Abstract
Background
An interesting finding in the epidemiology of human immunodeficiency virus (HIV) infection is that certain mutations in genes coding for chemokines, and their receptors and ligands, may confer resistance or susceptibility to HIV-1 infection and acquired immunodeficiency syndrome (AIDS) progression. The mutation most frequently studied is stromal cell-derived factor (SDF)1-3′A, a single nucleotide polymorphism in the 3′ untranslated region at the 801 position of the SDF1 gene, which seems to be associated with susceptibility or resistance to diseases, including AIDS. We examined the frequency of the above polymorphisms in the Tunisian population, and evaluated their contribution to a protective genetic background against HIV infection and progression.
Methods and materials
One hundred forty blood samples from HIV-infected patients from the Cellular Immunology Research Laboratory at the National Blood Transfusion Center were compared with those of 164 random blood donors from the same center. Genotyping was initially performed by polymerase chain reaction (PCR) analysis. SDF1 PCR product genomic regions were further subjected to restriction fragment length polymorphism analysis for genotype determination. Screening for the SDF1 polymorphism in the HIV-infected population yielded 56 heterozygous (40%), 52 mutation homozygous (37.1%), and 32 wild-type homozygous (22.8%) subjects. In contrast, in our healthy population, we found 70/164 heterozygous (42.6%), nine mutation homozygous (5.4%), and 85 wild-type homozygous (51.8%) subjects. The allele frequencies in the HIV-infected and healthy populations were f(SD1 3′A) = 57.1%, f(SDF1) = 42.8%, f(SDF1 3′A) = 26.8%, and f(SDF1) = 73.1%, respectively. The allelic and genotypic frequencies of the SDF1 3′A in our population show significantly higher distribution profiles compared with those observed in other Caucasian, European, and African American populations. Our results were examined by χ2 test and appear to confirm an association between polymorphism and AIDS progression. A higher odds ratio (>1) was found for the SDF1-3′A allele than for the wild-type allele (<1).
Conclusion
This result seems to confirm that the SDF1-3′A allele is associated with acceleration and progression from HIV infection to AIDS in the Tunisian population.
Keywords: human immunodeficiency virus, SDF1 polymorphism, Tunisia
Introduction
The discovery that chemokines can block human immunodeficiency virus (HIV) replication, and that their receptors play an important role in fusion of HIV to target cells, has led to the expectation that chemokines might hold the key to understanding the pathogenesis of HIV. In fact, the chemokine receptors, together with the CD4 molecule, serve as necessary cofactors for HIV entry into CD4 cells.1 The chemokine ligand 12 (CXCL12), better known as stromal cell-derived factor 1 (SDF1), inhibits infection of T cell line-tropic or syncytium-inducing viruses, usually found during late-stage HIV infection. Two chemokine receptors, CXCR4 and CCR5, are the major coreceptors required for entry of T cell line-tropic and macrophage-tropic strains of HIV-1, respectively, into CD4 cells.2,3 In addition, the expression of the SDF1 mutation, a single nucleotide polymorphism (designated SDF1-3′UTR-801G-A) at position 801 in the 3′-untranslated region (UTR) of the SDF1 gene, the only known ligand of CXCR4,4 may inhibit transmission of T cell line-tropic HIV strains, and may be implicated in mechanisms of resistance to acquired immunodeficiency syndrome (AIDS) progression throughout the disease stages.5–7 However, other studies have not observed any progression-retarding effect, and have reported its association with a faster decline in CD4+ T cells and progression to AIDS.8,9
This mutation has been widely studied during recent years in many populations, as well as worldwide, but this study is the first relevant research done in Tunisia. The global, ethnic, and regional distribution of these HIV/AIDS mutations varies significantly, giving each population a different genetic profile for HIV infection and AIDS progression. In the present study, we investigated the frequency of SDF1-3′A polymorphism in the Tunisian population and compared its distribution with that in other populations. It must be noted that the odds ratio (OR) was estimated for our population in order to evaluate natural resistance, and, therefore, these indices are of special importance for the prevention of AIDS.
Materials and methods
Sample collection
This study was conducted in 2003–2008 in 1047 Tunisian patients with HIV disease on antiretroviral therapy who made visits to the Laboratory of Cellular Immunology in order to have their CD3, CD4, and CD8 immune cell numbers monitored by flow cytometry.
The patients comprised 727 (69.43%) men of median age 37 years and 320 (30.56%) women of median age 34 years. These patients were sent to our laboratory by the Infectious Diseases Services of many hospitals, ie, La Rabta, Bourguiba, F Hached, A Othmana, as well as some private institutions. All information collected from this group of patients enabled us to follow the chronologic evolution of the disease in order to correlate it with numbers of CD4 immune cell lines in different stages of infection. HIV blood samples were also collected from 140 prospective patients, including 93 men of median age 36 years and 47 women of median age 34 years, in order to study the SDF1 polymorphism compared with that in 164 healthy donors.
Isolation of genomic DNA and genotyping
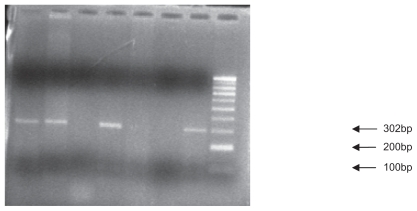
Five milliliters of blood was collected in an ethylenediamine tetraacetic acid tube (15%), and DNA was extracted by a chemical method using TKM solution.10 SDF1 has three splice variants, among which the beta variant may have a guanine to adenine (G-A) mutation. Genotyping of the SDF1 3′A mutation was performed by polymerase chain reaction (PCR) using pairs of external primers (SDF1 F 5′-CAGTCAACCTGGGCAAAGCC-3′ and SDF1 – R 5′-AGGTTTGGTCCTGAGAGTCC-3′) framing the region surrounding the polymorphic sites, and restriction fragment length polymorphism (RFLP) was done by using a restriction enzyme “MspI”. The SDF1 3′A variant was detected by electrophoresis on 1.5% agarose gel. Ethidium bromide staining was used to reveal the fragment under ultraviolet light. PCR products showed a band at base pair 302.11
If the 3′A allele existed at position 801, no digestion occurred and only one 302 base pair band in the agarose gel emerged, whereas if the wild-type allele (G) existed at this site, two shorter fragments, including a 200 and 100 base pair resulted.11
Statistical analysis
Allelic and genotypic frequencies were evaluated by tools used for population genetic analyses, ie, SPSS version 16.0 (SPSS Inc., Chicago, IL), as well as by calculating OR and P values. In fact, we had to do an association study between SDF1 mutation and disease progression in order to identify genes or alleles associated with susceptibility or resistance to HIV infection. The gene may be a risk factor for infection, if among the gene expressions (alleles) one increases the risk of disease rather than having a protector effect. The χ2 test enables calculation of the P value, which is the probability of a different allelic distribution observed between the two groups occurring at random. The association was considered significant for P < 0.05. The OR values can be >1> if OR > 1: accelerator allele; if OR< 1: protector allele, and if OR = 1: neutral allele.
Results
General representation of Tunisian patients living with HIV
In order to survey the influence of antiretroviral treatment on the dynamic rate of CD4 cells, blood samples from HIV patients were sent to our laboratory for flow cytometry analysis. In total, 1047 patients were analyzed, with a 2:27 gender ratio. Our study was conducted during the period 2003–2008. Table 1 represents the distribution of the total number of patients, and their gender ratio and median age, which enabled study of the new HIV patient rate shown in Table 2.
Table 1.
Age and gender distribution of patients with human immunodeficiency virus
| 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | |
|---|---|---|---|---|---|---|
| Median age (years) | 34.76 | 35.7 | 36.26 | 37.09 | 43.71 | 35.81 |
| Men (n) | 153 | 184 | 213 | 177 | 275 | 302 |
| Women (n) | 76 | 74 | 91 | 142 | 114 | 183 |
| Total patients | 229 | 258 | 304 | 319 | 389 | 485 |
Table 2.
Rate of increase in numbers of patients with new human immunodeficiency virus in the Tunisian population
| 2005 | 2006 | 2007 | 2008 | |
|---|---|---|---|---|
| Women (n) | 25 | 28 | 67 | 85 |
| Men (n) | 70 | 98 | 129 | 170 |
| Total new patients | 96 | 118 | 196 | 256 |
| Rate increase (%) | 18.51% | 22.91% | 66.1% | 30.61% |
SDF1 polymorphism study
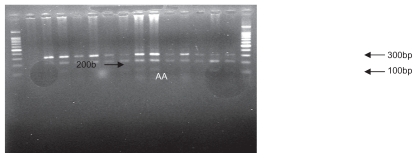
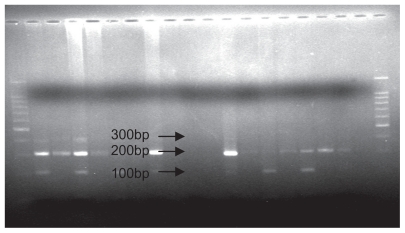
The mixture of the PCR product and “MspI” restriction enzyme was stored at 37°C for one night. The restriction enzyme cut in the 801 position in the 3′ UTR of the SDF1 gene. The digestion products were analyzed by 1.5% agarose gel electrophoresis and visualized by ultraviolet fluorescence after staining with ethidium bromide. SDF1 wild-type alleles (SDF1/SDF1 or G/G) yielded 100 and 200 base pair products, whereas mutation allele homozygotes (SDF1 3′A/SDF1 3′A or A/A) and heterozygotes (SDF1 3′A/SDF1 or A/G) yielded, respectively, 302 base pairs and 302, 200, and 100 base pair products.12 A 100 base pair molecular ruler was used as a marker. Figure 1 displays the PCR product, and Figures 2 and 3 show all the possible genotypes involving PCR and digestion products.
Figure 1.
Polymerase chain reaction product: The primers produce polymerase chain reaction products of either 302 base pairs.
Figure 2.
Polymerase chain reaction restriction fragment length polymorphism results of SDF1 followed by enzymatic digestion on 1.5% agarose gel (genotypes observed, A/A and A/G).
Figure 3.
Polymerase chain reaction restriction fragment length polymorphism analysis results of SDF1 followed by enzymatic digestion on 1.5% agarose gel (genotype observed, G/G).
SDF1 3′A in HIV patients versus healthy donors
Patient genotypes were identified using electrophoresis profiles. We counted 52 of 140 patients who were homozygous (37.1%), 56 who were heterozygous (40%), and 32 who were homozygous for the wild-type allele (22.8%). However, in the donor population, we counted only 9/164 patients who were homozygous for the mutation (5.48%), 70 who were heterozygous (42.6%), and 85 who were wild-type homozygous (51.8%). Table 3 summarizes the genotypic frequency in the healthy and infected populations, as well as the X2 test and P value, and Table 4 illustrates the allelic frequencies.
Table 3.
The frequency of different SDF1 genotypes, χ2 test, and P value
| Genotypes | HIV patients | Donors | P value | ||
|---|---|---|---|---|---|
| n | Frequency | n | Frequency | ||
| SDF1 3′A/SDF1 3′A (A/A) | 52 | 0.371 (37.1%) | 9 | 0.054 (5.4%) | <0.05 |
| SDF1 3′A/SDF1 (A/G) | 56 | 0.40 (40%) | 70 | 0.426 (42.6%) | >0.05 |
| SDF1/SDF1 (G/G) | 32 | 0.228 (22.8%) | 85 | 0.518 (51.8%) | <0.05 |
Abbreviation: HIV, human immunodeficiency virus.
Table 4.
Allelic frequency distribution, χ2 test, P value and odds ratio (OR)
| Allelic frequencies | HIV patients | Donors | P value | OR |
|---|---|---|---|---|
| F (SDF1 3′A) or f(A) | 0,571 (57,1%) | 0,428 (42,8%) | P < 0,05 | >1 |
| F (SDF1) or f(G) | 0,268 (26,8%) | 0,731 (73,1%) | P < 0,05 | <1 |
Abbreviation: HIV, human immunodeficiency virus.
Table 4 demonstrates that the SDF1 3′A allele frequency is higher in the HIV population (57.1%) compared with the healthy donors (42.8%) in contrast with the wild-type allele which is more frequent in the healthy population (73.1%), and low in the HIV patient cohort (26.8%). This finding was highly significant (P < 0.05) according to the χ2 test. This table also summarizes the allelic and genotypic frequencies of the SDF1 wild-type allele and SDF1 3′A alleles reported in different countries compared with the Tunisian cohort.
Polymorphism distribution according to clinical stage of infection
Three different stages were observed in disease progression of HIV infection, ie, clinical stages A (primo infection), B (intermediate), and C (final). In the 140 infected patients, we counted 24 in clinical stage A, 25 in clinical stage B, and 70 in stage C. SDF1 3′A distribution was also carried out as a function of the clinical stages. Table 5 summarizes the results. The distribution study of SDF1 3′A in the initial stages shows nine homozygous for the mutated allele (A/A), nine heterozygous (A/G), and six homozygous for the wild-type allele (G/G). In the intermediate stage, we counted eight homozygous for the mutation, 12 heterozygous, and five homozygous for the wild-type allele. In contrast, for the final stage, 30 patients homozygous for the mutation were detected, with 26 heterozygous and 14 homozygous for the wild-type allele.
Table 5.
Allelic and genotypic frequencies of SDF1, wild-type allele, and SDF1 3′A reported in different countries compared with the Tunisian cohort
| Countries | HIV patients | n | Controls | n | References | ||
|---|---|---|---|---|---|---|---|
| Allelic frequencies (%) | SDF1 3′A | SDF1 | SDF1 3′A | SDF1 | |||
| Japan | 34.47 | – | 393 | – | – | – | Ikegawa et al15 |
| Poland | 14.6 | – | 1063 | – | – | – | Lewandowska et al24 |
| Italy | 15.0 | – | 37 | – | – | – | Su et al18 |
| Pavia | 3.6 | – | 55 | – | – | – | Guerini et al17 |
| Southern Spain | 24.5 | – | 100 | – | – | – | Royo et al25 |
| Spain | 32.32 | 67.68 | 164 | 8.33 | 91.67 | 60 | Soriano et al26 |
| Russia | 22.2 | – | 171 | – | – | – | Ryabov et al27 |
| Greece | 29.75 | – | 200 | – | – | – | Apostolakis et al16 |
| South India | 17–35 | – | 525 | – | – | – | Ramana et al |
| North India | 20.4 | – | 500 | – | – | – | Verma et al21 |
| Caucasian | 21.1 | – | 1835 | – | – | – | Winkler et al13 |
| African Americans | 5.7 | – | 859 | – | – | – | |
| Asian | 25.7 | – | 37 | – | – | – | |
| Brazilian Caucasians | 18.84–23 | – | 469 | – | – | – | EMV Reiche et al (2006) |
| Brazilian non-Caucasians | 14–20 | – | 225 | – | – | – | |
| Asia (Oceanian) | 50–70 | – | – | – | – | – | Arezana-Seisdedos et al14 |
| Tunisia | 57.1 | 42.8 | 140 | 26.8 | 73.1 | 164 | Present study |
Abbreviations: n, total number; (–), data not available; HIV, human immunodeficiency virus.
SDF1 polymorphism distribution by clinical stage and CD4+ T cell number
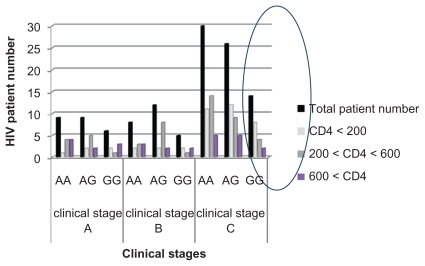
The relationship between SDF1 polymorphism, immune cell numbers, and clinical stage progression is shown in Figure 4.
Figure 4.
SDF1 polymorphism distribution according to the clinicals stages and the CD4+ T cells numbers.
Discussion
This is the first study undertaken to determine the different aspects of the Tunisian HIV-infected population and the frequency of the SDF1 3′A allele in a large sample of symptomatic HIV-infected subjects compared with healthy donors.
Analysis of the clinical, therapeutic, and biologic data for 2003–2008 demonstrates many features peculiar to the Tunisian cohort. For each year, we determined the median age of our patients, which shows a range between 34 and 43 years, giving a total median age of 37 years. This observation suggests that the HIV-infected Tunisian population is young compared with other cohorts12 and also shows that the principal cause of disease transmission could be unprotected sexual activity. It is also important to note the large difference between the numbers of male and female HIV patients, with a gender ratio of 2:27. The rate of new cases in our population is increasing year by year, from 18.51% in 2005, to 22.91% in 2006, to 66.1% in 2007, and to 30.61% in 2008.
In the second part of this study, we tried to determine the role of SDF1 polymorphism in HIV infection and the role of the SDF1 3′A allele in delay or susceptibility to HIV progression. The reported frequency of the homozygous genotype for the SDF1 3′A allele observed in the HIV-infected population is 37.1% and only 5.4% in the healthy population. In addition, our study also demonstrates that, compared with the homozygous wild-type SDF1 allele, there is a frequency of 22.8% in the HIV-infected population and 51.8% in the healthy one. These results are comparable with those of a previous study in the Caucasian population,13 in Oceania, and in Asia.14 Moreover, analysis of the genotype frequency data confirmed that our results are highly significant (P ≤ 0.05). Consequently, an association probably exists between HIV infection and SDF1 polymorphism.
No significant difference in the frequency of the heterozygous genotype between the HIV-infected population (40%) and healthy donors (42.6%) has been reported. Another particularity of the Tunisian population is that the SDF1 3′A allele frequency differed significantly between our HIV-infected patients (57.1%) and our donor group (42.8%), whereas the wild-type allele of SDF1 is more frequent in the donor population (73.1%) than in the HIV-infected population (26.8%). This result is comparable with that in a previous study in a Caucasian population,13 and in Asia,14 Japan,15 and Greece.16 The contrast is observed in Italy17,18 and in the African American population.13 The χ2 test confirms that our results are highly significant (P ≤ 0.05), so the SDF1 3′A mutation could be associated with progression or delay of HIV disease. For this reason, we investigated SDF1 polymorphism according to clinical stage of infection. The results showed that many of the 140 HIV-infected patients were in the final stages of infection (70/140); in contrast, there were 24 in clinical stage A and 25 in clinical stage B.
In addition, the homozygous mutation for the SDF1 3′A allele differs among patients in various stages of HIV infection, and most of the patients in the final stage are homozygous for the mutated allele 3′A (30/70) which seems to be the most frequent, and is pronounced every time upon progression to the later stage of infection.13,16,19–21 This result is in agreement with other studies confirming that SDF1-3′A homozygosity is associated with a faster CD4 T cell decrease and a rapid progression to AIDS.8.22,26 However, different investigations confirm that, in contrast, SDF1 3′A/SDF1 3′A has a protective effect, and there was a marked slowing in progression to AIDS. Furthermore, the gradation suggests that the protection afforded by the mutation is more marked in the later stages of HIV infection, and is possibly related to inhibition of T cell-tropic HIV strains.13
Finally, we used a cross-sectional approach to analyze for the presence of SDF13′A, and correlated it with clinical stage and number of CD4+ T cells. The results show that most patients in the final stages of infection carry the mutated allele, which is present when the CD4+ T cells decrease below 200/mm3 (the CD4+ threshold). Furthermore, the χ2 test shows that the SDF1 3′A OR is >1, which suggests that, in our population, the 3′A allele may accelerate to the final stage of infection, in contrast with the wild-type allele (SDF1) which seems to be protective (OR < 1). This result is in agreement with another study carried out in the Sub-Saharan African population which confirmed that the mutation was associated with an increased risk for HIV infection.23
In molecular terms, many studies confirm that the SDF-1 3′A mutation leads to increased levels of SDF1 mRNA, and demonstrate also that this effect is a consequence of enhanced mRNA stability.29 This observation is supported by other studies showing that homozygosity is associated with delayed progression to AIDS, and, because the 3′ UTR is also involved in regulating the stability of mRNA, it has been suggested that this homozygosity might involve overproduction of SDF1 that could act by inhibiting the appearance of X4 strains and by competing with the HIV-1 virulent X4 and R5X4 strains for binding to the CXCR4 coreceptor of the target cells.13,30
In our study we identified a correlation between SDF1 genotyping results and progression of the clinical stages of HIV infection in the Tunisian population according to the number of CD4+ T cells. We found an association between the presence of the SDF1 3′A allele and risk of HIV in a Tunisian cohort, and observed a statistically significant difference in genotype and allele frequencies between patients and a healthy control group. There were significantly more patients than healthy individuals who were homozygous for SDF1 3′A. The progression-delaying effect of the SDF1 3′A genotype was not confirmed in our population, but we did demonstrate significant expression of the 3′A allele in the later stages of infection, that may be related to the evolution of HIV infection and suggests an accelerator role of this allele for progression to AIDS. In fact, most of our AIDS patients carry the 3′A mutation, with an important decrease in CD4 immune cells in the late stage of disease. This may be related to overproduction of chemokines and immune cells which exhausts the already deficient immune system, causing a faster decline in CD4+ T cells and progression to later stages of disease. This impression should be confirmed by further molecular studies, as well as quantification of viral charge and plasma SDF1 levels in order to perform a molecular analysis. It would be most important to include other researchers’ results in association with SDF1 polymorphism based on the roles of further chemokines, and their receptors and interactions in HIV infection and AIDS progression.
Conclusion
This study highlights the clinic, epidemiologic, and biologic characteristics of HIV patients in a Tunisian population, identifies genetic polymorphism of SDF1, and compares them with a healthy control population. Our results show the accelerative effect of the SDF-1 3′A allele to the late stage of infection and disease progression, in contrast with the wild-type allele, which seems to be protective. No progression- delaying effects have been detected for this mutation in our population, but it is associated with a faster decline in CD4+ T cells and progression to AIDS.
Table 6.
SDF1 polymorphism distribution according to clinical stages
| Genotypes | Clinical stages | ||
|---|---|---|---|
| A | B | C | |
| A/A | 9 | 8 | 30 |
| A/G | 9 | 12 | 26 |
| G/G | 6 | 5 | 14 |
| Total | 24 | 25 | 70 |
Acknowledgment
We wish to thank the cell immunology laboratory staff for their invaluable technical assistance, and those who helped us directly or indirectly to carry out this project.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Dung H, Liu R, Ellmeier W, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 2.Caruz A, Samson M, Alonso JM, et al. Genomic organization and promoter chracterization of human CXCR4 gene. FEBS Lett. 1998;426:271–278. doi: 10.1016/s0014-5793(98)00359-7. [DOI] [PubMed] [Google Scholar]
- 3.Reiche EMV, Wanabe MA, Bonametti AM, et al. Stromal cell-derived factor 1 (SDF1) genetic polymorphism in a sample of healthy individuals, seronegative individuals exposed to human immunodeficiency virus type 1 (HIV-1) and patients infected with HIV-1 from the Brazilian population. Int J Immunogenet. 2006;33:127–133. doi: 10.1111/j.1744-313X.2006.00583.x. [DOI] [PubMed] [Google Scholar]
- 4.Wang J, Babcock GJ, Choe H, Farzan M, Sodroski J, Gabuzda D. N-linked glycosylation in the CXCR4 N-terminus inhibits binding to HIV-1 envelope glycoproteins. Virology. 2004;324:140–150. doi: 10.1016/j.virol.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: Functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 6.Kaur G, Tuen M, Virland D, et al. Antigen stimulation induces HIV envelope gp120-specific CD4(+) T cells to secrete CCR5 ligands and suppress HIV infection. Virology. 2007;369:214–255. doi: 10.1016/j.virol.2007.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdelwahab SF, Cocchi F, Bagley KC, Gallo KR, Devico RC, Lewis A. HIV suppressive factors are secreted by CD4+ T cells during primary immune responses. Proc Natl Acad Sci U S A. 2003;100:15006–15010. doi: 10.1073/pnas.2035075100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mummidi S, Modi W, Gonzales E, et al. Genealogy of the CCR5 locus and chemokines system gene variants associated with altered rate of HIV-1 disease progression. Nat Med. 1998;4:786–793. doi: 10.1038/nm0798-786. [DOI] [PubMed] [Google Scholar]
- 9.Ioannidis JPA, Rosenberg PS, Goedert JJ, et al. Effects of CCR5-D32, CCR2-64I and SDF-1 3′A alleles on HIV-1 disease progression: An international meta-analysis of individual patient data. Ann Intern Med. 2001;135:782–795. doi: 10.7326/0003-4819-135-9-200111060-00008. [DOI] [PubMed] [Google Scholar]
- 10.Hofstetter JR, Zhang A, Mayeda AR, Guscar T, Nurnberger JL, Jr, Lahiri DK. Genomic DNA from mice: A comparison of recovery methods and tissue sources. Biochem Mol Med. 1997;62:197–202. doi: 10.1006/bmme.1997.2637. [DOI] [PubMed] [Google Scholar]
- 11.Razmkhah M, Doroudchi M, Ghayumi SM, Erfani N, Ghaderi A. Stromal cell derived factor-1 gene and susceptibility of Iranian patients with lung cancer. Lung Cancer. 2005;49:311–315. doi: 10.1016/j.lungcan.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 12.Rollot Trad F, Launay O, Le Baut V, et al. Le VIH... le virus et les personnes infectées prennent de l’âge. La Revue de Médecine Interne. 2008;29(Suppl 3):S297. [Google Scholar]
- 13.Winkler C, Modi W, Smith MW, et al. Genetic restriction of AIDS pathogenesis by an SDF1 chemokine gene variant. Science. 1998;279:389–390. doi: 10.1126/science.279.5349.389. [DOI] [PubMed] [Google Scholar]
- 14.Arezana-Seisdedos F, Parmentier M. Genetics of resistance to HIV infection role of coreceptors and coreceptors ligands. Semin Immunol. 2006;18:387–403. doi: 10.1016/j.smim.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Ikegawa M, Yuan J, Matsumoto K, et al. Elevated plasma stromal cell derived factor 1 protein level in the progression of HIV type 1 infection/AIDS. AIDS Res Hum Retroviruses. 2001;17:587–595. doi: 10.1089/088922201300119680. [DOI] [PubMed] [Google Scholar]
- 16.Apostolakis S, Baritaki S, Karmbovitis E, Spandidos DA. Distribution HIV/AIDS protective SDF1, CCR5 and CCR2 gene variants with a Cretan population. J Clin Virol. 2005;34:310–314. doi: 10.1016/j.jcv.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Guerini FR, Debulbe S, Zanzottera M, et al. Analysis of CCR5, CCR2, SDF1 and RANTES gene polymorphisms in subjects with HIV-related PML and not determined leukoencephalopathy. Biomed Pharmacother. 2008;62:26–30. doi: 10.1016/j.biopha.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 18.Su B, Jin L, Hu F, Xiao J, Luo J, Lu D. Distribution of two HIV-1 resistant polymorphisms (SDF1 3′A and CCR2-64I) in East Asian and world populations and its implication in AIDS epidemiology. Am J Hum Genet. 1999;65:1047–1053. doi: 10.1086/302568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voevodin A, Samilchuk E, Dashi S, et al. Frequencies of SDF1 chemokine, CCR5 and CCR2 chemokine receptor gene alleles conferring resistance to human immunodeficiency virus type 1 and AIDS in Kuwaitis. J Med Virol. 1999;58:54–58. doi: 10.1002/(sici)1096-9071(199905)58:1<54::aid-jmv8>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 20.Ramamoorti N, Kumarvelu J, Shanmugasundaram GK, Rani K, Banerjea AC. High frequency of G to A transition mutation in the stromal cell derived factor-1 in India, a chemokine that blocks HIV (X4) infection: Multiple proteins bind to 3′untranslated region of SDF-1 RNA. Genes Immun. 2001;2:408–410. doi: 10.1038/sj.gene.6363800. [DOI] [PubMed] [Google Scholar]
- 21.Verma R, Gupta RB, Singh K, et al. Distribution of CCR5delta32, CCR2-64I and SDF-3′A and plasma levels of SDF-1 in HIV seronegative North Indians. J Clin Virol. 2007;38:198–203. doi: 10.1016/j.jcv.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 22.Sei S, Boler AM, Nguyan GT, et al. Protective effect of CCR5 delta 32 heterozygosity is restricted by SDF1 genotype in children with HIV 1 infection. AIDS. 2001;15:1343–1352. doi: 10.1097/00002030-200107270-00003. [DOI] [PubMed] [Google Scholar]
- 23.Peterson DC, Glashoff RH, Shretha S, et al. Risk for HIV-1 infection associated with a common CXCL12 (SDF1) polymorphism and CXCR4 variation in African populations. J Acquir Immun Defic Syndr. 2006;40:521–526. doi: 10.1097/01.qai.0000186360.42834.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewandowska M, Franciszkieicz K, Prokop J, Ofori H, Jagodziski PP. Distribution of two HIV-1-resistant polymorphisms (SDF1 3′A and CCR2-64I alleles) in the Polish population. J Hum Genet. 2002;47:585–589. doi: 10.1007/s100380200089. [DOI] [PubMed] [Google Scholar]
- 25.Royo JL, Ruiz A, Borrego S, et al. Fluorescence resonance energy transfer analysis of CCR-V64I and SDF1 3′A polymorphisms: Prevalence in southern Spain HIV type 1 cohort and noninfected population. AIDS Res Hum Retroviruses. 2001;17:663–666. doi: 10.1089/088922201750236933. [DOI] [PubMed] [Google Scholar]
- 26.Soriano A, Martinez C, Garcia F, Plana M, Palou E, Lejeune M. Plasma stromal cell derived factor SDF-1 levels, SDF1 3′A genotype, and expression of CXCR4 on T lymphocytes: Their impact on resistance to human immunodeficiency virus type 1 infection and its progression. J Infect Dis. 2002;186:922–931. doi: 10.1086/343741. [DOI] [PubMed] [Google Scholar]
- 27.Ryabov GS, Kazennova EV, Bobkova MR, Bobkov AF. Prevalence of alleles associated with HIV resistance in Russia. Genet Test. 2004;8:73–76. doi: 10.1089/109065704323016067. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Moruja C, Rueda P, Torres C, Alcamí J, Luque F, Caruz A. Molecular phenotype of CXCL12beta 3′UTR G801A polymorphism (rs1801157) associated to HIV-1 disease progression. Curr HIV Res. 2009;186:384–389. doi: 10.2174/157016209788680543. [DOI] [PubMed] [Google Scholar]
- 29.Yonezawa A, Hori T, Takaori-Kondo A, Morita R, Uchiyama T. Replacement of V3 region of gp120 with SDF-1 preserves the infectivity of T-cell line-tropic human immunodeficiency virus type 1. J Virol. 2001;75:4258–4267. doi: 10.1128/JVI.75.9.4258-4267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]