Abstract
Maraviroc is the first CCR5 antagonist approved for the treatment of HIV-1 infection. It specifically inhibits the replication of R5 viruses by blocking viral entry. HIV-1 tropism can be estimated accurately and predict viral response to maraviroc. Genotypic tools are increasingly replacing phenotypic assays in most places. The favorable pharmacokinetic properties and the good safety profile of maraviroc may support an earlier use of the drug in HIV-1 infection, as well as favor its consideration as part of switch strategies in patients under suppressive antiret-roviral regimens containing less-well-tolerated drugs. Moreover, a particular immune benefit of maraviroc might encourage its use as part of intensification strategies in HIV-infected patients with impaired CD4 gains despite prolonged suppression of HIV replication with antiretroviral therapy. However, the long-term consequences of using maraviroc must be carefully checked, given its particular mechanism of action, blocking a physiologic cell receptor.
Keywords: maraviroc, tropism, HIV, antiretroviral therapy, treatment strategies
Introduction
Although most HIV+ patients respond well to current antiretroviral regimens, a subset show treatment failure generally due to selection of drug resistance. For this reason, new therapeutic options for HIV+ patients continue to be needed. The development and approval of new antiretroviral drugs targeting different steps of the HIV replication cycle is one of the best ways to ensure this goal. These new drugs are active against HIV variants resistant to former compounds and generally exhibit better safety profiles. An attractive step in the HIV replication cycle to be targeted is viral entry.1,2 The fusion inhibitor enfuvirtide3 and the CCR5 antagonist maraviroc4 (Celsentri® [UK], Selzentry® [US]; Pfizer) are so far the only entry inhibitors approved for clinical use. Maraviroc is the first CCR5 antagonist and the unique oral HIV entry inhibitor. It was approved by the US Food and Drug Administration on June 2007 for the treatment of antiretroviral-experienced patients failing prior regimens. Maraviroc exclusively inhibits the replication of R5-tropic HIV variants by an allosteric mechanism after binding to the transmembrane CCR5 co-receptor cavity. This review goes over the main features of maraviroc, in an attempt to define its optimal use in clinical practice.
Mechanism of action of CCR5 antagonists
CCR5 antagonists are the newest agents that have entered the HIV armamentarium and the second class of entry inhibitors to gain regulatory approval. During the HIV entry process, the CD4-gp120 interaction induces conformational changes in the viral envelope that expose a chemokine receptor binding site and consequently allows the CD4-gp120 complex to interact with a chemokine co-receptor, typically CCR5 or CXCR4. The CD4-gp120 complex binds to either co-receptor through interactions with the V3 region of gp120, though other HIV gp120 regions such as V1/V2, C4 and the bridging sheet are also involved.5 The use of CCR5 or CXCR4 co-receptors by HIV-1 is mainly determined by the amino acid sequence of the V3 region of gp120.6,7 Accordingly, HIV isolates are classified as either R5-tropic, X4-tropic, or as dual/mixed tropic, depending on their co-receptor use.8 The term dual/mixed refers to isolates that may contain true dual tropic viruses (particles that can use either or both chemokine co-receptors) or mixtures of viruses that exclusively use CCR5 and others that use CXCR4, thus giving the virus population a dual tropic character due to the mixed tropism of the individual viral variants.
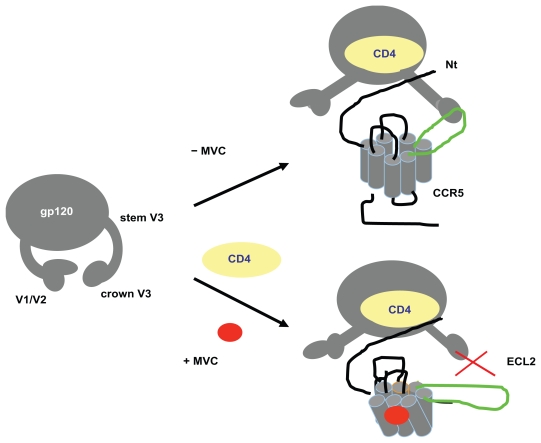
Maraviroc is an allosteric inhibitor of the CCR5 chemokine co-receptor, orally bioavailable with potent in vitro activity (IC90 ~2 nM). Maraviroc binds to the trans-membrane co-receptor cavity, within the 2, 3, 6 and 7 helix.9,10 Following binding, CCR5 coreceptor conformational changes occur, especially in the ECL2 region, which ultimately mean that maraviroc can no longer interact with the V3 crown of the HIV envelope (Figure 1).
Figure 1.
Mechanism of action of maraviroc (MVC).
HIV tropism testing in the clinic
CCR5 antagonists do not display activity against CXCR4-using HIV variants. Consequently, the presence of detectable X4 or R5/X4 dual-tropic viruses has been associated with therapeutic failure using maraviroc.11–13 Therefore, assessment of HIV-1 tropism is required before recommending treatment with CCR5 antagonists. Several assays have been developed to determine HIV tropism in clinical samples.14,15 The Trofile ® phenotypic assay (Monogram Biosciences, South San Francisco, CA), which is based on the recombinant virus technology,16 has been extensively utilized to provide tropism information in clinical trials, showing good correlation with virological outcomes, and accordingly it has been the most widely used to date.15–17
The Trofile® assay identifies X4 strains with a sensitivity of 10% when using clonal mixtures, but does not differentiate between dually tropic viruses and mixtures of X4 and R5 strains, reporting results as “dually or mixed” (D/M) virus.16 Monogram Biosciences has developed an enhanced sensitivity tropism assay (ESTA), which is 10- to 100-fold more sensitive for detecting X4 minor populations when using clonal mixtures.17 ESTA has been available since June 2008 and has replaced the original Trofile® assay used in the pivotal clinical trials.
Although phenotypic assays such as Trofile® are considered reliable for assessing HIV tropism, they remain far from perfect as diagnostic tests for clinical purposes. They are labor intensive, expensive and require special laboratory facilities and expertize. They are not widely available, and in the case of Trofile® specimens must be shipped to the reference laboratory in the United States. Moreover, up to 15% of specimens are non-reportable even when testing samples with plasma HIV-RNA > 1000 copies/mL. In recent years, efforts have been made to explore alternative testing approaches, mainly using genotypic predictors of viral tropism, as a guide to the use of maraviroc in clinical practice.
The reliability of genotypic tools to determine HIV tropism in clinical samples compared with phenotypic assays has been examined in multiple studies, mainly conducted in Europe and Canada. Some of these comparisons showed relatively poor concordances, mainly due to low sensitivity (< 45%) in detecting X4 variants by genotypic algorithms;18 however, more recent studies have demonstrated improved sensitivity when using certain genotypic tools and/or using phenotypic assays other than Trofile® as the reference “gold standard”.19–22 Different strategies to improve the sensitivity of genotypic methods to detect X4 variants have been examined, including simple modifications in the interpretation algorithms,23 or adding structural/biochemical properties of the V3 loop and clinical parameters such as CD4 and CD8 counts, and plasma viremia in the final report interpretation.24 Finally, another approach has combined the results given by different genotypic algorithms to produce a “pooled” X4 sensitive tropism prediction.25
The validation of genotypic tropism prediction methods, however, do not require perfect concordance with the Trofile® (or ESTA) assay, but rather evidence of a similar ability to correctly identify patients who will benefit from the use of maraviroc. In this context, recent studies have shown that the use of genotypic tropism prediction tools, based on V3 sequence data, have an ability similar to that of Trofile® to predict virological response to maraviroc and therefore can reliably guide clinical practice.26,27
The pyrosequencing technology may enable the composition of viral quasispecies to be explored more deeply.28,29 It may provide a unique opportunity to enhance the sensitivity for identif ication of elusive minority variants, including minimally present X4 viruses.30 Ultradeep sequencing, however, is a sophisticated and expensive method, available in only a few research facilities. Moreover, the analysis of large amount of sequencing data generated for each sample remains quite challenging. Additionally, the interpretation of results is related to the optimal sensitivity threshold for X4 variants that may be clinically relevant. Preliminary data suggest that the proportion of X4 viruses beyond 5% (between 1% and 10%) may have a clinical impact, but not below this threshold, and therefore deeper exploration may not be needed in terms of clinical application in routine diagnostic settings.
The potential for using maraviroc without knowing the result of a preceding tropism assay is also under debate. This approach could be of especial interest in antiretroviral-naïve patients with good CD4 counts in whom R5-tropic variants are predominant (74%–82%),31 or in subjects infected with HIV-1 subtypes with a low propensity for CXCR4 use, such as clade C.32
Efficacy and safety of maraviroc in clinical trials
Antiretroviral-naïve patients
The in vivo antiviral activity of maraviroc was initially described in a monotherapy study which compared several once-daily (QD) and twice-daily (BID) doses of the drug (25, 50, 100, 150, 300 mg) vs placebo over 10 days in HIV-infected drug-naïve volunteers infected with R5-tropic viruses. Reductions in plasma HIV-RNA ≥ 1 log copies/mL were observed with all doses of 100 mg or greater.33
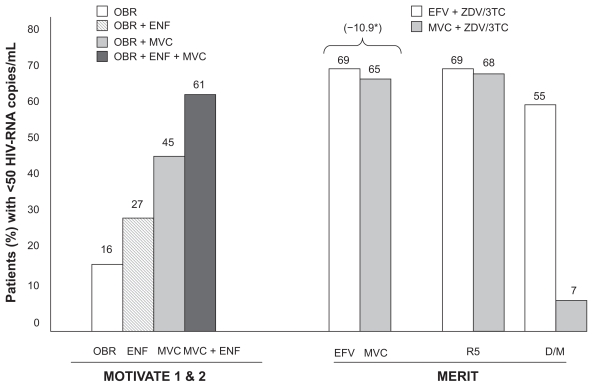
The MERIT trial evaluated the safety and efficacy of maraviroc (300 mg BID) vs efavirenz (600 mg QD), each in combination with zidovudine and lamivudine, in drug-naïve HIV-1 patients. The trial failed to demonstrate non-inferiority of either QD or BID maraviroc arms compared to efavirenz using the attainment of plasma HIV-RNA < 50 copies/mL at week 48 as the primary endpoint. 34 However, the non-inferiority was demonstrated for viral load suppression < 400 HIV-RNA copies/mL. Subsequently, the MERIT trial was analyzed in detail to understand the virological correlates of treatment failure. In 13 patients receiving maraviroc (3.8%) a switch in viral tropism was observed from R5 to D/M between the screening and baseline. In this subset of patients a reduced response to maraviroc compared to efavirenz was observed (54.6% vs 7.1% achieved <50 HIV-RNA copies/mL, respectively), while for patients harboring R5 viruses, similar responses were observed between maraviroc and efavirenz groups (69.3% vs 68%, respectively) (Figure 2).35 Therefore, the presence of X4-variants within the quasispecies population at a rate just below the sensitivity threshold of the old Trofile® assay at baseline seemed to be an important predictor of failure on maraviroc in drug-naïve patients.
Figure 2.
Efficacy of maraviroc in MOTIVATE and MERIT trials: 48-week results.
*Non-inferiority margin in –10%.
Abbreviations: OBR, optimized background regimen; ENF, enfuvirtide; MVC, maraviroc; EFV, efavirenz; ZDV, zidovudine.
Consistent with this hypothesis, a re-analysis of the MERIT trial using ESTA reclassified as D/M nearly 15% of viruses from samples originally scored as having R5 by Trofile®. Following this new assignment, the proportion of patients achieving <50 HIV-RNA copies/mL at 48 weeks was the same (68%) in both maraviroc and efavirenz arms.36
More recently, the 96-week results of the MERIT trial were reported only for the subset of patients with baseline R5 using ESTA, and maraviroc remained non-inferior to efavirenz in terms of plasma HIV-RNA suppression below 50 copies/mL (59% vs 62%, respectively); however, more patients discontinued efavirenz due to side effects while more failed virologically in the maraviroc arm.37
Since the proportion of patients harboring R5 variants in this drug-naïve population was 80%,36 the use of maraviroc as first-line therapy, especially in patients presenting with non-advanced immune deficiency, may be considered as an attractive option. In contrast, in advanced stages of HIV infection approximately half of the patients may harbor X4 viruses as a dominant or just significant population.13
Antiretroviral-experienced patients
In the MOTIVATE 1 (conducted in the US and Canada) and MOTIVATE 2 (in Europe, Australia and the US) trials (Maraviroc plus Optimized Background Therapy in Viremic, ART-Experienced Patients), triple-class-resistant patients harboring R5-tropic virus at baseline were randomized to receive maraviroc 150 or 300 mg QD or BID or placebo, each combined with an optimized background regimen (OBR). In these double-blind, placebo-controlled studies, maraviroc plus an OBR demonstrated significantly greater virological and immunological efficacy and similar safety profile compared with an OBR alone during 48 weeks. Nearly twice the percentage of patients on maraviroc BID plus OBR vs placebo plus OBR achieved the primary end-point of plasma HIV-RNA 50 copies/mL compared with those receiving an OBR alone (45% vs 16%, respectively)38 (Figure 2).
Maraviroc demonstrated a better virologic efficacy in treatment-experienced subjects when combined with ≥ 1 active agent in the OBR. Overall the MOTIVATE trials demonstrated the beneficial effect of combining both entry inhibitors maraviroc and enfuvirtide. The proportion of patients that achieved HIV-RNA < 50 copies/mL was substantially higher in patients who received enfuvirtide for the first time along with maraviroc than those who received enfuvirtide plus OBR, or those who received an OBR alone (61% vs 27% vs16 %) (Figure 2). Similar data were obtained in patients who received maraviroc along with a protease inhibitor (PI) for the first-time and lacking PI-resistant mutations. Altogether these data point out the importance of including additional fully active agents when using maraviroc in heavily antiretroviral-experienced patients.38,39
The A4001029 study compared the activity of maraviroc vs placebo in treatment-experienced individuals with dual/mixed (D/M) or X4 viruses, who also received an OBR.40 This study showed no significant differences in plasma HIV-RNA outcomes in the maraviroc group compared to subjects who received placebo, highlighting the limited antiviral activity of maraviroc when confronting CXCR4-tropic viruses.
Recently Valdez et al41 showed that a weighted optimized background treatment susceptibility score, rather than low-level X4 viruses at baseline (as defined as a change in Trofile® test result from R5 at screening to DM at baseline), was the strongest predictor of virological response at 48 weeks in the MOTIVATE trials. This finding highlights the contribution to virus suppression of other antiretroviral drugs, such as most nucleoside analogues or PIs, for which partial activity may be recognized when confronting viruses with only a few drug resistance mutations. It is the activity of the accompanying drugs that may enable maraviroc to benefit patients with a low proportion of X4 variants.
In this regard, a re-analysis of the maraviroc A4001029 study, in which all enrolled patients had baseline evidence of X4 or D/M viruses by Trofile®, demonstrated by ultradeep sequencing that there is an inverse relationship between the proportion of plasma variants and the extent of virological responses to maraviroc.42 Overall, patients with a low prevalence of X4 variants (<10%) showed a substantial viral load decline (−2.6 log HIV-RNA copies/mL at week 8), regard-less of the result provided by Trofile®. If these results are confirmed, patients with <10% X4 variants might benefit from maraviroc therapy.
Based on these findings, it may be proposed that attempts at categorically excluding presence of X4 strains at very low frequency within the viral population may lead to the unnecessary exclusion of a therapeutic option that could still provide at least partial activity.
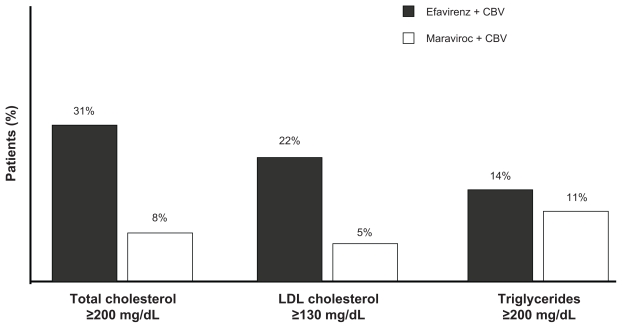
Maraviroc has demonstrated an excellent safety profile in clinical trials. Data from the MERIT trial showed that fewer patients discontinued maraviroc than efavirenz due to adverse events (4.2% vs 13.6%, respectively).34 Moreover, lipid abnormalities occurred less frequently in patients taking maraviroc than efavirenz34 (Figure 3). Maraviroc can be given once a day (600 mg) as two pills,43 causes minimal side effects and displays easily manageable drug interactions. No evidence of an increased incidence of malignancies was reported in the MOTIVATE trials. The favorable pharmacokinetic properties and the safety profile of maraviroc may support an earlier use of the drug in HIV-1 infection beyond the initial approval for antiretroviral-experienced patients.
Figure 3.
Lipid profile in the MERIT trial.
Novel scenarios for maraviroc use
Simplification strategies
The long-term use of the most commonly prescribed antiretroviral drugs has been associated with a broad range of adverse events, including metabolic abnormalities,44 increased cardiovascular risk,45,46 lipodistrophy,47 hepatotoxicity,48 gastrointestinal disturbances49 and neuropsychiatric conditions.50 In contrast, maraviroc has demonstrated an excellent safety profile in clinical trials.34 Maraviroc has also demonstrated a favorable pharmacokinetic profile when given once a day (600 mg) as two pills,42 displaying easily manageable drug interactions. Its convenience and safety profiles may further support consideration of maraviroc as part of switch strategies in patients having suppressed HIV replication under regimens that are less well tolerated.
Immunological non-responders
In the MOTIVATE trials, a greater mean CD4 gain was observed in patients on maraviroc who experienced virological failure compared to the placebo arm (mean [95% confidence interval (CI)]: +64 [47 to 82], +74 [56 to 92] and +24 [10 to 40] cells/mm3, respectively).38,39 Based on this observation, treatment with maraviroc might provide an immunological benefit beyond its direct antiviral activity. This phenomenon has already been reported in HIV patients failing the fusion inhibitor enfuvirtide;50–53 however, the underlying mechanisms are still unknown. The particular immune benefit provided by maraviroc therapy may support its use as part of intensification strategies for the subset of HIV+ individuals with low CD4 counts showing an impaired CD4 gain despite prolonged suppression of HIV replication with highly active antiretroviral therapy (HAART). Although preliminary results have so far being modest in this specific situation,54,55 further studies including larger number of patients and longer follow-up are warranted.
In HIV patients with undetectable viremia, determination of viral tropism in plasma is not feasible. Two alternative strategies may be proposed in this situation. Firstly, viral tropism should be checked in stored plasma specimens collected before initiating HAART. Secondly, viral tropism should be assessed in current proviral DNA. Emerging information from studies examining the dynamics of viral tropism during prolonged HIV suppression under HAART and about the extent of correlation between plasma and cell compartments support any of these approaches.56,57 Shifts in viral tropism under prolonged suppressive HAART seem to be very rare and correlation between plasma RNA and proviral DNA is good, although X4 variants can be recognized more often in proviral DNA than in plasma. As a result, maraviroc might be confidently used as part of simplification or intensification strategies as long as viral tropism excluded X4 variants in retrospective plasma specimens, or following testing of current proviral DNA.
Conclusions
Maraviroc is the first CCR5 antagonist approved for the treatment of HIV-1 infection, which exclusively inhibits the replication of R5 viruses. HIV-1 tropism can accurately be estimated and predict viral response to maraviroc. Genotypic tools are increasingly replacing the initial phenotypic assays. The favorable pharmacokinetic properties and the good safety profile of maraviroc may support an earlier use of the drug in HIV-1 infection, as well as favor its consideration as part of switch strategies in patients under suppressive HAART with less-well-tolerated drugs. However, long-term adverse events using this drug for long periods must be carefully checked, given the particular mechanism of action of maraviroc (blocking a physiologic cell receptor). A particular immune benefit of maraviroc might encourage the use of the drug as part of intensification strategies in HIV+ patients with impaired CD4 gains despite prolonged suppression of HIV replication under HAART. Finally, the performance of maraviroc in resource-limited settings must be examined, as the majority of persons infected with HIV live in such settings.
Acknowledgments
This work was funded in part by grants from Fundación Investigación y Educación en SIDA (IES), Red de Investigación en SIDA (RIS, ISCIII-RETIC-RD06), and Fondo de Investigación Sanitaria (CP08/00214).
Footnotes
Disclosures
The authors disclose no conflicts of interest.
References
- 1.Briz V, Poveda E, Soriano V. HIV entry inhibitors: mechanisms of action and resistance pathways. J Antimicrob Chemother. 2006;57:619–627. doi: 10.1093/jac/dkl027. [DOI] [PubMed] [Google Scholar]
- 2.Esté J, Telenti A. HIV entry inhibitors. Lancet. 2007;370:81–88. doi: 10.1016/S0140-6736(07)61052-6. [DOI] [PubMed] [Google Scholar]
- 3.Roberston D. US FDA approves new class of HIV therapeutics. Nat Biotechnol. 2003;21:470–471. doi: 10.1038/nbt0503-470. [DOI] [PubMed] [Google Scholar]
- 4.Anonymous. FDA approves maraviroc tablets. AIDS Patient Care STDs. 2007;21:702. doi: 10.1089/apc.2007.9970. [DOI] [PubMed] [Google Scholar]
- 5.Comier E, Dragic T. The crown and stem of the V3 loop play distinct roles in HIV type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J Virol. 2002;76:8953–8957. doi: 10.1128/JVI.76.17.8953-8957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan S, Speck R, Power C, et al. V3 recombinants indicate a central role for CCR5 as coreceptor in tissue infection by HIV type 1. J Virol. 1999;73:2350–2358. doi: 10.1128/jvi.73.3.2350-2358.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen M, Van’t Wout A. Predicting HIV-1 coreceptor usage with sequence analysis. AIDS Rev. 2003;19:145–149. [PubMed] [Google Scholar]
- 8.Berger E, Doms R, Fenyö E, Korber BT, Littman DR, Moore JP, et al. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 9.Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, Macartney M, et al. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-HIV type 1 activity. Antimicrob Agents Chemother. 2005;49:4721–4732. doi: 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castonguay L, Wengg Y, Adolfsen W, Di Salvo, Kilburn R, Caldwell C, et al. Binding of 2-aryl-4(piperidin-1yl)butanmines and 1, 3, 4-trisubstitued pyrrolidines to human CCR5: a molecular modelling-guide mutagenesis study of the binding pocket. Biochemistry. 2003;42:1544–1550. doi: 10.1021/bi026639s. [DOI] [PubMed] [Google Scholar]
- 11.Saag M, Goodrich J, Fatkenheuer G, Clotet B, Clumeck N, Sullivan J, et al. A double-blind, placebo-controlled trial of maraviroc in treatment-experienced patients infected with non-R5 HIV-1. J Infect Dis. 2009;199:1638–1647. doi: 10.1086/598965. [DOI] [PubMed] [Google Scholar]
- 12.Fatkenheuer G, Nelson M, Lazzarin A, Konourina I, Hoepelman A, Lampiris H, et al. Subgroup analyses of maraviroc in previously treated R5 HIV-1 infection. N Engl J Med. 2008;359:1442–1455. doi: 10.1056/NEJMoa0803154. [DOI] [PubMed] [Google Scholar]
- 13.Vandekerckhove L, Verhofstede C, Vogelaers D. Maraviroc: spectives for use in antiretroviral-naïve HIV-1-infected patients. J Antimicrob Chemother. 2009;63:1087–1096. doi: 10.1093/jac/dkp113. [DOI] [PubMed] [Google Scholar]
- 14.Poveda E, Briz V, Quinones-Mateu M, Soriano V. HIV tropism: diagnostic tools and implications for disease progression and treatment with entry inhibitors. AIDS. 2006;20:1359–1367. doi: 10.1097/01.aids.0000233569.74769.69. [DOI] [PubMed] [Google Scholar]
- 15.Rose J, Rhea A, Weber J, Quiñones-Mateu M. Current tests to evaluate HIV-1 coreceptor tropism. Current Opin HIV AIDS. 2009;4:136–142. doi: 10.1097/COH.0b013e328322f973. [DOI] [PubMed] [Google Scholar]
- 16.Whitcomb J, Huang W, Fransen S, Limoli K, Toma J, Wrin T, et al. Development and characterization of a novel single-cycle recombinant-virus assay to determine HIV type 1 coreceptor tropism. Antimicrob Agents Chemother. 2007;51:566–575. doi: 10.1128/AAC.00853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trinh L, Han D, Huang W, Wrin T, Larson J, Kiss L, et al. Technical validation of an enhanced sensitivity Trofile HIV coreceptor tropism assay for selecting patients for therapy with entry inhibitors targeting CCR5. Antivir Ther. 2008;13(Suppl 3):A128. [Google Scholar]
- 18.Low A, Dong W, Chan D, Sing T, Swanstrom R, Jensen M, et al. Current V3 genotyping algorithms are inadequate for predicting X4 co-receptor usage in clinical isolates. AIDS. 2007;21:F17–F24. doi: 10.1097/QAD.0b013e3282ef81ea. [DOI] [PubMed] [Google Scholar]
- 19.Poveda E, Briz V, Roulet V, González MM, Faudon J, Skrabal K, et al. Correlation between a phenotypic assay and three bioinformatics tools for determining HIV coreceptor use. AIDS. 2007;21:1487–1490. doi: 10.1097/QAD.0b013e32826fb741. [DOI] [PubMed] [Google Scholar]
- 20.Raymond S, Delobel P, Mavigner M, Cazabat M, Souyris C, Sandres-Sauné K, et al. Correlation between genotypic predictions based on V3 sequences and phenotypic determination of HIV-1 tropism. AIDS. 2008;22:F11–F17. doi: 10.1097/QAD.0b013e32830ebcd4. [DOI] [PubMed] [Google Scholar]
- 21.Garrido C, Roulet V, Chueca N, Poveda E, Aguilera A, Skrabal K, et al. Evaluation of eight different bioinformatics tools to predict viral tropism in different HIV-1 subtypes. J Clin Microbiol. 2008;61:694–698. doi: 10.1128/JCM.01611-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Mendoza C, Van Baelen K, Poveda E, Rondelez E, Zahonero N, Stuyer L, et al. Performance of a population-based HIV-1 tropism phenotypic assay and correlation with V3 genotypic prediction tools in recent HIV-1 seroconverters. J Acquir Immune Defic Syndr. 2008;48:241–244. doi: 10.1097/QAI.0b013e3181734f0e. [DOI] [PubMed] [Google Scholar]
- 23.Poveda E, Seclén E, Gonzalez M, García F, Chueca N, Aguilera A, et al. Design and validation of new genotypic tools for easy and reliable estimation of HIV tropism before using CCR5 antagonists. J Antimicrob Chemother. 2009;63:1006–1010. doi: 10.1093/jac/dkp063. [DOI] [PubMed] [Google Scholar]
- 24.Strang A, Cameron J, Booth C, Garcia-Diaz A, Geretti AM. Genotypic prediction of viral co-receptor tropism: correlation with enhanced Trofile. 7th European HIV Drug Resistance Workshop; Stockholm, Sweden. March 25–27 2009; [abstract 80] [Google Scholar]
- 25.Chueca N, Martin L, Alvarez M, Peña A, Guillot V, Garcia-Casas V, et al. A combination of bioinformatics tools can be accurately used for the screening of coreceptor usage in clinical samples. Antivir Ther. 2008;13(Suppl 3):A106. [Google Scholar]
- 26.Harrigan PR, McGovern R, Dong W, Thielen A, Jensen M, Mo T, et al. Screening for HIV tropism using population-based V3 genotypic analysis: a retrospective virological outcome analysis using stored plasma screening samples from MOTIVATE-1. Antivir Ther. 2009;14(Suppl):A17. [Google Scholar]
- 27.Obermeier M, Carganico A, Berg T, Hintsche B, Koppe S, Moll A, et al. The Berlin Maraviroc cohort – influence of genotypic tropism testing results on therapeutic outcomes. 7th European HIV Drug Resistance Workshop; Stockholm, Sweden. March 25–27 2009; [abstract 79] [Google Scholar]
- 28.Tsibris A, Korber B, Arnaout R, Russ C, Lo C, Leitner T, et al. Quantitative deep sequencing reveals dynamic HIV-1 escape and large population shifts during CCR5 antagonist therapy in vivo. PLoS One. 2009;4:e5683. doi: 10.1371/journal.pone.0005683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Archer J, Braverman M, Taillon B, Desany B, James I, Harrigan PR, et al. Detection of low-frequency pre-therapy chemokine (CXC motif) receptor 4 (CXCR4)-using HIV-1 with ultra-deep pyrosequencing. AIDS. 2009;23:1209–1218. doi: 10.1097/QAD.0b013e32832b4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Däumer M, Kaiser R, Klein R, Lengauer T, Thiele B, Thielen A. Inferring viral tropism from genotype with massively parallel sequencing: qualitative and quantitative analysis. [abstract] Antivir Ther. 2008;13(Suppl 3):A101. [Google Scholar]
- 31.Brumme Z, Goodrich J, Mayer H, Brumme C, Henrick B, Wynhoven B, et al. Molecular and clinical epidemiology of CXCR4-using HIV-1 in a large population of antiretroviral-naive individuals. J Infect Dis. 2005;192:466–474. doi: 10.1086/431519. [DOI] [PubMed] [Google Scholar]
- 32.Ping L, Nelson J, Hoffman I, Schock J, Lamers S, Goodman M, et al. Characterization of V3 sequence heterogeneity in subtype C HIV type 1 isolates from Malawi: underrepresentation of X4 variants. J Virol. 1999;73:6271–6281. doi: 10.1128/jvi.73.8.6271-6281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fatkenheuer G, Pozniak A, Johnson M, Johnson M, Plettenberg A, Staszewski S, et al. Efficacy of short-term monotherapy with maraviroc, a new CCR5 antagonist, in patients infected with HIV-1. Nat Med. 2005;11:1170–1172. doi: 10.1038/nm1319. [DOI] [PubMed] [Google Scholar]
- 34.Saag M, Ive P, Heera J, Tawadrous M, DeJesus E, Clumeck N, et al. A multicenter, randomized, double-blind, comparative trial of a novel CCR5 antagonist, maraviroc vs efavirenz, both in combination with Combivir (zidovudine/lamivudine), for the treatment of antiretroviral naive patients infected with R5 HIV-1: week 48 results of the MERIT study. 4th IAS Conference on HIV pathogenesis, treatment and prevention; Sydney, Australia. 2007. Abstract WESS104. [Google Scholar]
- 35.Heera J, Saag M, Ive P, Whitcomb J, Lewis M, McFadyen L, et al. Virological correlates associated with treatment failure at week 48 in the phase 3 study of Maraviroc in treatment-naïve patients. 15th Conference on Retroviruses and Opportunistic Infections; Boston, MA. 2008. Abstract 40LB. [Google Scholar]
- 36.Saag M, Heera J, Goodrich J, DeJesus E, Clumeck N, Cooper D, et al. Reanalysis of the MERIT study with the enhanced Trofile Assay (MERIT-ES). 48th ICAAC Annual/IDSA 46th Annual Meeting; October 25–28 2008; Washington, DC. Abstract H-1269. [Google Scholar]
- 37.Nelson M on behalf of the MERIT study team. 96-week results of the MERIT trial. International AIDS Conference; Cape Town. July 2009; [abstract MOPEBO4O] [Google Scholar]
- 38.Gulick R, Lalezari J, Goodrich J, Clumeck N, De Jesus E, Horban A, et al. Maraviroc for previously treated patients with R5 HIV-1 infection. N Engl J Med. 2008;359:1429–1441. doi: 10.1056/NEJMoa0803152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fätkenheuer G, Nelson M, Lazzarin A, Konourina I, Hoepelman AI, Lampiris H, et al. Subgroup analyses of maraviroc in previously treated R5 HIV-1 infection. N Engl J Med. 2008;359:1442–1455. doi: 10.1056/NEJMoa0803154. [DOI] [PubMed] [Google Scholar]
- 40.Saag M, Goodrich J, Fatkenheuer G, Clotet B, Clumeck N, Sullivan J, et al. A double-blind, placebo-controlled trial of maraviroc in treatment-experienced patients infected with non-R5 HIV-1. J Infect Dis. 2009;199:1638–1647. doi: 10.1086/598965. [DOI] [PubMed] [Google Scholar]
- 41.Valdez H, Lewis M, Delogne C, Simpson P. Weighted OBT susceptibility score (wOBTSS) is a stronger predictor of virologic response at 48 weeks than baseline tropism result in MOTIVATE 1 and 2. Program and abstract of the 48th Annual ICAAC/IDSA 46th Annual Meeting; October 25–28 2008; Washington, DC. Abstract H-1221. [Google Scholar]
- 42.Swenson L, Dong W, Mo T, Woods C, Thielen A, Jensen M, et al. Quantification of HIV Tropism by “deep” sequencing shows a broad distribution of prevalence of X4 variants in clinical samples that is associated with virological outcome. 16th Conference on Retroviruses and Opportunistic Infections; 8–11 February 2009; Montreal, Canada. Abstract 680. [Google Scholar]
- 43.Abel S, Back D, Vourvahis M. Maraviroc: pharmacokinetics and drug interactions. Antivir Ther. 2009;14:607–618. [PubMed] [Google Scholar]
- 44.Barbaro G, Iacobellis G. Metabolic syndrome associated with HIV and highly active antiretroviral therapy. Curr Diab Rep. 2009;9:37–42. doi: 10.1007/s11892-009-0008-7. [DOI] [PubMed] [Google Scholar]
- 45.D:A:D Study Group. Sabin C, Worm S, Weber R, Reiss P, El-Sadr W, et al. Use of nucleoside reverse transcriptase inhibitors and risk of myocardial infarction in HIV-infected patients enrolled in the D:A:D study: a multicohort collaboration. Lancet. 2008;371:1417–1426. doi: 10.1016/S0140-6736(08)60423-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lundgren J, Reiss P, Worm S, Weber R, El-Sadr W, De Wit S, et al. Risk of myocardial infarction with exposure to specific ARV from the PI, NNRTI, and NRTI drug classes: the D:A:D study. 16th Conference on Retroviruses and Opportunistic Infections; 8–11 February 2009; Montreal, Canada. Abstract 44LB. [Google Scholar]
- 47.Giralt M, Domingo P, Gallart J, Rodriguez de la Concepción ML, Alegre M, Domingo J, et al. HIV-1 infection alters gene expression in adipose tissue, which contributes to HIV-1/HAART-associated lipodystrophy. Antivir Ther. 2006;11:729–740. [PubMed] [Google Scholar]
- 48.Soriano V, Puoti M, Garcia-Gasco P, Rockstroh J, Benhamou Y, Barreiro P, et al. Antiretroviral drugs and liver injury. AIDS. 2008;22:1–13. doi: 10.1097/QAD.0b013e3282f0e2fd. [DOI] [PubMed] [Google Scholar]
- 49.Hill A, Balkin A. Risk factors for gastrointestinal adverse events in HIV treated and untreated patients. AIDS Rev. 2009;11:30–38. [PubMed] [Google Scholar]
- 50.Poveda E, Rodés B, Labernardière JL, Benito J, Toro C, González-Lahoz J, et al. Evolution of genotypic and phenotypic resistance to enfuvirtide in HIV-infected patients experiencing prolonged virologic failure. J Med Virol. 2004;74:21–28. doi: 10.1002/jmv.20141. [DOI] [PubMed] [Google Scholar]
- 51.Wei X, Decker J, Liu H, Zhang Z, Arani R, Kilby J, et al. Emergence of resistant HIV-1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother. 2002;46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aquaro S, D’Arrigo R, Svicher V, Di Perri G, Lo Caputo S, Visco-Comandini U, et al. Specific mutations in HIV-1 gp41 are associated with immunological success in HIV-1 infected patients receiving enfuvirtide treatment. J Antimicrob Chemother. 2006;58:714–722. doi: 10.1093/jac/dkl306. [DOI] [PubMed] [Google Scholar]
- 53.Poveda E, Briz V, Soriano V. Enfuvirtide, the first fusion inhibitor to treat HIV infection. AIDS Rev. 2005;7:139–147. [PubMed] [Google Scholar]
- 54.Lanzafame M, Lattuada E, Vento S. Maraviroc and CD4+ cell count recovery in patients with virologic suppression and blunted CD4+ cell response. AIDS. 2009;27:869. doi: 10.1097/QAD.0b013e3283262aa0. [DOI] [PubMed] [Google Scholar]
- 55.Stepanyuk O, Chiang T, Dever L, Paez S, Smith S, Perez G, et al. Impact of adding maraviroc to antiretroviral regimens in patients with full viral suppression but impaired CD4 recovery. AIDS. 2009;23:1911–1913. doi: 10.1097/QAD.0b013e32832f3c65. [DOI] [PubMed] [Google Scholar]
- 56.Seclén E, González MM, Soriano V, Poveda E. Dynamics of viral tropism in HIV-infected patients under prolonged HIV-RNA suppression under HAART. 7th European HIV Drug Resistance Workshop; Stockholm. 25–27 March 2009; Abstract 26. [Google Scholar]
- 57.Waters L, Scourfield A, Marcano M, Gazzard B, Nelson M. The evolution of co-receptor tropism in patients interrupting suppressive HAART. 16th Conference on Retroviruses and Opportunistic Infections; 8–11 February 2009; Montreal, Canada. Abstract 439a. [Google Scholar]