Abstract
Traditional methods for epigenomic analysis provide a static picture of chromatin, which is actually a highly dynamic assemblage. Recent approaches have allowed direct measurements of chromatin dynamics, providing deeper insights into processes such as transcription, DNA replication and epigenetic inheritance.
Chromatin and epigenetic control
A eukaryotic genome can produce many cell types with widely different morphologies and functions. Given that the diverse cell types of a multicellular organism all contain the same DNA, there must be information in addition to the DNA sequence itself that controls which genes are expressed in a particular cell type. This extra layer of information was termed 'epigenetic control' by Nanney in 1958 [1]. Epigenetic control in eukaryotes occurs in the context of nucleosome particles, which can occlude or allow access to DNA by the proteins that bind specific sequences and precisely regulate active processes, including transcription and replication. Understanding the molecular basis for epigenetic control is a central goal of chromatin research.
The eukaryotic genome is tightly wrapped by histones to form nucleosomes, which must be densely packed to fit within the confines of the nucleus, overall up to approximately 1 million-fold compaction of DNA relative to an extended double helix. Despite these tight constraints, nucleosomes must be able to allow the DNA sequences to be accessible to DNA-binding proteins and to the action of 'molecular machines' such as DNA and RNA polymerases, ATP-dependent nucleosome remodelers and topoisomerases. Nuclear organization involves multiple levels of chromatin packaging, including compartments, territories and self-organizing nuclear bodies, which might appear to be static at a gross cytological level, but which must be sufficiently dynamic to allow for access of regulatory factors to the DNA (Figure 1). Although the precise nature of chromatin beyond the level of single nucleosomes is unclear [2], some principles are beginning to emerge, such as the fractal globule large-scale organization of chromosomes, which allows them to decondense and recondense without becoming entangled [3].
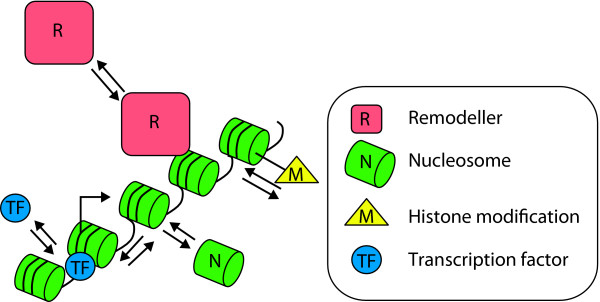
Figure 1.
Dynamic chromatin. Chromatin consists of arrays of nucleosomes (N) with a number of dynamic features such as nucleosome position, histone-variant composition of nucleosomes, post-translational modifications of histones, as well as the binding of transcription factors, chromatin-remodeling complexes, and modification binding proteins. Transcription factors (TFs) and remodeling complexes (R) are in equilibrium between the bound and unbound states, and nucleosomes can slide along DNA, be dislodged from DNA, and be reassembled. In addition, a wide variety of histone modifications (m) can be added and removed enzymatically. The right-angled arrow indicates the transcription start site.
Nucleosome particles consist of around 150 bp of DNA wrapped around an octameric histone core containing two copies of each of the four core histone proteins (H2A, H2B, H3 and H4) [4,5]. The properties of a nucleosome can be altered in various ways, including replacement of standard histones with specialized variant types, post-translational modification of histones, movement of the particle relative to the underlying DNA sequence, and partial or complete removal of histones from the DNA. The regulation of chromatin structure to expose or occlude a particular DNA segment is controlled by the dynamic interplay between sequence-specific DNA-binding proteins, histone variants, histone-modifying enzymes, chromatin-associated proteins, histone chaperones and ATP-dependent nucleosome remodelers [6]. Collectively, these factors provide instructions that direct the transcriptional output of the genome, but exactly how this information is imparted and transmitted through cell division is unclear. Approaches to understanding chromatin-based regulation have included the identification of factors involved and mapping of chromatin proteins and histone modifications across the genome [6-8]. These approaches have taught us much about the control of transcription in particular and have provided a conceptual framework for further research. However, these methods give only a static snapshot of chromatin, whereas chromatin is actually a dynamic assemblage in which proteins are constantly associating and dissociating [9]. Therefore, understanding chromatin-based regulation has required the development and application of techniques that can capture these dynamic processes. This review will focus on epigenome dynamics at the level of the nucleosome and will explore how emerging technologies that allow time-dependent measurements are yielding deeper insights into the regulation of various genomic processes and the inheritance of gene-expression states.
Defining the epigenome through chromatin-mapping studies
Much of our understanding of the epigenome and its influence on regulating gene expression has come from genome-wide analyses of steady-state chromatin composition combined with genetic and biochemical studies that enable functional interpretation of these maps. To elucidate the primary structure of chromatin, many groups have sought to identify the locations of all nucleosomes across the genome and to understand the factors that dictate their locations. A popular mapping approach is to digest chromatin with micrococcal nuclease, which preferentially cleaves the DNA between nucleosomes, and then to infer nucleosome positions by analyzing the pool of sequences protected by nucleosomes [10]. These studies have collectively shown that certain fundamental rules of nucleosome positioning are common to many eukaryotes. The Saccharomyces cerevisiae genome has a large number of well-positioned nucleosomes covering approximately 80% of the genome, whereas metazoan and plant genomes have a smaller percentage of well-positioned nucleosomes [11-16]. However, all genomes examined show a characteristic distribution of nucleosomes around genes. There are often two well-positioned nucleosomes that flank the transcription start site (TSS) with a nucleosome-depleted region (NDR) in between [17]. Nucleosomes at the 5' ends of transcribed regions tend to be more precisely localized than those further downstream, and there is often another NDR at the 3' end [14,18]. The overall landscape of nucleosome locations and relative occupancy at a point in time seems to be dictated in part by intrinsic DNA sequence preferences of the nucleosomes themselves, and also by the action of nucleosome-remodeling complexes and competition between nucleosomes and sequence-specific DNA-binding proteins such as transcription factors [19-21].
Chromatin is further differentiated by variations in the characteristics and composition of nucleosomes. Biochemical studies of histones have shown that they are heavily modified post-translationally through the addition of acetyl, methyl, phosphoryl and ADP-ribose groups, as well as peptides such as ubiquitin and SUMO. Mapping of these modified histones has revealed distinct patterns of localization across the genome, and this has led to insights into genomic processes, including transcription as well as DNA replication and repair. It has emerged that certain histone modifications tend to co-occur, and each 'mark' can be broadly categorized as being associated with either actively transcribed genes, silenced genes or transposons [6-8]. Within these categories there are modifications, such as acetylation, that alter the physical properties of nucleosomes directly, and others such as methylation that can create binding sites for other proteins that have specific effects on chromatin-based processes. In terms of function, acetylation of the nucleosomes around TSSs seems to be required to support transcription, presumably by loosening the interaction between histones and DNA, while conversely, the deacetylation of nucleosomes throughout the body of the gene appears to repress spurious antisense transcription by increasing histone association with the DNA [22,23]. Chromatin modifications that are bound by specific effector proteins can either be involved in the repression of transcription, by mechanisms such as compaction of nucleosome arrays [24,25], or they can support transcription, by recruiting chromatin-remodeling complexes, modifying enzymes or other complexes involved in elongation or splicing [26,27]. Thus, histone modifications can affect access to DNA directly or indirectly, and also serve as a platform for the coordination of successive processes such as transcription and splicing [27].
Nucleosomes are also differentiated by the substitution of canonical histones with the universal variants H2A.Z and H3.3 [28]. These variants are replication-independent in their assembly, and so must be inserted by disruption of existing nucleosomes. H2A.Z is inserted by the Swr1 ATP-dependent nucleosome-remodeling complex into nucleosome cores by partial unwrapping and replacement of an H2A/H2B dimer with an H2A.Z/H2B dimer. To insert H3.3 into the central (H3/H4)2 tetramer, a nucleosome must be completely unwrapped, a process that amounts to dynamic eviction of the histone core and replacement with two dimeric units of H3.3/H4 [29]. H3.3 replacement requires a histone chaperone, such as HirA or DAXX, and various ATP-dependent nucleosome-remodeling complexes, including Chd1 and Atrx [30-32]. H2A.Z and H3.3 show partially overlapping distributions: H2A.Z is often enriched at the -1 nucleosome position relative to the TSS and in gene-body nucleosomes near the 5' end [33], whereas H3.3 is low in promoter nucleosomes and is enriched in essentially all gene-body nucleosomes, with its occupancy positively correlated with the level of transcription [34]. Nucleosomes containing H2A.Z but not H3.3 are relatively stable, whereas those that contain both variants may be prone to disassembly in vivo [35] (although not in vitro [36]). Unstable double-variant nucleosomes are found at TSSs and so may regulate exposure of promoter DNA [37]. Thus, both the replication-independent replacement of canonical histones with histone variants, and the altered properties of double-variant nucleosomes that sometimes result, indicate that the nucleosomes that package genes are inherently dynamic. The emerging picture of the epigenome is one in which the composition of chromatin in terms of histone modifications, variants and chromatin-associated proteins dictates the intrinsic stability of nucleosomes as well as their propensity to be disrupted or moved by chromatin-remodeling enzymes and the transcription machinery. In this way, access to the underlying DNA is regulated [38].
Measuring epigenome dynamics
Given the evidence that many regions of chromatin are in a state of flux, various approaches have been developed to measure chromatin dynamics directly by using tools such as microscopy, mass spectrometry (MS), immunoprecipitation of inducible tagged proteins, and metabolic labeling of newly synthesized proteins (Table 1). The application of these methods has led to unexpected new insights into the regulation of various genomic processes such as transcription, DNA replication and the inheritance of patterns of gene expression.
Table 1.
Comparison of methods for measuring chromatin dynamics
| Method | Utility | Benefits | Drawbacks |
|---|---|---|---|
| Fluorescence recovery after photobleaching (FRAP) | Measurement of chromatin protein binding kinetics | 1. Can be used for nucleosomes as well as other chromatin binding proteins 2. Allows observation of protein location within the nucleus |
1. Cannot determine the specific genomic sites that are bound 2. Requires an epitope-tagged protein that may not behave exactly like the native form |
| MS-based kinetic methods | Measurements of histone modification kinetics | Can be used for nucleosomes as well as other chromatin-binding proteins | Cannot determine the kinetics at specific genomic sites |
| Inducible transgene-based methods | Measurement of nucleosome turnover kinetics as well as binding of other chromatin proteins | Can be used for nucleosomes as well as other chromatin-binding proteins | 1. Requires an epitope-tagged protein 2. Time lag during induction limits time resolution |
| Recombination-induced tag exchange (RITE) | Measurement of nucleosome turnover kinetics as well as binding of other chromatin proteins | Can be used for nucleosomes as well as other chromatin-binding proteins | 1. Requires an epitope-tagged protein that may not behave exactly like the native form 2. Time lag during recombination limits time resolution |
| Covalent attachment of tags to capture histones and identify turnover (CATCH-IT) | Measurement of nucleosome turnover kinetics | 1. No transgenes or antibodies are required 2. Excellent time resolution 3. Can be used on many different cell types |
Only (H3/H4)2 tetramer incorporation kinetics can be measured easily |
Fluorescence recovery after photobleaching
One approach to observing chromatin dynamics in vivo is fluorescence recovery after photobleaching (FRAP) and related cytological methods. In FRAP, a discrete region of a nucleus containing a fluorescently labeled chromatin protein is subjected to laser photobleaching, and the amount of time required for the bleached region to regain fluorescence is measured (Figure 2a). The time required for fluorescence recovery is a measure of the residence time of the protein on chromatin; therefore, this technique can be used to infer the binding kinetics of chromatin proteins [39]. FRAP has the advantage that any protein that can be tagged can be analyzed, and information on the nuclear distribution of each protein can also be obtained. However, in contrast to methods that use genomics tools as a readout, FRAP does not provide information on the specific site to which the factor of interest binds. Furthermore, like all methods that rely on epitope-tagged proteins, there is the possibility that the protein will not behave like the native form, producing artifactual results.
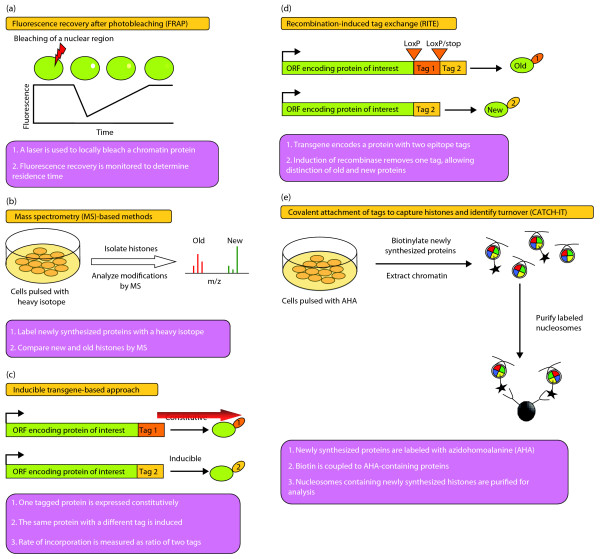
Figure 2.
Methods for investigating chromatin dynamics. (a) In fluorescence recovery after photobleaching (FRAP), a laser is used to bleach the fluorescence of a chromatin protein in a discrete region of the nucleus. The time required for the fluorescent protein to move back into the bleached region and restore fluorescence is proportional to the residence time of the protein on chromatin. The shorter the residence time of a protein, the faster fluorescence is recovered in the bleached region. (b) Mass spectrometry (MS) can be used to study the dynamics of post-translational modifications in the total histone pool by briefly labeling newly synthesized histones with a heavy isotope, such as 15N or 13C. Histone modifications can then be determined by MS for both the old and new histones, based on the mass difference between these two pools of histones. (c) In the inducible transgene-based approach, the protein to be assayed is expressed from two different transgenes. One transgene is expressed constitutively and carries an epitope tag while the other is inducible and carries a different epitope tag. Induction of the second transgene allows the measurement of binding kinetics by comparing the relative abundances of the two tags at a given genomic location. (d) Recombination-induced tag exchange (RITE) can be used to measure the dynamics of chromatin binding and nucleosome assembly by enabling old and new proteins expressed from the same transgene to be distinguished. This method uses a single transgene that encodes two different epitope tags. The first tag is flanked by loxP recombination sites and incorporates a stop codon, whereas the downstream tag is in-frame but comes after the stop codon. Normally, the protein encoded by the transgene has only the first tag, but after induction of Cre recombinase the first tag is removed from the transgene and now the encoded protein contains only the second tag. Dynamics of a RITE-tagged protein can be inferred by comparing the relative abundances of the old and new versions of the protein at a given genomic location. (e) Covalent attachment of tags to capture histones and identify turnover (CATCH-IT) can be used to estimate rates of disassembly and reassembly, or turnover, of native nucleosomes across the genome. In this method, newly synthesized proteins are labeled by treating cells with the methionine analog azidohomoalanine (AHA). Nuclei are isolated from AHA-treated cells and biotin is coupled to AHA-containing nuclear proteins through a reaction of the azide group of AHA with an alkyne linked to biotin. Chromatin is then digested to mononucleosomes, the nucleosomes are extracted, and nucleosomes containing biotinylated histones are purified via streptavidin. Stringent washes are used to remove H2A/H2B dimers and all other DNA-binding proteins from the purified nucleosomes. Microarray analysis or deep sequencing of the purified DNA allows the rates of nucleosome turnover across the genome to be estimated on the basis of the extent of newly synthesized H3/H4 dimer incorporation at each site.
The FRAP method has been used to measure residence times of a wide variety of proteins, including nuclear hormone receptors, transcription factors, chromatin-remodeling enzymes and the histones themselves. The results of these experiments have consistently shown that hormone receptors, transcription factors and remodeling enzymes have residence times on chromatin on the order of seconds [40]. By contrast, photobleaching studies of core histones revealed that these proteins have residence times much longer than most chromatin-associated proteins, on the order of tens of minutes to hours, and that the residence time of H2A and H2B on chromatin is shorter than that of H3 and H4 [41]. These results were confirmed by experiments in which epitope-tagged histones were introduced into the slime mold Physarum, and chromatin immunoprecipitation (ChIP) was used to track their incorporation into several active genes over time [42]. Collectively, these results argue strongly that complexes that regulate transcription are unstable, and show that histones also dissociate from DNA over timescales that can be shorter than the cell cycle.
Kinetic methods based on mass spectrometry
Dozens of different histone post-translational modifications have been identified by MS, and, in recent years, MS has also been used to determine the different combinations in which they occur [43,44]. To exploit this powerful discriminating tool for measuring nucleosome dynamics, MS has been combined with Meselson-Stahl type incorporation of heavy isotopes in protocols designed to distinguish newly synthesized histones from pre-existing histones (Figure 2b). By synchronizing cells and releasing them into S-phase with the addition of an amino acid labeled with a heavy isotope, such as 15N-labeled arginine or 13C-labeled methione, peptides from newly synthesized histones can be distinguished from their counterparts from old histones using MS [45,46]. This allows for changes in modification to be scored and quantified during chromatin assembly. Consistent with classical studies, histone acetylation and deacetylation was found to be highly dynamic. In the case of histone methylation, mono-methylation occurred on most H3 residues (lysine (K)4, K9, K27 and K36) soon after synthesis and incorporation, whereas di- and tri-methylation occurred more slowly over the course of the cell cycle. An exception is H3K79, which was found to undergo mono-methylation continuously on both old and new nucleosomes with very little conversion to the di-methylated form [47]. This strategy for following global histone modification changes should become increasingly powerful as MS-based technologies improve. Together with genomic-based methods described below, MS-based kinetics promises to provide a wealth of information on the dynamics of histone marks and how they might be involved in governing nucleosome dynamics.
Inducible transgene-based methods
In the past few years, the rate of replication-independent nucleosome disassembly and reassembly, or turnover, has been measured at high resolution across the genome of budding yeast (S. cerevisiae) by using inducible epitope-tagged histones as a means of estimating relative nucleosome turnover rates. In this method, cells have two transgenic sources of a particular histone protein: one that is constitutively expressed and has an epitope tag, and another that is inducible and has a different epitope tag. By arresting cells in G1 phase and inducing the second tagged histone, ChIP assays can be performed separately for each tag at multiple time points after induction. Analysis of the resulting DNA by microarrays then allows estimation of nucleosome turnover rates across the genome by comparing the ratio of signals from the two tagged histones (Figure 2c). In addition to measuring nucleosome turnover kinetics, this approach could also be used to measure the dynamics of other chromatin proteins. One caveat to this approach is that there is a time lag during induction of the transgene and synthesis of the encoded protein, which limits the temporal resolution of kinetic measurements.
Several inducible transgene studies have been carried out using histone H3, and these collectively showed that nucleosome turnover was highest both upstream and downstream of the TSS, whereas turnover in the body of the gene was relatively low regardless of expression level, except at very highly expressed genes [48,49]. Using a similar approach, in which only the inducible epitope-tagged histone was used, Jamai et al. [50] found that, in contrast to H3, H2B turns over rapidly within promoters and across gene bodies irrespective of expression level, implying that the turnover of H2A/H2B dimers is a distinct process from the turnover of (H3/H4)2 tetramers. Results from these studies also indicated that nucleosome turnover is very high at known chromatin boundary elements flanking silenced regions, leading to the suggestion that nucleosome turnover might help to prevent the spread of silent chromatin and silencing of nearby genes [48].
Recombination-induced tag exchange
Recombination-induced tag exchange (RITE) allows one to distinguish between old and new proteins encoded by the same transgene (Figure 2d). In this method, a transgene encoding the protein of interest is engineered to contain an epitope tag and a stop codon flanked by loxP recombination sites, with a second in-frame epitope tag after the stop codon. When Cre recombinase activity is induced, the sequence encoding the first tag is removed, resulting in all subsequent transcripts encoding a protein with only the second tag. This allows the tracking of proteins produced before and after recombinase induction.
Using this approach, Verzijlbergen et al. [51] tagged H3 in yeast and showed that nucleosome turnover occurs not only in G1 phase but also in G2/M phase, and that the rate is dependent on the expression level of a given gene. The RITE method was also used to examine replacement of proteasomal subunits in this study, suggesting that it will be useful for studying the dynamics not only of histones but also of other chromatin proteins.
Covalent attachment of tags to capture histones and identify turnover
As an alternative to using transgenes, we developed a general method for estimating nucleosome turnover rates by metabolically labeling newly synthesized histones with an amino-acid analog that could be coupled to an affinity tag. This technique is called covalent attachment of tags to capture histones and identify turnover, or CATCH-IT (Figure 2e). This method has an advantage over previously discussed genomic methods in that no transgenes are required and the behavior of the native protein itself is being measured. Furthermore, the temporal resolution is very high as there is no time lag associated with transgene induction and protein synthesis. However, essentially all proteins will be labeled, and therefore CATCH-IT analysis is limited to (H3/H4)2 tetramers as these remain associated with DNA under conditions that remove all other proteins.
Using CATCH-IT on Drosophila S2 cells, we found that nucleosome turnover landscapes were very similar to those previously reported for steady-state incorporation of H3.3 [34], and that turnover rates were highest in gene bodies, with the rate being correlated to expression level. By contrast, turnover within promoters was relatively low and seemed to be mostly independent of expression level [52]. Interestingly, nucleosome turnover measurements at sites bound by the origin recognition complex, which dictates the location and timing of replication origin firing [53,54], showed very high turnover rates compared with those of surrounding regions, indicating that regulated nucleosome turnover might also play an important part in the selection of replication origins and replication timing. This would help to explain the lack of DNA sequence conservation between replication origins [55].
Examination of nucleosome turnover at epigenetic regulatory elements bound by either trithorax group (trxG) activators or Polycomb group (PcG) silencer proteins [56] in S2 cells revealed that the rates of turnover were different between these two classes of sites. Turnover rates were higher at trxG sites than at PcG sites, suggesting that differences in nucleosome turnover rates provide the mechanistic basis for the opposing activities of these regulatory proteins on gene expression. We hypothesize that trxG proteins promote turnover to allow greater access of sequence-specific regulators, whereas PcG proteins slow turnover to reduce access. In fact, the PRC1 complex, which is responsible for PcG silencing, causes compaction of the chromatin fiber in vitro [24]. Recent in vivo support for this paradigm of developmental silencing has come from the finding that the PRC1 complex silences Hox genes by compacting chromatin [25], which presumably results in reduced nucleosome turnover.
Inheritance of chromatin states
A point of contention in the field of epigenetics is the basis of the inheritance of a chromatin state through cell division. One view is that histone modifications are informational, by virtue of a 'histone code' analogous with the genetic code [57]. However, it is now clear that nucleosomes are reconstituted from newly synthesized histones many times during a cell cycle, essentially erasing histone modifications and making it unlikely that the modifications themselves are capable of transmitting information [52]. An alternative hypothesis is that histone modifications and secondary effector proteins that recognize them collectively modulate the intrinsic stability of a given nucleosome as well as its propensity to be remodeled. These characteristics in turn determine how likely a nucleosome is to be disassembled or to change position, and thereby expose the underlying DNA to sequence-specific regulators that control genome output. Rather than constituting an informational code, histone modifications, variants, nucleosome remodelers and other chromatin-associated proteins could be considered as components of a dynamic system that regulates nucleosome turnover and, consequently, DNA exposure to sequence-specific regulators.
The perpetuation of gene-expression states during the cell cycle and through cell divisions may be based on a competition between the binding of sequence-specific regulators and the reassembly of nucleosomes onto a particular DNA segment [58]. For example, an active gene-expression state could be initiated and maintained by binding of an activator that brings along factors that promote nucleosome turnover to favor further activator binding. The active state could then be transmitted through cell division by a process based on the perpetuation of nucleosome turnover, which in some instances would be driven by the continued binding of activators, such as general transcription factors, through mitosis [59]. The silent state would then be inherited by default via silencing complexes that reduce nucleosome turnover through chromatin compaction [24], thereby reducing access of activators to DNA. Thus, nucleosome turnover would simply govern the level of exposure of a gene regulatory site and thereby determine whether the gene will remain active or silent during development [60].
Prospects for the future
New approaches to studying chromatin dynamics have made it clear that chromatin and its associated proteins are in a constant state of flux and are not as stable as once thought. Thus, chromatin dynamics need to be taken into account when interpreting ChIP and other static measures of the epigenome. The combination of static and dynamic mapping of chromatin features along with mechanistic studies has the potential to provide a deep understanding of the epigenome.
Although we are only beginning to understand the mechanisms that maintain the epigenome, it has become clear that chromatin dynamics play a central role in the regulation of genome function. We look forward to the development of new methods for measuring dynamics at each structural level of chromatin from the primary fiber to secondary folding and on to the three-dimensional arrangement of the genome in the nucleus. Continuing technological progress is needed to generate a mechanistic description of the relationship between chromatin dynamics and transcriptional regulation, as well as other genomic processes such as DNA replication, repair, and recombination. This will bring us closer to the holy grail of understanding how the epigenome programs the genome to give the specific patterns of gene expression that define a given cell type and how it allows the stable perpetuation of phenotype.
Acknowledgements
We thank Melissa Conerly and Paul Talbert for helpful suggestions on improving the manuscript. This work was supported by NIH grant 1R21DA025758 to SH, NIH Postdoctoral Fellowship 1F32GM083449 to RBD, and the Howard Hughes Medical Institute.
References
- Nanney DL. Epigenetic control systems. Proc Natl Acad Sci USA. 1958;44:712–717. doi: 10.1073/pnas.44.7.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock CL, Ghosh RP. Chromatin higher-order structure and dynamics. Cold Spring Harb Perspect Biol. 2010;2:a000596. doi: 10.1101/cshperspect.a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD, Thomas JO. Chromatin structure; oligomers of the histones. Science. 1974;184:865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Rando OJ, Chang HY. Genome-wide views of chromatin structure. Annu Rev Biochem. 2009;78:245–271. doi: 10.1146/annurev.biochem.78.071107.134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager GL, Elbi C, Becker M. Protein dynamics in the nuclear compartment. Curr Opin Genet Dev. 2002;12:137–141. doi: 10.1016/S0959-437X(02)00278-2. [DOI] [PubMed] [Google Scholar]
- Kent NA, Mellor J. Chromatin structure snap-shots: rapid nuclease digestion of chromatin in yeast. Nucleic Acids Res. 1995;23:3786–3787. doi: 10.1093/nar/23.18.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G-C, Liu Y-J, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Tan FJ, McCullough HL, Riordan DP, Fire AZ. Flexibility and constraint in the nucleosome core landscape of Caenorhabditis elegans chromatin. Genome Res. 2006;16:1505–1516. doi: 10.1101/gr.5560806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, Gilmour DS, Albert I, Pugh BF. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh T-Y, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, Casero D, Bernal M, Huijser P, Clark AT, Krämer U, Merchant SS, Zhang X, Jacobsen SE, Pellegrini M. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman-Livaja M, Rando OJ. Nucleosome positioning: how is it established, and why does it matter? Dev Biol. 2010;339:258–266. doi: 10.1016/j.ydbio.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarragudi A, Miyake T, Li R, Morse RH. Comparison of ABF1 and RAP1 in chromatin opening and transactivator potentiation in the budding yeast Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:9152–9164. doi: 10.1128/MCB.24.20.9152-9164.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR. Chromatin remodeling complexes: strength in diversity, precision through specialization. Curr Opin Genet Dev. 2005;15:185–190. doi: 10.1016/j.gde.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Reinke H, Horz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell. 2003;11:1599–1607. doi: 10.1016/S1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, Bickmore WA. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- Sims RJ, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K. Assembly of variant histones into chromatin. Annu Rev Cell Dev Biol. 2005;21:133–153. doi: 10.1146/annurev.cellbio.21.012704.133518. [DOI] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/S0092-8674(03)01064-X. [DOI] [PubMed] [Google Scholar]
- Konev AY, Tribus M, Park SY, Podhraski V, Lim CY, Emelyanov AV, Vershilova E, Pirrotta V, Kadonaga JT, Lusser A, Fyodorov DV. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science. 2007;317:1087–1090. doi: 10.1126/science.1145339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, Wen D, Chapgier A, DeKelver RC, Miller JC, Lee YL, Boydston EA, Holmes MC, Gregory PD, Greally JM, Rafii S, Yang C, Scambler PJ, Garrick D, Gibbons RJ, Higgs DR, Cristea IM, Urnov FD, Zheng D, Allis CD. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Madhani HD. Patterning chromatin: form and function for H2A.Z variant nucleosomes. Curr Opin Genet Dev. 2006;16:119–124. doi: 10.1016/j.gde.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakar A, Gupta P, Ishibashi T, Finn R, Silva-Moreno B, Uchiyama S, Fukui K, Tomschik M, Ausio J, Zlatanova J. H2A.Z and H3.3 histone variants affect nucleosome structure: biochemical and biophysical studies. Biochemistry. 2009;48:10852–10857. doi: 10.1021/bi901129e. [DOI] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- Voss TC, Hager GL. Visualizing chromatin dynamics in intact cells. Biochim Biophys Acta. 2008;1783:2044–2051. doi: 10.1016/j.bbamcr.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair RD, Scaffidi P, Elbi C, Vecerová J, Dey A, Ozato K, Brown DT, Hager G, Bustin M, Misteli T. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol Cell Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H, Cook PR. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiriet C, Hayes JJ. Replication-independent core histone dynamics at transcriptionally active loci in vivo. Genes Dev. 2005;19:677–682. doi: 10.1101/gad.1265205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia BA, Pesavento JJ, Mizzen CA, Kelleher NL. Pervasive combinatorial modification of histone H3 in human cells. Nat Methods. 2007;4:487–489. doi: 10.1038/nmeth1052. [DOI] [PubMed] [Google Scholar]
- Young NL, DiMaggio PA, Plazas-Mayorca MD, Baliban RC, Floudas CA, Garcia BA. High throughput characterization of combinatorial histone codes. Mol Cell Proteomics. 2009;8:2266–2284. doi: 10.1074/mcp.M900238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf AN, Barth TK, Imhof A. Establishment of histone modifications after chromatin assembly. Nucleic Acids Res. 2009;37:5032–5040. doi: 10.1093/nar/gkp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zee BM, Levin RS, Xu B, LeRoy G, Wingreen NS, Garcia BA. In vivo residue-specific histone methylation dynamics. J Biol Chem. 2010;285:3341–3350. doi: 10.1074/jbc.M109.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet SM, Li M, Thomas PM, Durbin KR, Kelleher NL. Kinetics of re-establishing H3 K79 methylation marks in global human chromatin. J Biol Chem. 2010. [DOI] [PMC free article] [PubMed]
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- Rufiange A, Jacques P-E, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Jamai A, Imoberdorf RM, Strubin M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol Cell. 2007;25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- Verzijlbergen KF, Menendez-Benito V, van Welsem T, van Deventer SJ, Lindstrom DL, Ovaa H, Neefjes J, Gottschling DE, van Leeuwen F. Recombination-induced tag exchange to track old and new proteins. Proc Natl Acad Sci USA. 2010;107:64–68. doi: 10.1073/pnas.0911164107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncker BP, Chesnokov IN, McConkey BJ. The origin recognition complex protein family. Genome Biol. 2009;10:214. doi: 10.1186/gb-2009-10-3-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine HK, Gordân R, Powell SK, Hartemink AJ, MacAlpine DM. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 2010;20:201–211. doi: 10.1101/gr.097873.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM. In search of the holy replicator. Nat Rev Mol Cell Biol. 2004;5:848–855. doi: 10.1038/nrm1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- Sarge KD, Park-Sarge OK. Mitotic bookmarking of formerly active genes: keeping epigenetic memories from fading. Cell Cycle. 2009;8:818–823. doi: 10.4161/cc.8.6.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad K, Henikoff S. Modulation of a transcription factor counteracts heterochromatic gene silencing in Drosophila. Cell. 2001;104:839–847. doi: 10.1016/S0092-8674(01)00281-1. [DOI] [PubMed] [Google Scholar]