23.Waheed AA, Jones TL. Hsp90 interactions and acylation target the G protein Galpha 12 but not Galpha 13 to lipid rafts. J. Biol. Chem. 2002;277:32409–32412. doi: 10.1074/jbc.C200383200. [DOI] [PubMed] [Google Scholar]
- 24.Voyno-Yasenetskaya TA, Pace AM, Bourne HR. Mutant alpha subunits of G12 and G13 proteins induce neoplastic transformation of Rat-1 fibroblasts. Oncogene. 1994;9:2559–2565. [PubMed] [Google Scholar]
- 25.Chan AM, Fleming TP, McGovern ES, Chedid M, Miki T, Aaronson SA. Expression cDNA cloning of a transforming gene encoding the wild-type G alpha 12 gene product. Mol. Cell. Biol. 1993;13:762–768. doi: 10.1128/mcb.13.2.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gohla A, Schultz G, Offermanns S. Role for G(12)/G(13) in agonist-induced vascular smooth muscle cell contraction. Circ. Res. 2000;87:221–227. doi: 10.1161/01.res.87.3.221. [DOI] [PubMed] [Google Scholar]
- 27.Kranenburg O, Poland M, van Horck FP, Drechsel D, Hall A, Moolenaar WH. Activation of RhoA by lysophosphatidic acid and Galpha12/13 subunits in neuronal cells: induction of neurite retraction. Mol. Biol. Cell. 1999;10:1851–1857. doi: 10.1091/mbc.10.6.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukuhara S, Chikumi H, Gutkind JS. Leukemia-associated Rho guanine nucleotide exchange factor (LARG) links heterotrimeric G proteins of the G(12) family to Rho. FEBS Lett. 2000;485:183–188. doi: 10.1016/s0014-5793(00)02224-9. [DOI] [PubMed] [Google Scholar]
- 29.Fukuhara S, Murga C, Zohar M, Igishi T, Gutkind JS. A novel PDZ domain containing guanine nucleotide exchange factor links heterotrimeric G proteins to Rho. J. Biol. Chem. 1999;274:5868–5879. doi: 10.1074/jbc.274.9.5868. [DOI] [PubMed] [Google Scholar]
- 30.Kourlas PJ, Strout MP, Becknell B, Veronese ML, Croce CM, Theil KS, Krahe R, Ruutu T, Knuutila S, Bloomfield CD, Caligiuri MA. Identification of a gene at 11q23 encoding a guanine nucleotide exchange factor: evidence for its fusion with MLL in acute myeloid leukemia. Proc. Natl Acad. Sci. USA. 2000;97:2145–2150. doi: 10.1073/pnas.040569197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hacker U, Perrimon N. DRhoGEF2 encodes a member of the Dbl family of oncogenes and controls cell shape changes during gastrulation in Drosophila. Genes Dev. 1998;12:274–284. doi: 10.1101/gad.12.2.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrett K, Leptin M, Settleman J. The Rho GTPase and a putative RhoGEF mediate a signaling pathway for the cell shape changes in Drosophila gastrulation. Cell. 1997;91:905–915. doi: 10.1016/s0092-8674(00)80482-1. [DOI] [PubMed] [Google Scholar]
- 33.Yau DM, Yokoyama N, Goshima Y, Siddiqui ZK, Siddiqui SS, Kozasa T. Identification and molecular characterization of the G alpha12-Rho guanine nucleotide exchange factor pathway in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA. 2003;100:14748–14753. doi: 10.1073/pnas.2533143100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hart MJ, Sharma S, elMasry N, Qiu RG, McCabe P, Polakis P, Bollag G. Identification of a novel guanine nucleotide exchange factor for the Rho GTPase. J. Biol. Chem. 1996;271:25452–25458. doi: 10.1074/jbc.271.41.25452. [DOI] [PubMed] [Google Scholar]
- 35.Wells CD, Liu MY, Jackson M, Gutowski S, Sternweis PM, Rothstein JD, Kozasa T, Sternweis PC. Mechanisms for reversible regulation between G13 and Rho exchange factors. J. Biol. Chem. 2002;277:1174–1181. doi: 10.1074/jbc.M105274200. [DOI] [PubMed] [Google Scholar]
- 36.Bhattacharyya R, Wedegaertner PB. Characterization of G alpha 13-dependent plasma membrane recruitment of p115RhoGEF. Biochem. J. 2003;371:709–720. doi: 10.1042/BJ20021897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H, Wang L, Kao S, Whitehead IP, Hart MJ, Liu B, Duus K, Burridge K, Der CJ, Su L. Functional interaction between the cytoplasmic leucine-zipper domain of HIV-1 gp41 and p115-RhoGEF. Curr. Biol. 1999;9:1271–1274. doi: 10.1016/s0960-9822(99)80511-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snabaitis AK, Muntendorf A, Wieland T, Avkiran M. Regulation of the extracellular signal-regulated kinase pathway in adult myocardium: differential roles of G(q/11), Gi and G(12/13) proteins in signalling by alpha1-adrenergic, endothelin-1 and thrombin-sensitive protease-activated receptors. Cell Signal. 2005;17:655–664. doi: 10.1016/j.cellsig.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 39.Jaffe AB, Hall A, Schmidt A. Association of CNK1 with Rho guanine nucleotide exchange factors controls signaling specificity downstream of Rho. Curr. Biol. 2005;15:405–412. doi: 10.1016/j.cub.2004.12.082. [DOI] [PubMed] [Google Scholar]
- 40.Bourguignon LY, Singleton PA, Zhu H, Diedrich F. Hyaluronan-mediated CD44 interaction with RhoGEF and Rho kinase promotes Grb2-associated binder-1 phosphorylation and phosphatidylinositol 3-kinase signaling leading to cytokine (macrophage-colony stimulating factor) production and breast tumor progression. J. Biol. Chem. 2003;278:29420–29434. doi: 10.1074/jbc.M301885200. [DOI] [PubMed] [Google Scholar]
- 41.Chen Z, Singer WD, Sternweis PC, Sprang SR. Structure of the p115RhoGEF rgRGS domain-Galpha13/i1 chimera complex suggests convergent evolution of a GTPase activator. Nat. Struct. Mol. Biol. 2005;12:191–197. doi: 10.1038/nsmb888. [DOI] [PubMed] [Google Scholar]
- 42.Hajicek N, Kukimoto-Niino M, Mishima-Tsumagari C, Chow CR, Shirouzu M, Terada T, Patel M, Yokoyama S, Kozasa T. Identification of critical residues in G(alpha)13 for stimulation of p115RhoGEF activity and the structure of the G(alpha)13-p115RhoGEF regulator of G protein signaling homology (RH) domain complex. J. Biol. Chem. 2011;286:20625–20636. doi: 10.1074/jbc.M110.201392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Z, Guo L, Sprang SR, Sternweis PC. Modulation of a GEF switch: autoinhibition of the intrinsic guanine nucleotide exchange activity of p115-RhoGEF. Protein Sci. 2011;20:107–117. doi: 10.1002/pro.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kristelly R, Gao G, Tesmer JJ. Structural determinants of RhoA binding and nucleotide exchange in leukemia-associated Rho guanine-nucleotide exchange factor. J. Biol. Chem. 2004;279:47352–47362. doi: 10.1074/jbc.M406056200. [DOI] [PubMed] [Google Scholar]
- 45.Vazquez-Prado J, Miyazaki H, Castellone MD, Teramoto H, Gutkind JS. Chimeric G alpha i2/G alpha 13 proteins reveal the structural requirements for the binding and activation of the RGS-like (RGL)-containing Rho guanine nucleotide exchange factors (GEFs) by G alpha 13. J. Biol. Chem. 2004;279:54283–54290. doi: 10.1074/jbc.M410594200. [DOI] [PubMed] [Google Scholar]
- 46.Kreutz B, Hajicek N, Yau DM, Nakamura S, Kozasa T. Distinct regions of Galpha13 participate in its regulatory interactions with RGS homology domain-containing RhoGEFs. Cell Signal. 2007;19:1681–1689. doi: 10.1016/j.cellsig.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 47.Mao J, Yuan H, Xie W, Wu D. Guanine nucleotide exchange factor GEF115 specifically mediates activation of Rho and serum response factor by the G protein alpha subunit Galpha13. Proc. Natl Acad. Sci. USA. 1998;95:12973–12976. doi: 10.1073/pnas.95.22.12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wells CD, Gutowski S, Bollag G, Sternweis PC. Identification of potential mechanisms for regulation of p115 RhoGEF through analysis of endogenous and mutant forms of the exchange factor. J. Biol. Chem. 2001;276:28897–28905. doi: 10.1074/jbc.M102913200. [DOI] [PubMed] [Google Scholar]
- 49.Chikumi H, Barac A, Behbahani B, Gao Y, Teramoto H, Zheng Y, Gutkind JS. Homo- and hetero-oligomerization of PDZ-RhoGEF, LARG and p115RhoGEF by their C-terminal region regulates their in vivo Rho GEF activity and transforming potential. Oncogene. 2004;23:233–240. doi: 10.1038/sj.onc.1207012. [DOI] [PubMed] [Google Scholar]
- 50.Eisenhaure TM, Francis SA, Willison LD, Coughlin SR, Lerner DJ. The Rho guanine nucleotide exchange factor Lsc homo-oligomerizes and is negatively regulated through domains in its carboxyl terminus that are absent in novel splenic isoforms. J. Biol. Chem. 2003;278:30975–30984. doi: 10.1074/jbc.M303277200. [DOI] [PubMed] [Google Scholar]
- 51.Holinstat M, Mehta D, Kozasa T, Minshall RD, Malik AB. Protein kinase Calpha-induced p115RhoGEF phosphorylation signals endothelial cytoskeletal rearrangement. J. Biol. Chem. 2003;278:28793–28798. doi: 10.1074/jbc.M303900200. [DOI] [PubMed] [Google Scholar]
- 52.Peng J, He F, Zhang C, Deng X, Yin F. Protein kinase C-alpha signals P115RhoGEF phosphorylation and RhoA activation in TNF-alpha-induced mouse brain microvascular endothelial cell barrier dysfunction. J. Neuroinflammation. 2011;8:28. doi: 10.1186/1742-2094-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc. Natl Acad. Sci. USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guilluy C, Bregeon J, Toumaniantz G, Rolli-Derkinderen M, Retailleau K, Loufrani L, Henrion D, Scalbert E, Bril A, Torres R M, Offermanns S, Pacaud P, Loirand G. The Rho exchange factor Arhgef1 mediates the effects of angiotensin II on vascular tone and blood pressure. Nat. Med. 2010;16:183–190. doi: 10.1038/nm.2079. [DOI] [PubMed] [Google Scholar]
- 55.Girkontaite I, Missy K, Sakk V, Harenberg A, Tedford K, Potzel T, Pfeffer K, Fischer KD. Lsc is required for marginal zone B cells, regulation of lymphocyte motility and immune responses. Nat. Immunol. 2001;2:855–862. doi: 10.1038/ni0901-855. [DOI] [PubMed] [Google Scholar]
- 56.Harenberg A, Girkontaite I, Giehl K, Fischer KD. The Lsc RhoGEF mediates signaling from thromboxane A2 to actin polymerization and apoptosis in thymocytes. Eur. J. Immunol. 2005;35:1977–1986. doi: 10.1002/eji.200425769. [DOI] [PubMed] [Google Scholar]
- 57.Rubtsov A, Strauch P, Digiacomo A, Hu J, Pelanda R, Torres RM. Lsc regulates marginal-zone B cell migration and adhesion and is required for the IgM T-dependent antibody response. Immunity. 2005;23:527–538. doi: 10.1016/j.immuni.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 58.Francis SA, Shen X, Young JB, Kaul P, Lerner DJ. Rho GEF Lsc is required for normal polarization, migration, and adhesion of formyl-peptide-stimulated neutrophils. Blood. 2006;107:1627–1635. doi: 10.1182/blood-2005-03-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Green JA, Suzuki K, Cho B, Willison LD, Palmer D, Allen CD, Schmidt TH, Xu Y, Proia RL, Coughlin SR, Cyster JG. The sphingosine 1-phosphate receptor S1P maintains the homeostasis of germinal center B cells and promotes niche confinement. Nat. Immunol. 2011;12:672–680. doi: 10.1038/ni.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zizer E, Beilke S, Bauerle T, Schilling K, Mohnle U, Adler G, Fischer KD, Wagner M. Loss of Lsc/p115 protein leads to neuronal hypoplasia in the esophagus and an achalasia-like phenotype in mice. Gastroenterology. 2010;139:1344–1354. doi: 10.1053/j.gastro.2010.06.041. [DOI] [PubMed] [Google Scholar]
- 61.Nagase T, Ishikawa K, Nakajima D, Ohira M, Seki N, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. Prediction of the coding sequences of unidentified human genes. VII. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 1997;4:141–150. doi: 10.1093/dnares/4.2.141. [DOI] [PubMed] [Google Scholar]
- 62.Whitehead IP, Khosravi-Far R, Kirk H, Trigo-Gonzalez G, Der CJ, Kay R. Expression cloning of lsc, a novel oncogene with structural similarities to the Dbl family of guanine nucleotide exchange factors. J. Biol. Chem. 1996;271:18643–18650. doi: 10.1074/jbc.271.31.18643. [DOI] [PubMed] [Google Scholar]
- 63.Glaven JA, Whitehead IP, Nomanbhoy T, Kay R, Cerione RA. Lfc and Lsc oncoproteins represent two new guanine nucleotide exchange factors for the Rho GTP-binding protein. J. Biol. Chem. 1996;271:27374–27381. doi: 10.1074/jbc.271.44.27374. [DOI] [PubMed] [Google Scholar]
- 64.Rumenapp U, Blomquist A, Schworer G, Schablowski H, Psoma A, Jakobs KH. Rho-specific binding and guanine nucleotide exchange catalysis by KIAA0380, a dbl family member. FEBS Lett. 1999;459:313–318. doi: 10.1016/s0014-5793(99)01270-3. [DOI] [PubMed] [Google Scholar]
- 65.Jackson M, Song W, Liu MY, Jin L, Dykes-Hoberg M, Lin CI, Bowers WJ, Federoff HJ, Sternweis PC, Rothstein JD. Modulation of the neuronal glutamate transporter EAAT4 by two interacting proteins. Nature. 2001;410:89–93. doi: 10.1038/35065091. [DOI] [PubMed] [Google Scholar]
- 66.Togashi H, Nagata K, Takagishi M, Saitoh N, Inagaki M. Functions of a rho-specific guanine nucleotide exchange factor in neurite retraction. Possible role of a proline-rich motif of KIAA0380 in localization. J. Biol. Chem. 2000;275:29570–29578. doi: 10.1074/jbc.M003726200. [DOI] [PubMed] [Google Scholar]
- 67.Derewenda U, Oleksy A, Stevenson AS, Korczynska J, Dauter Z, Somlyo AP, Otlewski J, Somlyo AV, Derewenda ZS. The crystal structure of RhoA in complex with the DH/PH fragment of PDZRhoGEF, an activator of the Ca(2+) sensitization pathway in smooth muscle. Structure. 2004;12:1955–1965. doi: 10.1016/j.str.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 68.Yamada T, Ohoka Y, Kogo M, Inagaki S. Physical and functional interactions of the lysophosphatidic acid receptors with PDZ domain-containing Rho guanine nucleotide exchange factors (RhoGEFs) J. Biol. Chem. 2005;280:19358–19363. doi: 10.1074/jbc.M414561200. [DOI] [PubMed] [Google Scholar]
- 69.Hirotani M, Ohoka Y, Yamamoto T, Nirasawa H, Furuyama T, Kogo M, Matsuya T, Inagaki S. Interaction of plexin-B1 with PDZ domain-containing Rho guanine nucleotide exchange factors. Biochem. Biophys. Res. Commun. 2002;297:32–37. doi: 10.1016/s0006-291x(02)02122-8. [DOI] [PubMed] [Google Scholar]
- 70.Perrot V, Vazquez-Prado J, Gutkind JS. Plexin B regulates Rho through the guanine nucleotide exchange factors leukemia-associated Rho GEF (LARG) and PDZ-RhoGEF. J. Biol. Chem. 2002;277:43115–43120. doi: 10.1074/jbc.M206005200. [DOI] [PubMed] [Google Scholar]
- 71.Swiercz JM, Kuner R, Behrens J, Offermanns S. Plexin-B1 directly interacts with PDZ-RhoGEF/LARG to regulate RhoA and growth cone morphology. Neuron. 2002;35:51–63. doi: 10.1016/s0896-6273(02)00750-x. [DOI] [PubMed] [Google Scholar]
- 72.Longenecker KL, Lewis ME, Chikumi H, Gutkind JS, Derewenda ZS. Structure of the RGS-like domain from PDZ-RhoGEF: linking heterotrimeric g protein-coupled signaling to Rho GTPases. Structure. 2001;9:559–569. doi: 10.1016/s0969-2126(01)00620-7. [DOI] [PubMed] [Google Scholar]
- 73.Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AlF4–activated G(i alpha1): stabilization of the transition state for GTP hydrolysis. Cell. 1997;89:251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- 74.Chen Z, Singer WD, Danesh SM, Sternweis PC, Sprang SR. Recognition of the activated states of Galpha13 by the rgRGS domain of PDZRhoGEF. Structure. 2008;16:1532–1543. doi: 10.1016/j.str.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen Z, Singer WD, Wells CD, Sprang SR, Sternweis PC. Mapping the Galpha13 binding interface of the rgRGS domain of p115RhoGEF. J. Biol. Chem. 2003;278:9912–9919. doi: 10.1074/jbc.M212695200. [DOI] [PubMed] [Google Scholar]
- 76.Sprang SR, Chen Z, Du X. Structural basis of effector regulation and signal termination in heterotrimeric Galpha proteins. Adv. Protein Chem. 2007;74:1–65. doi: 10.1016/S0065-3233(07)74001-9. [DOI] [PubMed] [Google Scholar]
- 77.Oleksy A, Opalinski L, Derewenda U, Derewenda ZS, Otlewski J. The molecular basis of RhoA specificity in the guanine nucleotide exchange factor PDZ-RhoGEF. J. Biol. Chem. 2006;281:32891–32897. doi: 10.1074/jbc.M606220200. [DOI] [PubMed] [Google Scholar]
- 78.Cierpicki T, Bielnicki J, Zheng M, Gruszczyk J, Kasterka M, Petoukhov M, Zhang A, Fernandez EJ, Svergun DI, Derewenda U, Bushweller JH, Derewenda ZS. The solution structure and dynamics of the DH-PH module of PDZRhoGEF in isolation and in complex with nucleotide-free RhoA. Protein Sci. 2009;18:2067–2079. doi: 10.1002/pro.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen Z, Medina F, Liu MY, Thomas C, Sprang SR, Sternweis PC. Activated RhoA binds to the pleckstrin homology (PH) domain of PDZ-RhoGEF, a potential site for autoregulation. J. Biol. Chem. 285:21070–21081. doi: 10.1074/jbc.M110.122549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zheng M, Cierpicki T, Momotani K, Artamonov MV, Derewenda U, Bushweller JH, Somlyo AV, Derewenda ZS. On the mechanism of autoinhibition of the RhoA-specific nucleotide exchange factor PDZRhoGEF. BMC Struct. Biol. 2009;9:36. doi: 10.1186/1472-6807-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Katoh H, Aoki J, Yamaguchi Y, Kitano Y, Ichikawa A, Negishi M. Constitutively active Galpha12, Galpha13, and Galphaq induce Rho-dependent neurite retraction through different signaling pathways. J. Biol. Chem. 1998;273:28700–28707. doi: 10.1074/jbc.273.44.28700. [DOI] [PubMed] [Google Scholar]
- 82.Gohla A, Harhammer R, Schultz G. The G-protein G13 but not G12 mediates signaling from lysophosphatidic acid receptor via epidermal growth factor receptor to Rho. J. Biol. Chem. 1998;273:4653–4659. doi: 10.1074/jbc.273.8.4653. [DOI] [PubMed] [Google Scholar]
- 83.Chikumi H, Fukuhara S, Gutkind JS. Regulation of G protein-linked guanine nucleotide exchange factors for Rho, PDZ-RhoGEF, and LARG by tyrosine phosphorylation: evidence of a role for focal adhesion kinase. J. Biol. Chem. 2002;277:12463–12473. doi: 10.1074/jbc.M108504200. [DOI] [PubMed] [Google Scholar]
- 84.Ying Z, Giachini FR, Tostes RC, Webb RC. PYK2/PDZ-RhoGEF links Ca2+ signaling to RhoA. Arterioscler. Thromb. Vasc. Biol. 2009;29:1657–1663. doi: 10.1161/ATVBAHA.109.190892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ohtsu H, Mifune M, Frank GD, Saito S, Inagami T, Kim-Mitsuyama S, Takuwa Y, Sasaki T, Rothstein JD, Suzuki H, Nakashima H, Woolfolk EA, Motley ED, Eguchi S. Signal-crosstalk between Rho/ROCK and c-Jun NH2-terminal kinase mediates migration of vascular smooth muscle cells stimulated by angiotensin II. Arterioscler. Thromb. Vasc. Biol. 2005;25:1831–1836. doi: 10.1161/01.ATV.0000175749.41799.9b. [DOI] [PubMed] [Google Scholar]
- 86.Barac A, Basile J, Vazquez-Prado J, Gao Y, Zheng Y, Gutkind JS. Direct interaction of p21-activated kinase 4 with PDZ-RhoGEF, a G protein-linked Rho guanine exchange factor. J. Biol. Chem. 2004;279:6182–6189. doi: 10.1074/jbc.M309579200. [DOI] [PubMed] [Google Scholar]
- 87.Suzuki N, Tsumoto K, Hajicek N, Daigo K, Tokita R, Minami S, Kodama T, Hamakubo T, Kozasa T. Activation of leukemia-associated RhoGEF by Galpha13 with significant conformational rearrangements in the interface. J. Biol. Chem. 2009;284:5000–5009. doi: 10.1074/jbc.M804073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Aurandt J, Vikis HG, Gutkind JS, Ahn N, Guan KL. The semaphorin receptor plexin-B1 signals through a direct interaction with the Rho-specific nucleotide exchange factor, LARG. Proc. Natl Acad. Sci. USA. 2002;99:12085–12090. doi: 10.1073/pnas.142433199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Taya S, Inagaki N, Sengiku H, Makino H, Iwamatsu A, Urakawa I, Nagao K, Kataoka S, Kaibuchi K. Direct interaction of insulin-like growth factor-1 receptor with leukemia-associated RhoGEF. J. Cell. Biol. 2001;155:809–820. doi: 10.1083/jcb.200106139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kitzing TM, Sahadevan AS, Brandt DT, Knieling H, Hannemann S, Fackler OT, Grosshans J, Grosse R. Positive feedback between Dia1, LARG, and RhoA regulates cell morphology and invasion. Genes Dev. 2007;21:1478–1483. doi: 10.1101/gad.424807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Grabocka E, Wedegaertner PB. Disruption of oligomerization induces nucleocytoplasmic shuttling of leukemia-associated rho Guanine-nucleotide exchange factor. Mol. Pharmacol. 2007;72:993–1002. doi: 10.1124/mol.107.035162. [DOI] [PubMed] [Google Scholar]
- 92.Aittaleb M, Gao G, Evelyn CR, Neubig RR, Tesmer JJ. A conserved hydrophobic surface of the LARG pleckstrin homology domain is critical for RhoA activation in cells. Cell Signal. 2009;21:1569–1578. doi: 10.1016/j.cellsig.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guilluy C, Swaminathan V, Garcia-Mata R, O'Brien ET, Superfine R, Burridge K. The Rho GEFs LARG and GEF-H1 regulate the mechanical response to force on integrins. Nat. Cell Biol. 2011;13:722–727. doi: 10.1038/ncb2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hamazaki Y, Kojima H, Mano H, Nagata Y, Todokoro K, Abe T, Nagasawa T. Tec is involved in G protein-coupled receptor- and integrin-mediated signalings in human blood platelets. Oncogene. 1998;16:2773–2779. doi: 10.1038/sj.onc.1201799. [DOI] [PubMed] [Google Scholar]
- 95.Jiang Y, Ma W, Wan Y, Kozasa T, Hattori S, Huang XY. The G protein G alpha12 stimulates Bruton's tyrosine kinase and a rasGAP through a conserved PH/BM domain. Nature. 1998;395:808–813. doi: 10.1038/27454. [DOI] [PubMed] [Google Scholar]
- 96.Mao J, Xie W, Yuan H, Simon MI, Mano H, Wu D. Tec/Bmx non-receptor tyrosine kinases are involved in regulation of Rho and serum response factor by Galpha12/13. EMBO J. 1998;17:5638–5646. doi: 10.1093/emboj/17.19.5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bos JL. The ras gene family and human carcinogenesis. Mutat. Res. 1988;195:255–271. doi: 10.1016/0165-1110(88)90004-8. [DOI] [PubMed] [Google Scholar]
- 98.Boettner B, Van Aelst L. The role of Rho GTPases in disease development. Gene. 2002;286:155–174. doi: 10.1016/s0378-1119(02)00426-2. [DOI] [PubMed] [Google Scholar]
- 99.Sahai E, Marshall CJ. RHO-GTPases and cancer. Nat. Rev. Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 100.Rujkijyanont P, Beyene J, Wei K, Khan F, Dror Y. Leukaemia-related gene expression in bone marrow cells from patients with the preleukaemic disorder Shwachman-Diamond syndrome. Br. J. Haematol. 2007;137:537–544. doi: 10.1111/j.1365-2141.2007.06608.x. [DOI] [PubMed] [Google Scholar]
- 101.Ong DC, Ho YM, Rudduck C, Chin K, Kuo WL, Lie DK, Chua CL, Tan PH, Eu KW, Seow-Choen F, Wong CY, Hong GS, Gray JW, Lee AS. LARG at chromosome 11q23 has functional characteristics of a tumor suppressor in human breast and colorectal cancer. Oncogene. 2009;28:4189–4200. doi: 10.1038/onc.2009.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wirth A, Benyo Z, Lukasova M, Leutgeb B, Wettschureck N, Gorbey S, Orsy P, Horvath B, Maser-Gluth C, Greiner E, Lemmer B, Schutz G, Gutkind JS, Offermanns S. G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat. Med. 2008;14:64–68. doi: 10.1038/nm1666. [DOI] [PubMed] [Google Scholar]
- 103.Medlin MD, Staus DP, Dubash AD, Taylor JM, Mack CP. Sphingosine 1-phosphate receptor 2 signals through leukemia-associated RhoGEF (LARG), to promote smooth muscle cell differentiation. Arterioscler. Thromb. Vasc. Biol. 2010;30:1779–1786. doi: 10.1161/ATVBAHA.110.209395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lappano R, Maggiolini M. G protein-coupled receptors: novel targets for drug discovery in cancer. Nat. Rev. Drug Discov. 2011;10:47–60. doi: 10.1038/nrd3320. [DOI] [PubMed] [Google Scholar]