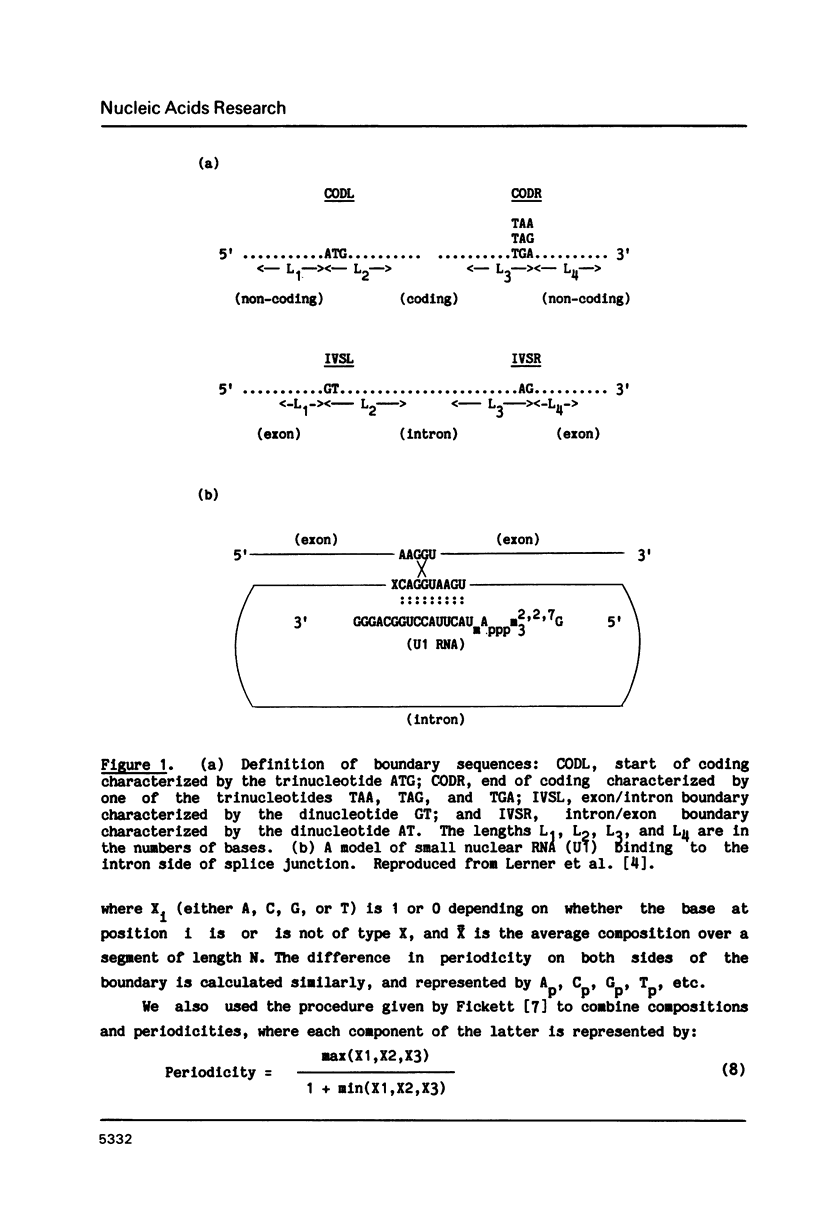
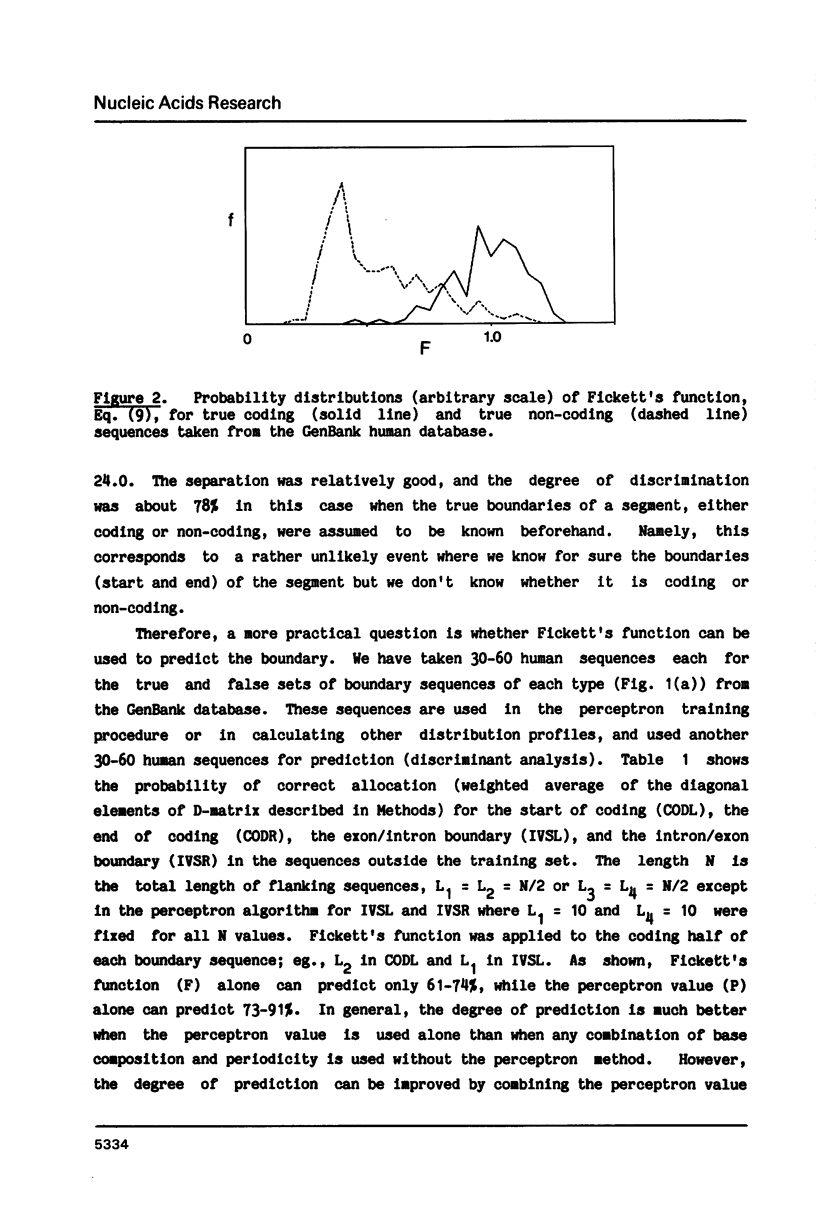
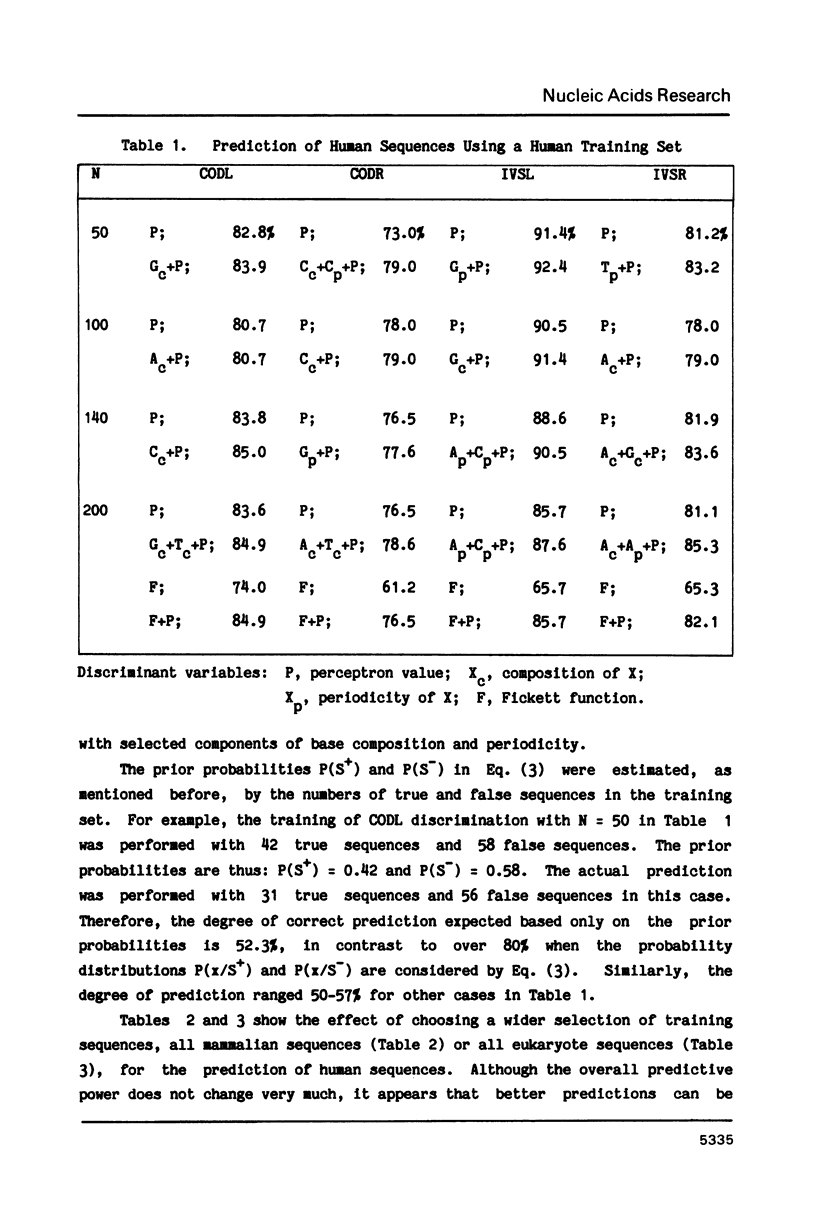
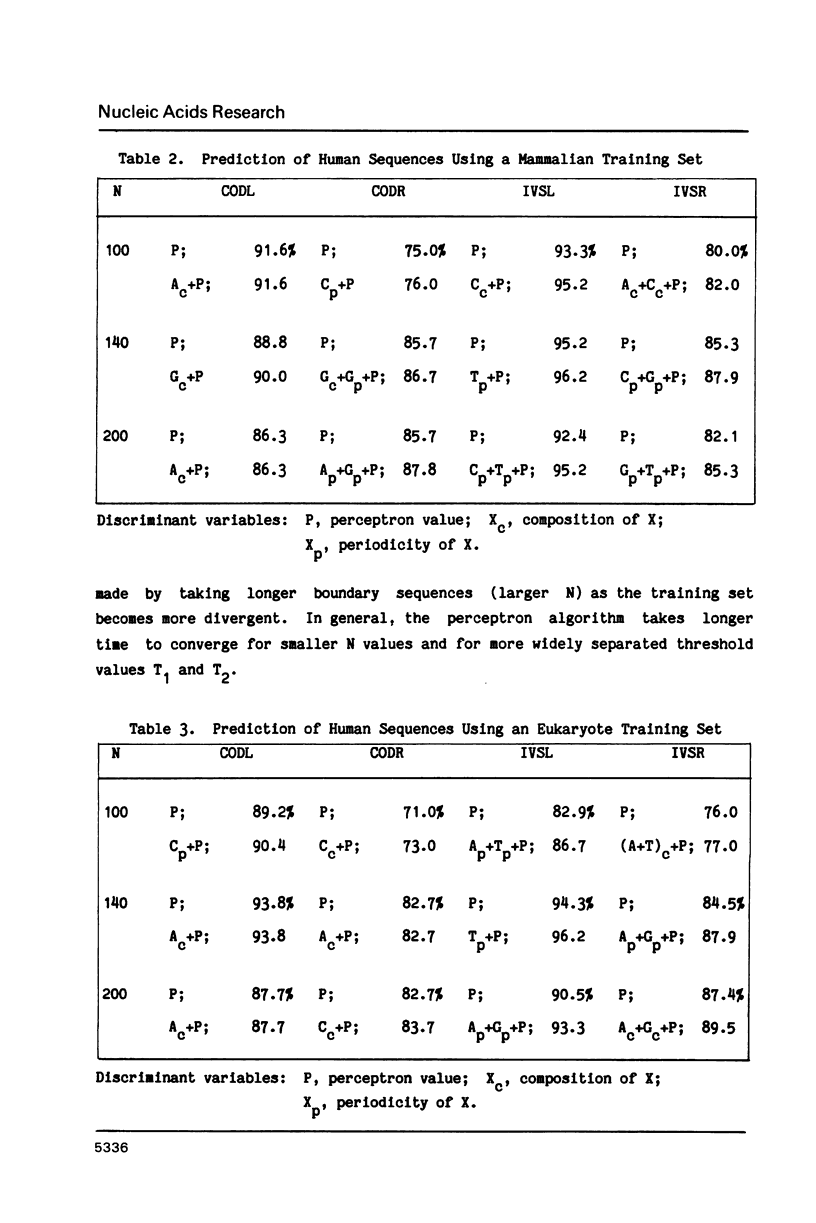
Abstract
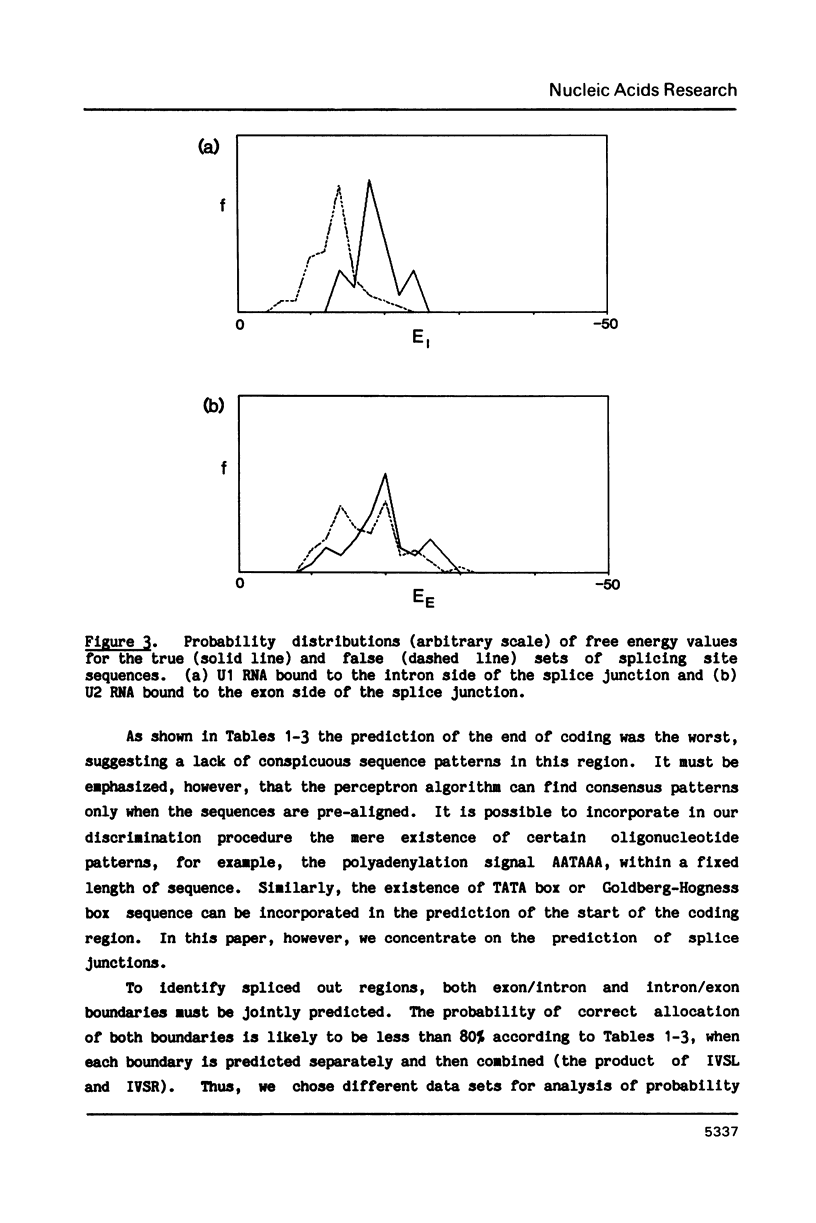
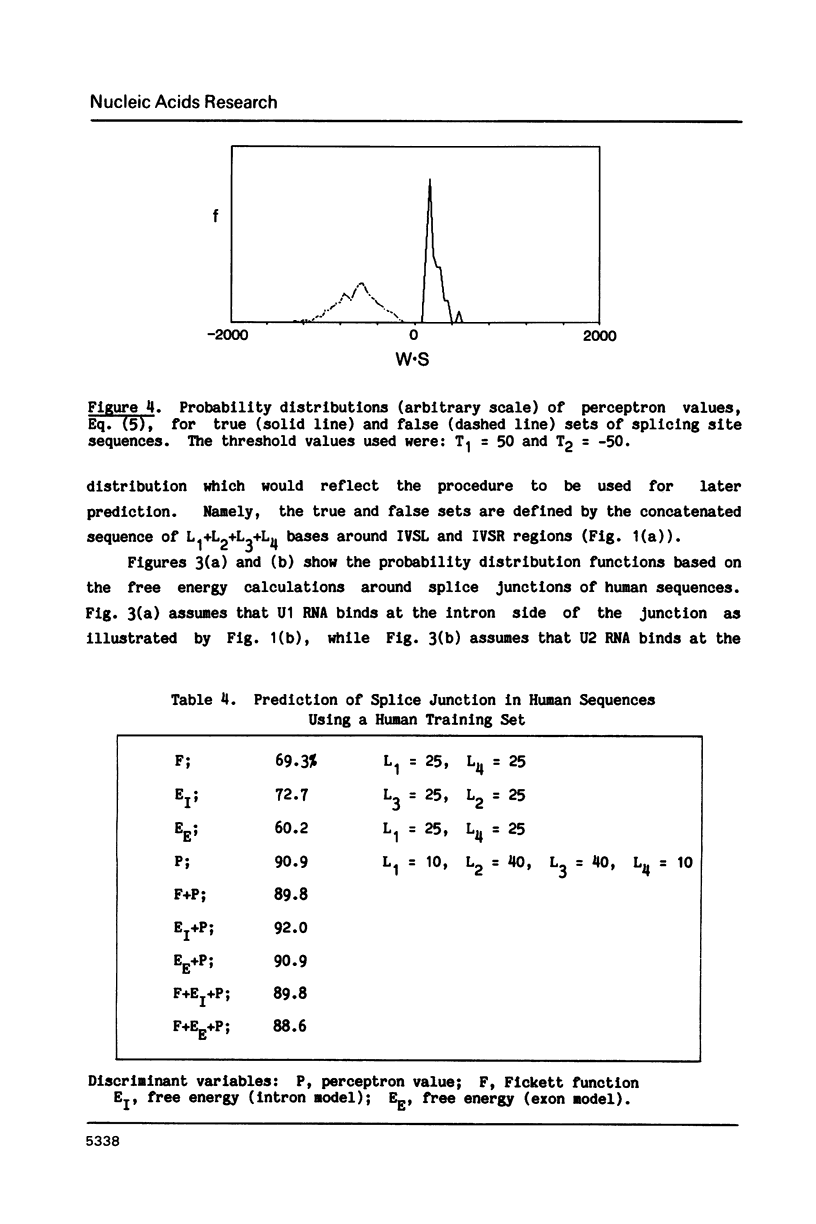
A general method based on the statistical technique of discriminant analysis is developed to distinguish boundaries of coding and non-coding regions in nucleic acid sequences. In particular, the method is applied to the prediction of splicing sites in messenger RNA precursors. Information used for discrimination includes consensus sequence patterns around splice junctions, free energy of snRNA and mRNA base pairing, and statistical differences between coding and non-coding regions such as periodic appearance of specific bases in coding regions reflecting the non-random usage of degenerate codons. Given the reading frame of an exon (but not the exon/intron boundaries), the method will predict the following exon, namely, the intron to be excised out. When applied to human sequences in the GenBank database, the method correctly identified 80% of true splice junctions.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Branlant C., Krol A., Ebel J. P., Lazar E., Gallinaro H., Jacob M., Sri-Widada J., Jeanteur P. Nucleotide sequences of nuclear U1A RNAs from chicken, rat and man. Nucleic Acids Res. 1980 Sep 25;8(18):4143–4154. doi: 10.1093/nar/8.18.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Cech T. R. RNA splicing: three themes with variations. Cell. 1983 Oct;34(3):713–716. doi: 10.1016/0092-8674(83)90527-5. [DOI] [PubMed] [Google Scholar]
- Fickett J. W. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res. 1982 Sep 11;10(17):5303–5318. doi: 10.1093/nar/10.17.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein P., Kanehisa M., DeLisi C. Prediction of protein function from sequence properties. Discriminant analysis of a data base. Biochim Biophys Acta. 1984 Jun 28;787(3):221–226. doi: 10.1016/0167-4838(84)90312-1. [DOI] [PubMed] [Google Scholar]
- Lerner M. R., Boyle J. A., Mount S. M., Wolin S. L., Steitz J. A. Are snRNPs involved in splicing? Nature. 1980 Jan 10;283(5743):220–224. doi: 10.1038/283220a0. [DOI] [PubMed] [Google Scholar]
- Ohshima Y., Itoh M., Okada N., Miyata T. Novel models for RNA splicing that involve a small nuclear RNA. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4471–4474. doi: 10.1073/pnas.78.7.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy R., Henning D., Epstein P., Busch H. Primary and secondary structure of U2 snRNA. Nucleic Acids Res. 1981 Nov 11;9(21):5645–5658. doi: 10.1093/nar/9.21.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A. Speculations on RNA splicing. Cell. 1981 Mar;23(3):643–646. doi: 10.1016/0092-8674(81)90425-6. [DOI] [PubMed] [Google Scholar]
- Shepherd J. C. Method to determine the reading frame of a protein from the purine/pyrimidine genome sequence and its possible evolutionary justification. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1596–1600. doi: 10.1073/pnas.78.3.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):505–519. doi: 10.1093/nar/12.1part2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L., Ehrenfeucht A. Use of the 'Perceptron' algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2997–3011. doi: 10.1093/nar/10.9.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]



