Abstract
Technological advances in molecular and in silico research have enabled significant progress towards personalized transplantation medicine. It is now possible to conduct comprehensive biomarker development studies of transplant organ pathologies, correlating genomic, transcriptomic and proteomic information from donor and recipient with clinical and histological phenotypes. Translation of these advances to the clinical setting will allow assessment of an individual patient's risk of allograft damage or accommodation. Transplantation biomarkers are needed for active monitoring of immunosuppression, to reduce patient morbidity, and to improve long-term allograft function and life expectancy. Here, we highlight recent pre- and post-transplantation biomarkers of acute and chronic allograft damage or adaptation, focusing on peripheral blood-based methodologies for non-invasive application. We then critically discuss current findings with respect to their future application in routine clinical transplantation medicine. Complement-system-associated SNPs present potential biomarkers that may be used to indicate the baseline risk for allograft damage prior to transplantation. The detection of antibodies against novel, non-HLA, MICA antigens, and the expression of cytokine genes and proteins and cytotoxicity-related genes have been correlated with allograft damage and are potential post-transplantation biomarkers indicating allograft damage at the molecular level, although these do not have clinical relevance yet. Several multi-gene expression-based biomarker panels have been identified that accurately predicted graft accommodation in liver transplant recipients and may be developed into a predictive biomarker assay.
Biomarkers for personalized transplantation medicine
In 2010, 28,663 transplantations were performed in the United States. Currently, more than 100,000 US patients are waiting for an organ transplant, and each month approximately 4,000 patients are added (Organ Procurement and Transplantation Network data as of April 2011). A significant number of patients on the waiting list are added due to functional failure of a first transplant, reflecting our current inability to ensure long-term allograft function and survival and representing a major problem in transplantation medicine.
The major reason for late allograft loss is chronic allograft damage (CAD), seen as the progressive decline of graft function > 1 year post-transplantation. The underlying mechanisms of CAD are poorly understood and need to be unraveled if graft function and treatment are to be successful. The definition of valid pre- and post-transplantation biomarkers will facilitate personalized transplantation medicine, leading to long-term graft survival and decreasing numbers of patients on the waiting list.
Identification of biomarkers will aid the understanding of underlying mechanisms by indicating damage early post-transplantation when pathological changes are taking place at the molecular level. This will enable us to better predict the likelihood of an individual's allograft survival and assist the development of currently unavailable treatments for CAD. Biomarkers will also allow better matching of donor and recipient and the assessment of an individual's risk for graft injury. Current methods for diagnosing graft injury require invasive biopsies and detect pathological changes at advanced and often irreversible stages of allograft damage. The use of more sensitive and specific methodologies based on donor and recipient genotyping, and transcriptional and proteomic profiling to differentiate and detect early stages of organ injury would bridge this gap. This highlights the importance of -omics-based approaches for the improvement of transplant practice.
Nowadays, biomarker studies increasingly integrate information from multiple platforms, such as genotype analyses of single-nucleotide polymorphisms (SNPs), epigenetic studies and analyses of mRNA, microRNA (miRNA), as well as protein, peptide, antibody and metabolite profiling. High-throughput analyses are becoming more accessible, affordable and customizable, and rapid developments in analytical tools now allow integrated meta-analyses of different datasets across different experiments, platforms and technologies [1-4]. Functional biomarker studies require a discovery and several validation stages, including horizontal and vertical meta-analyses and prospective validation. By this means, several potential biomarkers have been identified. However, advances towards regulatory application, approval and clinical implementation have been slow and costly, partly because of the difficulties faced in externally and prospectively validating these biomarkers.
Here, we concentrate on recent advances made in transplantation biomarker medicine, focusing on the key stages of the biomarker development process. We highlight both laboratory test-based and clinically applied pre- and post-transplantation genomic, transcriptomic and proteomic biomarkers of acute and chronic allograft injury and graft accommodation. We point out the advantages and pitfalls of trying to identify non-invasive blood-based biomarkers and present recent approaches to overcoming related obstacles. Finally, we critically discuss the current status of transplant biomarker research along the road to clinical application.
Identification of clinically relevant biomarkers
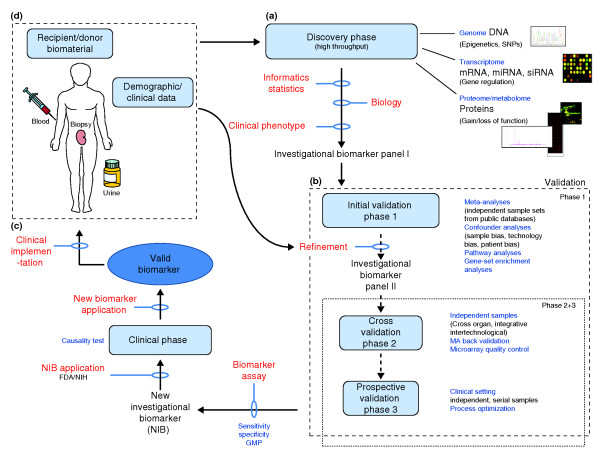
The number of biomarker studies performed so far with respect to solid organ transplantation exceeds 15,000, yet the number of resulting US Food and Drug Administration (FDA) approved biomarker-based diagnostic tests in transplantation stands at two, one being a functional immune assay and the other a non-invasive test based on blood gene expression for predicting the absence of acute allograft rejection (AR) after heart transplantation [5]. Needless to say, the path from discovery and validation of a biomarker in the academic laboratory to its approval for the clinic is torturous. Well-thought-out validation and prospective feasibility studies are needed to move the biomarker discovery process towards FDA application, approval and clinical implementation (Figure 1).
Figure 1.
Outline of the biomarker development process in the US from clinic to bench and back to clinic. As in drug development, the key phases are the discovery and validation phases, which involve complex FDA-regulated processes. (a) High-throughput, often in silico technologies are used to discover genomic, transcriptomic, proteomic or integrative investigational biomarkers, which are then (b) redefined in several validation phases using independent samples, technologies, and horizontal and vertical meta-analyses. (c) A clinically applicable biomarker assay based on good manufacturing practice (GMP) can be developed after prospective studies have confirmed the investigational biomarker. The FDA has to approve clinical studies, and only after successful completion and additional FDA regulation can the biomarker be considered valid and (d) be implemented into the clinic.
The initial key steps in biomarker development are the discovery phase and the validation phase. In the discovery phase, usually high-throughput technologies on multiple molecular platforms and subsequent biostatistical analyses identify a first biomarker panel, which often comprises several hundreds of candidates. The platforms and molecular techniques used in this phase, such as DNA, RNA, miRNA microarray or antigen-based proto-arrays, usually generate large quantities of data; these methodologies have recently been reviewed by us in detail [6]. Mandatory data deposition in the public domain, such as into the Gene Expression Omnibus (GEO), increasingly allows the use of publicly available data for the biomarker discovery phase and the use of new patient samples for the validation phase. Pathway and network analyses enable integration of experimental data into biological and cellular contexts, and by studying cellular crosstalk and molecular interactions, pathological pathways can be better elucidated [1-4]. In the near future, data obtained by next-generation sequencing, copy number variation analyses and SNP arrays will be added.
The discovery phase is followed by one, or most frequently, two or three validation phases to increase sensitivity and specificity. The first validation phase analyzes the initial biomarker panel in independent samples, leading to a refined set often consisting of 50 to 100 candidates. Meta-analyses improve the sensitivity and specificity of the initial candidate set, integrating results from different, often publicly available datasets. Horizontal approaches investigate the same molecular platform in different organs [7-10], and vertical meta-analyses involve integration between different platforms, as in proteogenomic studies [11-13]. The advantages of meta-analyses are increased sample sizes and reduced experimental work, which help to increase the specificity and sensitivity of the initial biomarker. For example, a putative gene-based fingerprint in peripheral blood for kidney transplant tolerance was identified using this approach [14]. Information from the statistical analysis of microarrays (SAM) and predictive analysis of microarray (PAM) techniques identified an initial biomarker set, which was then cross-validated in independent samples and further refined in sample data from different microarray platforms [15].
However, the comparability of data from different laboratories has to be ensured and different laboratory procedures, inter-center variations and array performance on different days and when performed by different people have to be corrected for. For this purpose, the microarray quality control (MAQC) studies [16,17] were initiated. These consisted of two phases aiming to provide quality control tools, develop data analysis guidelines and assess limitations and capabilities of various predictive biomarker models. As a result, common practices for the development and validation of microarray-based classifier models were defined and guidelines for global gene expression analysis established. A third phase is underway, focusing on next-generation sequencing techniques.
After the initial validation and refinement, the biomarker panel needs to undergo prospective validation in the clinical setting to establish the sensitivity, specificity and negative and positive predictive values for clinical application. The organizational challenges and expense of conducting prospective observational or interventional studies on biomarkers are reflected by the fact that, so far, only few studies have reached this status in the biomarker development process [5,18,19]. Increased numbers of patients and samples need to be investigated for a long period, often for a minimum of 2 years, before clinically relevant conclusions can be made. These studies require skilled staff and financial resources as well as sufficient laboratory infrastructure. Most importantly, the health and safety of patients and transplant organs remain the first priority, and prospective studies often carry unpredicted risks.
Identifying confounders
Another step towards confirming the clinical usefulness of a biomarker is to identify and control for experimental confounders. Confounders include sample bias, technology bias and patient bias. A peripheral blood-based transcriptomic biomarker has the advantage of being minimally invasive and assessable on a frequent basis at reduced cost and risk compared to biopsied samples. Importantly, a peripheral transcriptomic biomarker might also be measurable early, when no or minimal allograft damage has taken place. However, most cellular components of peripheral blood respond quickly to exogenous stimuli, such as temperature changes or shear force, inducing changes in gene expression ex vivo. In this regard, a hypoxia-associated gene expression signature was detected in peripheral blood mononuclear cells (PBMCs) after delayed sample processing compared to immediate sample processing [20].
Different laboratory techniques for sample allocation and handling make comparison of results difficult, or even lead to controversial results [21-27]. This aspect becomes particularly important in multi-center studies or when using publicly available data from independently performed studies. Therefore, safe, quick and easy handling during sample procurement must be ensured to minimize the overall impact of ex vivo changes to gene expression. Currently there are no uniform sample procurement guidelines. Several studies have been addressing this issue [20,28,29].
The complex composition of samples useable for non-invasive tests, such as blood and urine, make the identification of valid biomarkers difficult. For example, the abundant presence of globin mRNA as well as the heterogeneous nature of blood are important internal confounding factors to be controlled for when trying to identify a blood-based biomarker. Globin mRNA leads to decreased percentage present calls, decreased call concordance and increased signal variation when analyzing whole-blood gene expression profiles by microarray. Debey et al. [30] presented a method of combined whole-blood RNA stabilization and globin mRNA reduction followed by genome-wide transcriptome analysis. We also reported [31] the interference of globin mRNA when using whole blood for the discovery of peripheral biomarkers of acute renal allograft rejection. A comparison of four different protocols for total RNA preparation, amplification and synthesis of complementary RNA or cDNA and array hybridization revealed that only a combination of globin mRNA reduction during handling together with a mathematical algorithm provided depletion of globin mRNA expression. This approach improved the detection of biological differences between blood samples collected from patients with biopsy-proven AR or stable graft function [31].
Another obstacle in identifying a blood-based biomarker is the heterogeneity of blood. A typical blood sample contains a large number of cell types, each with its own distinct expression profile [32]. Heterogeneity is further compounded by the frequency of the same cell type being different between individuals [33]. Consequently, a differential expression profile observed in whole blood between two phenotypes could be caused by either a change in frequency of a specific type of cell without a change in the expression profiles of each cell type or a change in the expression profile of a cell type while the frequency of the cell type remains constant. Although one way to address this issue is to isolate subsets of specific cell types (for example, using cytometry or laser capture microdissection) and profile them, such techniques are expensive, time consuming and limited by difficulties in obtaining sufficient purified tissue with adequate RNA, and they may affect cell physiology and gene expression [20,34]. To address these challenges, we and others have proposed several statistical approaches to deconvoluting gene expression profiles from heterogeneous tissues [35-37]. Using a deconvolution approach, we showed [35] that although whole-blood expression profiles did not reveal differential expression between patients with AR and those with stable transplant function, cell-type-specific expression profiles estimated by deconvolution of microarray data identified dramatic changes in two cell types that would have otherwise been completely missed. Differentially expressed genes in AR and stable transplant patients at a false discovery rate of 0.05 were identified between lymphocytes and neutrophils, as well as 137 upregulated genes in monocytes from the AR patients.
Laboratory test-based biomarkers in transplantation medicine
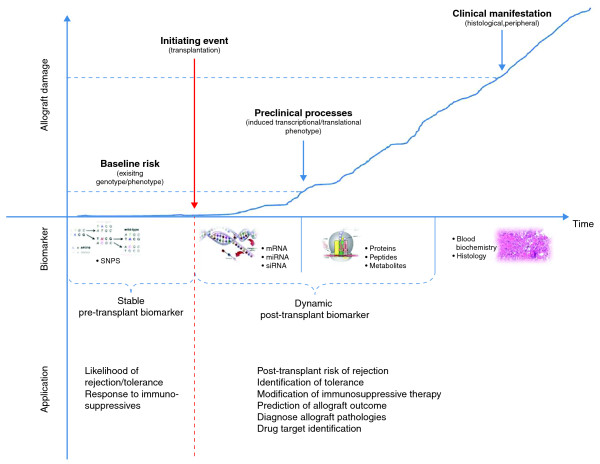
Currently, a match between the human leukocyte antigen (HLA) in the sera of the donor and the recipient is the best pre-transplant biomarker [38]. Yet even in the case of a total match, the risk of clinical or subclinical AR and or CAD cannot be excluded. Post-transplant biomarkers include functional parameters that are mainly measured at the protein level, such as serum creatinine. The current gold standard to differentially diagnose allograft pathologies is the histological assessment of invasive graft biopsies. The threshold indicating allograft damage by current post-transplant biomarkers is high and reached at a point when significant damage has already occurred (Figure 2). Therefore, biomarkers for predicting the risk of damage or for indicating preclinical damage at the molecular level are needed. Applications that require an invasive biopsy limit the clinical applicability of identified biomarkers, and functional monitoring assays that use non-invasive samples, such as peripheral blood or patient urine, are more favorable (for patients and economically).
Figure 2.
Biomarkers in transplantation medicine. The application of biomarkers in transplantation medicine is very sensitive to time. Allograft damage progresses with time after transplantation, and the earlier allograft damage is detected, the better the chances for long-term allograft function become. Transplantation is the process that initiates the changes that lead to allograft damage. Post-transplantation biomarkers are dynamic, and the current post-transplantation biomarkers have a high threshold, allowing clinical diagnoses only long after transplantation damage, when changes are clinically and histologically manifested. Novel post-transplantation biomarkers require high sensitivity and a low threshold to indicate allograft damage pre-clinically; examples include non-invasive transcriptomic or proteomic biomarkers that will be applied to diagnose pathologies, to predict rejection, functional outcome, or the individual patient's response to immunossupression. Other applications include targets for novel therapeutic interventions New pre-transplantation biomarkers are stable and are needed to indicate a patient's baseline risk for damage or graft accommodation after transplantation. New pre-transplantation biomarkers are also needed to predict graft rejection and/or accommodation or the response to immunosuppression.
Pre-transplantation biomarkers
Genomic analysis of donor and recipient peripheral blood DNA before transplantation has identified SNPs that indicate the risk or severity of allograft damage or predict allograft survival, and these markers are useful at the pre-transplantation stage [39]. Mutations in the innate immune system protein Toll-like receptor in donor and/or recipient blood were associated with reduced risk and severity of allograft rejection in liver, lung and kidney transplantation [40-45], and complement factor C3 mutations were predictive for renal allograft survival [46], further supporting the relevance of innate immunity for transplantation outcome. However, the success of SNP-based studies is often hindered by the need for large numbers of samples. Using samples across multiple centers might overcome this problem but results in inter-center variation. This variation has been successfully overcome by using statistical approaches, and a biomarker panel of ten SNPs for predicting AR was identified (Table 1a). Pre-transplantation transcriptome analyses have shown significant differences in C3 gene expression between living and deceased donors, and these differences were directly related to the length of cold ischemia. Cold ischemia during transplantation begins with the perfusion of the graft after procurement, which decreases the organ temperature due to the absence of blood supply and creates an environment of hypoxia. Cold ischemia for living donor transplantation was significantly shorter than that for deceased donor transplantation, and changes in C3 gene expression correlated with 2-year graft function [47].
Table 1.
Laboratory-based biomarkers
| Organ | Sample | Proposed mechanism | Biomarker | References | |
|---|---|---|---|---|---|
| (a) Pre-transplantation biomarkers | |||||
| Kidney, lung, liver | Blood (DNA) | Genetic variants in donor/recipient are associated with risk and severity of AR and with allograft survival | 15 SNPs, TLR, C3 | [39-44,46] | |
| Kidney | Biopsy (mRNA) | Expression profiles of innate immunity-related genes predict allograft survival | C3 | [47] | |
| Kidney | Serum (protein); biopsy (mRNA) | Novel immunogenic epitopes | Non-HLA antigens | [12,48-50] | |
| (b) Post-transplantation biomarkers: acute allograft rejection | |||||
| Kidney | Blood (PBMCs, mRNA), urine (mRNA) | Cytotoxic proteins indicate AR | FasL, GranzymeB, Perforine | [27,54,57,58] | |
| Kidney, lung, liver, heart | Blood (PBMCs), serum, BALF, urine (mRNA, protein) | Donor/recipient cytokine expression predicts/detects AR | CXCR, CXCL10 CXCL9 | [59-63] | |
| Kidney | Biopsy, blood (PBMCs, mRNA) | Alterations in miRNA are associated with AR | miR-142-5p, miR-155, miR-223 | [64-67] | |
| Kidney | Biopsy | Biomarkers for antibody-mediated rejection (diagnostic/predictive) | CD38, endothelial cell genes | [70,71] | |
| Kidney | Biopsy, serum (protein) | Antibodies against novel non-HLA antigens (diagnostic/predictive) | AT1R-AA, MICA, Duffy, Kidd, Agrin | [50,72-75] | |
| Kidney, heart | Biopsy, serum (mRNA, protein) | Integrative proteogenomic biomarkers predict and diagnose AR across organs | Novel non-HLA antigen PECAM1 | [12,76] | |
| Post-transplantation biomarkers: chronic allograft damage | |||||
| Kidney | Blood (mRNA), biopsy (mRNA), urine (mRNA) | Predictive peripheral genes and proteins for mild/moderate chronic allograft damage and chronic antibody-mediated damage | TRIB1, CCL2 | [13,77,82] | |
| Kidney, heart | Blood (protein), biopsy (mRNA), urine (protein) | Early diagnostic peripheral and urinary gene expression for IF/TA and anti-fibrotic target | KIM-1, CTGF | [78,79,85,86] | |
| Post-transplantation biomarkers: graft accommodation | |||||
| Liver, kidney | Blood (PBMCs, mRNA) | Peripheral gene expression identifies transplant recipients for discontinuation of immunosuppression | (a) Three classifiers of 2,3 and 7 genes; (b) 33-gene panel; (c) 343 genes | [88,89] | |
| Kidney | Blood (mRNA) | B-lymphocyte-related gene signature of tolerance in transplant patient PBMCs | (a) B-cell signature (IGKV1D-13, IGKV4-1, IGLL1); (b) B-cell signature, ratio of FOXP3/α-1,2-mannosidase |
[90,91] | |
AT1R-AA, agonistic antibodies against angiotensin type II receptor 1; BALF, bronchoalveolar fluid; CCL, CC chemokine ligand; FasL, Fas ligand; FOXP3, Forkhead box P3; IGKV, immunoglobulin kappa variable group; IGLL1, immunoglobulin lambda-like polypeptide 1; KIM-1, kidney injury molecule 1; TLR, Toll-like receptor; IF/TA, interstitial fibrosis/tubular atrophy.
Table 2.
FDA-approved biomarkers
| Organ | Sample | Proposed mechanism | Biomarker | References |
|---|---|---|---|---|
| Heart | Blood (PBMCs, mRNA) | Gene-expression-based diagnostic score distinguishes stable from acute heart allograft rejection patients and mild from severe AR | AlloMap (11-gene panel) |
[92,93] |
| Heart, liver, lung, kidney | Serum (protein) | T-cell activation status indicates risk of AR, under-/over-immunosuppression | ImmuKnow (T-cell-stimulation-dependent iATP levels) | [18,97-100] |
More recently, the detection of novel antigens located in allograft tissue that drive allograft damage has been another means to predict AR before the development of corresponding antibodies in the serum. Integrative proteogenomic analyses have identified tissue-specific novel non-HLAs that led to serological responses in renal transplant patients. Antibodies against MHC class I polypeptide related sequence A (MICA) in the recipients that recognized antigens specific to the renal pelvis and the renal cortex were identified [12]. The association of such novel non-HLA antigens with clinically relevant phenotypes could identify specific immunogenic epitopes in AR and CAD [12,48-50].
Post-transplantation biomarkers
Transplantation initiates the processes responsible for AR and CAD (Figure 2). Biomarkers of different subtypes of rejection injury in the graft itself that indicate damage at the molecular level are needed and could help distinguish rejection episodes with high versus low probability of full functional recovery after anti-rejection therapy [51]. Similarly, biomarkers for graft accommodation could lead to reduction of immunosuppressive drugs or identification of novel drug targets.
Biomarkers of acute allograft rejection
Advances in immunosuppressive therapy and improved patient monitoring have decreased the incidence of AR in solid organ transplantation. However, the lack of non-invasive biomarkers makes early diagnosis and optimized treatment regimens difficult, leading to approximately 10 to 30% of all transplant patients being diagnosed and treated for AR episodes within the first year after transplantation [52,53], on top of a high number of undetected subclinical episodes. AR represents a major risk factor for long-term allograft dysfunction.
Among the first non-invasive, gene-expression-based cellular AR biomarkers discovered were the lethal chemokine perforine, tumor necrosis factor α, transmembrane protein Fas ligand and the serine protease granzyme B, proteins involved in cytotoxic lymphocyte function [27,54] (Table 1a). Several whole-genome transcriptional studies using PBMCs or urine specimens from transplant patients showed that expression of these genes indicated cell-mediated AR. However, the results could not always be confirmed in gene expression studies using graft biopsies or geographically distinct sample sets. In addition, the differential expression of these potential markers in other renal diseases limited their feasibility as AR-specific biomarkers in kidney transplantation [21-23,55]. Only urinary cell transcriptional levels of perforin, granzyme B [56] and granulysin [57] were found to be diagnostic of biopsy-proven cell-mediated AR in renal transplant patients [58].
Other extensively studied potential biomarkers across liver, lung, kidney and heart transplants include chemokines and cytokines. These molecules lead to the differentiation, migration and proliferation of immune cells during AR. In this regard, the chemokines CXCL9 and CXCL10 and the chemokine receptor CXCR3 have been identified as potential biomarkers to predict AR and can be assessed in transplant patient serum, peripheral blood, urine and bronchoalveolar fluid. Other studies revealed their potential as novel therapeutic targets [59-63]. However, none of them has yet reached clinical trial status, and the relevance of these molecules needs to be determined in large cohort studies.
Other gene-expression-based AR biomarkers of increasing interest are miRNAs. These are small (about 19 to 25 nucleotides), naturally occurring noncoding RNAs that primarily repress the translation of mRNA or lead to its degradation [64]. miRNAs are potential biomarkers in renal transplant patient biopsies and stimulated PBMCs [65]. miR-155 has been found to be overexpressed in PBMCs from AR patients [65] and to enhance the development of inflammatory T cells [66]. miRNAs can influence AR, CAD and induction of tolerance [67].
Proteomic approaches identified urinary protein and peptide biomarkers that can correlate with AR. These studies provided a powerful means to distinguish for the first time between AR and BK virus nephropathy, two conditions that seem very similar when biopsied yet require opposing management strategies. A non-invasive urine-based test to distinguish between these entities is a major advance for the renal transplant field, especially with the increasing incidence of BK virus infection in transplant recipients [68,69].
Antibody-mediated AR occurs in a minority of transplant patients and is characterized by the recipient's B lymphocytes forming antibodies against donor antigens. Current diagnosis is based on the presence of donor-specific antibodies in the periphery and on immunostaining for CD20 and peritubular deposition of complement-activated factor C4d. Recently, C4d-negative antibody-mediated AR episodes have been reported and asymptomatic episodes were associated with poor allograft outcome. This potentially leads to higher numbers of actual antibody-mediated AR cases when assessed retrospectively, further strengthening the necessity for new biomarkers of rejection. Endothelial cell gene expression in kidney transplant biopsies has been positively associated with the presence of antibody-mediated AR [70] and the presence of infiltrating clusters of CD38-positive plasmablasts, which correlated better with antibody-mediated rejection than with intragraft C4d staining [71].
Antibody-based biomarkers have been identified by investigating non-HLA antigen responses after transplantation, which have a greater role in allograft outcome than previously thought and thus represent novel diagnostic and predictive biomarkers. Of note are the agonistic antibodies against the angiotensin II type 1 receptor (AT1R-AA) described in renal allograft recipients with severe vascular types of AR [72]. Antagonistic antibodies against MICA, the chemokine receptor Duffy, Kidd polymorphic blood group antigens and the most abundant heparin sulfate proteoglycan, Agrin, were associated with decreased allograft survival [50,73], chronic allograft damage [74] and the development of glomerulopathy [75].
In an integrative approach using transcriptomic and proteomic data, novel non-HLA antigens were identified as triggering de novo serological responses after transplantation in renal transplant recipients [12]. Interestingly, the antigens with the highest immunogenic power were located in the renal pelvis of the allograft. In another integrative study, genes coding for serum- and urine-detectable proteins that were differentially expressed in renal and cardiac biopsies from AR patients were tested for their potential as diagnostic protein biomarkers in a cross-organ, cross-platform study. Upregulated platelet endothelial cell adhesion molecule 1 (PECAM1) in biopsies, serum and urine identified renal AR with 89% sensitivity and 75% specificity in a cross-organ study using publicly available microarray data [76].
Biomarkers for chronic allograft injury
In contrast to AR, chronic allograft injury is a slow progressive disorder involving complex multistage molecular processes, which can be seen from gradual, accumulative changes that lead to declining allograft function after 1 year post-transplantation and finally often result in allograft loss. These processes remain poorly understood and studies are hampered by the slow rate of changes that only slowly reveal a measurable phenotype, and by increasing post-transplantation external biases introduced by immunosuppressive treatment, associated side-effects, patient compliance, lifestyles and subclinical processes, often resulting in inconclusive findings. As a result, biomarkers, and especially non-invasive biomarkers specific for chronic allograft injury, are sparse, and extremely sensitive methods are needed to detect relevant changes before they accumulate and become clinically detectable.
Non-invasive markers of CAD, including urinary and peripheral biomarkers, could not only be readily identified and validated at numerous time-points but would also allow regular monitoring over a long period of time at low cost and would be associated with low patient risk. In an attempt to correlate blood expression signatures with biopsy-proven chronic allograft damage, gene expression panels were identified that predicted mild and moderate/severe chronic allograft damage, and Tribbles-1 (TRIB1) was identified to predict chronic antibody-mediated rejection [13,77]. Well studied molecules in the pathogenesis of fibrosis, as seen in chronic allograft damage, are the transforming and connective tissue growth factors (transforming growth factor-β and connective tissue growth factor (CTGF)) [78,79]. CTGF was increased in transplant patient urine before histopathological and functional chronic dysfunction, revealing it as a potential early non-invasive biomarker [80] and as a potential antifibrotic target [81]. Urinary expression of the chemokine CCL2 at 6 months post-transplantation predicted the development of chronic allograft dysfunction at 24 months post-transplantation in 111 patients [82]. Kidney injury molecule 1 (KIM1), previously discovered as a proximal tubular biomarker of acute kidney injury [83,84], was associated with chronic allograft damage, including calcineurin inhibitor toxicity and interstitial fibrosis/tubular atrophy [85,86]. However, KIM1 expression also correlated with transplant-independent drug-induced nephrotoxicity [87] and renal cell carcinoma [84], revealing it as a marker of general renal injury [83].
Biomarkers for monitoring graft accommodation
Achieving an immunosuppression-free state, referred to as clinical operational tolerance, is the ultimate goal in transplantation. Current estimates report only 100 cases of clinical operational tolerance in renal transplants so far [88] and tolerance induction protocols, such as perioperative infusion of donor bone-marrow-derived stem cells or perioperative lymphocyte depletion, have failed and have led to graft loss in most cases. Specific biomarkers indicating immune quiescence and representing targets for novel tolerance induction protocols are needed. In a recent study [89], three gene-expression-based classifiers were identified, predicting liver tolerance and identifying liver transplant recipients for discontinuation of immunosuppression. Here, a combined approach of microarray discovery and quantitative reverse transcriptase (qRT)-PCR validation using PBMCs from a total of 44 tolerant and 48 non-tolerant patients was used [89] to determine a first gene expression signature of renal allograft tolerance consisting of 33 genes. This panel was able to predict the presence of a peripheral tolerant phenotype suggesting a pattern of reduced co-stimulatory signaling, immune quiescence, apoptosis and memory T cell responses [14].
Recently, two groups identified tolerance gene expression signatures in kidney transplant patients associated with B cells by applying the same microarray and qRT-PCR approach [90,91]. Genes identified by Newell et al. [90] were associated with clinical and phenotypic parameters and with increased expression of multiple B-cell differentiation genes. The tolerance signature identified by Sagoo et al. [91] was also related to B cells, consisting of ten individual genes with a high ratio of the forkhead box protein FOXP3 to α-1,2-mannosidase. Tolerant patients showed an expansion of peripheral blood B and natural killer lymphocytes, fewer activated CD4+ T cells, a lack of donor-specific antibodies and donor-specific hyporesponsiveness of CD4+ T cells. Similar studies on operational tolerance have also been done in liver transplant recipients [89]. Tolerance-associated gene-expression signatures seem to be promising, as validation studies have proven their relevance. Whether these signatures can be used to predict or monitor tolerance in transplant patients has to be assessed in prospective studies using larger numbers of patients, which will be difficult given the low incidence of tolerance.
FDA-approved biomarkers
A transcriptomic analysis of peripheral blood samples from heart allograft patients identified an 11-gene panel that discriminated patients with stable allograft function from patients with moderate or severe AR [92], which led to the development of the first FDA-approved non-invasive diagnostic test for acute heart allograft rejection (AlloMap, XDx). Applying a mathematical algorithm, gene expression was translated into a diagnostic score [93] that discriminated stable transplants from AR and mild from severe AR. Another approach has exploited the measurement of the ATP release that depends on T-cell stimulation (iATP) [94-96], hypothesizing that the activation status of T cells indicates patients at high risk of acute rejection or at high risk for over- or under-immunosuppression. The iATP levels led to the development of a therapeutic response assay, ImmuKnow (Cylex) [18,97-100]. Nevertheless, a new set of biomarkers is desperately needed to replace or complement these tests in order to improve clinical practice with regard to the function of transplanted organs. This will be achieved only with a biomarker panel - gene- or protein-based - that has high positive predictive value for injury (which is missing in the AlloMap panel) and has very high specificity and sensitivity for injury (which is missing in the Cylex test).
Conclusions
The ultimate goal of biomarker studies in transplantation is to find non-invasive biomarkers of transplant pathologies using patient urine or blood that indicate changes at the molecular level, before the development of a clinical phenotype, that predict allograft outcome or response to therapy, and that possibly reveal novel targets for therapeutic interventions. As a result of the technological advances in high-throughput methodologies, multiple biomarker studies have been performed, leading to numerous potential biomarkers being published. However, only very few have graduated from the laboratory and gained FDA approval.
Laboratory-dependent confounding factors include differences in sample processing and data analyses, making comparability of data difficult. Regulatory elements and analytical guidelines, as suggested by the NIH or the MACQ studies, have been introduced to increase the validity and robustness of identified biomarkers and to make studies more homogenous. Sample-dependent confounding factors, such as the abundance of globin mRNA in whole blood, have been identified and successfully overcome, and advances in analytical methods now allow horizontal and vertical meta-analyses.
Promising non-invasive biomarkers for acute rejection and operational tolerance have therefore been identified and now need prospective validation in large patient cohorts. Multi-center studies have been introduced: the US 'Clinical Trials in Organ Transplantation' (CTOT and CTOTC), the Canadian 'Biomarkers in Transplantation' (BIT) project and the European study of 'Reprogramming the Immune System for Establishment of Tolerance' (RISET).
In addition, we have gained deeper knowledge about the underlying pathogenic mechanisms of AR and CAD. The detection of novel non-HLA antibodies, C4d-negative antibody-mediated rejection, and the role of the innate immune system in acute rejection, as seen in the relevance of complement-system-associated molecules, will further biomarker development.
As seen for drug development studies, biomarker development studies need to become more uniform and standardized. Standard operating procedures for sample handling, experimental procedures and performance of data analyses need to be introduced, in addition to requirements for sample sizes, number and kind of validation studies.
Once transferred to the clinic, these recent advances will eventually lead to personalized transplantation medicine, including improved donor-recipient matching, individual immunosuppressive regimens, and individual risk assessment for AR or CAD and prediction of graft accommodation. These improvements will undoubtedly reduce the costs of health care dramatically. Finally, these changes will be reflected by increased allograft survival and decreased patient morbidity.
Abbreviations
AR: acute allograft rejection; CAD: chronic allograft damage; CTGF: connective tissue growth factor; FDA: Food and Drug Administration; HLA: human leukocyte specific antigen; iATP: intracellular ATP; MAQC: microarray quality control; MICA: MHC class I polypeptide related sequence A; miRNA: microRNA; PBMC: peripheral blood mononuclear cell; qRT-PCR: quantitative reverse transcriptase PCR; SNP: single nucleotide polymorphism.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Silke Roedder, Email: silke@stanford.edu.
Matthew Vitalone, Email: vitalone@stanford.edu.
Purvesh Khatri, Email: pkhatri@stanford.ed.
Minnie M Sarwal, Email: msarwal@stanford.edu.
References
- Khatri P, Sarwal MM. Using gene arrays in diagnosis of rejection. Curr Opin Organ Transplant. 2009;14:34–39. doi: 10.1097/MOT.0b013e32831e13d0. [DOI] [PubMed] [Google Scholar]
- Sigdel TK, Klassen RB, Sarwal MM. Interpreting the proteome and peptidome in transplantation. Adv Clin Chem. 2009;47:139–169. doi: 10.1016/s0065-2423(09)47006-9. [DOI] [PubMed] [Google Scholar]
- Sigdel TK, Sarwal MM. The proteogenomic path towards biomarker discovery. Pediatr Transplant. 2008;12:737–747. doi: 10.1111/j.1399-3046.2008.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying L, Sarwal M. In praise of arrays. Pediatr Nephrol. 2009;24:1643–1659. doi: 10.1007/s00467-008-0808-z. quiz 1655, 1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AlloMap; A non-invasive gene expression test (XDx Diagnostics). http://www.allomap.com/
- Naesens M, Sarwal MM. Molecular diagnostics in transplantation. Nat Rev Nephrol. 2010;6:614–628. doi: 10.1038/nrneph.2010.113. [DOI] [PubMed] [Google Scholar]
- Rodder S, Scherer A, Korner M, Eisenberger U, Hertig A, Raulf F, Rondeau E, Marti HP. Meta-analyses qualify metzincins and related genes as acute rejection markers in renal transplant patients. Am J Transplant. 2010;10:286–297. doi: 10.1111/j.1600-6143.2009.02928.x. [DOI] [PubMed] [Google Scholar]
- Rodder S, Scherer A, Raulf F, Berthier CC, Hertig A, Couzi L, Durrbach A, Rondeau E, Marti HP. Renal allografts with IF/TA display distinct expression profiles of metzincins and related genes. Am J Transplant. 2009;9:517–526. doi: 10.1111/j.1600-6143.2008.02512.x. [DOI] [PubMed] [Google Scholar]
- Kong X, Mas V, Archer KJ. A non-parametric meta-analysis approach for combining independent microarray datasets: application using two microarray datasets pertaining to chronic allograft nephropathy. BMC Genomics. 2008;9:98. doi: 10.1186/1471-2164-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park WD, Stegall MD. A meta-analysis of kidney microarray datasets: investigation of cytokine gene detection and correlation with RT-PCR and detection thresholds. BMC Genomics. 2007;8:88. doi: 10.1186/1471-2164-8-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling XB, Sigdel TK, Lau K, Ying L, Lau I, Schilling J, Sarwal MM. Integrative urinary peptidomics in renal transplantation identifies biomarkers for acute rejection. J Am Soc Nephrol. 2010;21:646–653. doi: 10.1681/ASN.2009080876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wadia P, Chen R, Kambham N, Naesens M, Sigdel TK, Miklos DB, Sarwal MM, Butte AJ. Identifying compartment-specific non-HLA targets after renal transplantation by integrating transcriptome and "antibodyome" measures. Proc Natl Acad Sci USA. 2009;106:4148–4153. doi: 10.1073/pnas.0900563106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian SM, Heilman R, Mondala TS, Nakorchevsky A, Hewel JA, Campbell D, Robison EH, Wang L, Lin W, Gaber L, Solez K, Shidban H, Mendez R, Schaffer RL, Fisher JS, Flechner SM, Head SR, Horvath S, Yates JR, Marsh CL, Salomon DR. Biomarkers for early and late stage chronic allograft nephropathy by proteogenomic profiling of peripheral blood. PLoS One. 2009;4:e6212. doi: 10.1371/journal.pone.0006212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouard S, Mansfield E, Braud C, Li L, Giral M, Hsieh SC, Baeten D, Zhang M, Ashton-Chess J, Braudeau C, Hsieh F, Dupont A, Pallier A, Moreau A, Louis S, Ruiz C, Salvatierra O, Soulillou JP, Sarwal M. Identification of a peripheral blood transcriptional biomarker panel associated with operational renal allograft tolerance. Proc Natl Acad Sci USA. 2007;104:15448–15453. doi: 10.1073/pnas.0705834104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH Scandling J Sigdel T Sarwal M A highly specific novel 3 gene-set can non-invasively predict operational renal allograft tolerance (#334). Am J Transplant 201010120089116 [Google Scholar]
- Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, Luo Y, Sun YA, Willey JC, Setterquist RA, Fischer GM, Tong W, Dragan YP, Dix DJ, Frueh FW, Goodsaid FM, Herman D, Jensen RV, Johnson CD, Lobenhofer EK, Puri RK, Schrf U, Thierry-Mieg J, Wang C, Wilson M, Wolber PK. et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Campbell G, Jones WD, Campagne F, Wen Z, Walker SJ, Su Z, Chu TM, Goodsaid FM, Pusztai L, Shaughnessy JD, Oberthuer A, Thomas RS, Paules RS, Fielden M, Barlogie B, Chen W, Du P, Fischer M, Furlanello C, Gallas BD, Ge X, Megherbi DB, Symmans WF, Wang MD, Zhang J, Bitter H, Brors B, Bushel PR, Bylesjo M. et al. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat Biotechnol. 2010;28:827–838. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam A, Morrissey PE, Brem AS, Fischer SA, Gohh RY, Yango AF, Monaco AP. Use of an immune function assay to monitor immunosuppression for treatment of post-transplant lymphoproliferative disorder. Pediatr Transplant. 2006;10:613–616. doi: 10.1111/j.1399-3046.2006.00510.x. [DOI] [PubMed] [Google Scholar]
- Kowalski RJ, Post DR, Mannon RB, Sebastian A, Wright HI, Sigle G, Burdick J, Elmagd KA, Zeevi A, Lopez-Cepero M, Daller JA, Gritsch HA, Reed EF, Jonsson J, Hawkins D, Britz JA. Assessing relative risks of infection and rejection: a meta-analysis using an immune function assay. Transplantation. 2006;82:663–668. doi: 10.1097/01.tp.0000234837.02126.70. [DOI] [PubMed] [Google Scholar]
- Debey S, Schoenbeck U, Hellmich M, Gathof BS, Pillai R, Zander T, Schultze JL. Comparison of different isolation techniques prior gene expression profiling of blood derived cells: impact on physiological responses, on overall expression and the role of different cell types. Pharmacogenomics J. 2004;4:193–207. doi: 10.1038/sj.tpj.6500240. [DOI] [PubMed] [Google Scholar]
- Veale JL, Liang LW, Zhang Q, Gjertson DW, Du Z, Bloomquist EW, Jia J, Qian L, Wilkinson AH, Danovitch GM, Pham PT, Rosenthal JT, Lassman CR, Braun J, Reed EF, Gritsch HA. Noninvasive diagnosis of cellular and antibody-mediated rejection by perforin and granzyme B in renal allografts. Hum Immunol. 2006;67:777–786. doi: 10.1016/j.humimm.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Alakulppi NS, Kyllonen LE, Partanen J, Salmela KT, Laine JT. Diagnosis of acute renal allograft rejection by analyzing whole blood mRNA expression of lymphocyte marker molecules. Transplantation. 2007;83:791–798. doi: 10.1097/01.tp.0000258726.13363.ab. [DOI] [PubMed] [Google Scholar]
- Graziotto R, Del Prete D, Rigotti P, Anglani F, Baldan N, Furian L, Valente M, Antonello A, Marchini F, D'Angelo A, Gambaro G. Perforin, Granzyme B, and fas ligand for molecular diagnosis of acute renal-allograft rejection: analyses on serial biopsies suggest methodological issues. Transplantation. 2006;81:1125–1132. doi: 10.1097/01.tp.0000208573.16839.67. [DOI] [PubMed] [Google Scholar]
- Shin GT, Kim SJ, Lee TS, Oh CK, Kim H. Gene expression of perforin by peripheral blood lymphocytes as a marker of acute rejection. Nephron Clin Pract. 2005;100:c63–c70. doi: 10.1159/000085050. [DOI] [PubMed] [Google Scholar]
- Simon T, Opelz G, Weimer R, Wiesel M, Feustel A, Ott RC, Susal C. The effect of ATG on cytokine and cytotoxic T-lymphocyte gene expression in renal allograft recipients during the early post-transplant period. Clin Transplant. 2003;17:217–224. doi: 10.1034/j.1399-0012.2003.00031.x. [DOI] [PubMed] [Google Scholar]
- Netto MV, Fonseca BA, Dantas M, Saber LT, Castro MC, Ferraz AS. Granzyme B, FAS-ligand and perforin expression during acute cellular rejection episodes after kidney transplantation: comparison between blood and renal aspirates. Transplant Proc. 2002;34:476–478. doi: 10.1016/S0041-1345(02)02601-5. [DOI] [PubMed] [Google Scholar]
- Vasconcellos LM, Schachter AD, Zheng XX, Vasconcellos LH, Shapiro M, Harmon WE, Strom TB. Cytotoxic lymphocyte gene expression in peripheral blood leukocytes correlates with rejecting renal allografts. Transplantation. 1998;66:562–566. doi: 10.1097/00007890-199809150-00002. [DOI] [PubMed] [Google Scholar]
- Bammler T, Beyer RP, Bhattacharya S, Boorman GA, Boyles A, Bradford BU, Bumgarner RE, Bushel PR, Chaturvedi K, Choi D, Cunningham ML, Deng S, Dressman HK, Fannin RD, Farin FM, Freedman JH, Fry RC, Harper A, Humble MC, Hurban P, Kavanagh TJ, Kaufmann WK, Kerr KF, Jing L, Lapidus JA, Lasarev MR, Li J, Li YJ, Lobenhofer EK, Lu X. et al. Standardizing global gene expression analysis between laboratories and across platforms. Nat Methods. 2005;2:351–356. doi: 10.1038/nmeth754. [DOI] [PubMed] [Google Scholar]
- Rainen L, Oelmueller U, Jurgensen S, Wyrich R, Ballas C, Schram J, Herdman C, Bankaitis-Davis D, Nicholls N, Trollinger D, Tryon V. Stabilization of mRNA expression in whole blood samples. Clin Chem. 2002;48:1883–1890. [PubMed] [Google Scholar]
- Debey S, Zander T, Brors B, Popov A, Eils R, Schultze JL. A highly standardized, robust, and cost-effective method for genome-wide transcriptome analysis of peripheral blood applicable to large-scale clinical trials. Genomics. 2006;87:653–664. doi: 10.1016/j.ygeno.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Li L, Ying L, Naesens M, Xiao W, Sigdel T, Hsieh S, Martin J, Chen R, Liu K, Mindrinos M, Davis R, Sarwal M. Interference of globin genes with biomarker discovery for allograft rejection in peripheral blood samples. Physiol Genomics. 2008;32:190–197. doi: 10.1152/physiolgenomics.00216.2007. [DOI] [PubMed] [Google Scholar]
- Min JL, Barrett A, Watts T, Pettersson FH, Lockstone HE, Lindgren CM, Taylor JM, Allen M, Zondervan KT, McCarthy MI. Variability of gene expression profiles in human blood and lymphoblastoid cell lines. BMC Genomics. 2010;11:96. doi: 10.1186/1471-2164-11-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, Relman DA, Brown PO. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci USA. 2003;100:1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feezor RJ, Baker HV, Mindrinos M, Hayden D, Tannahill CL, Brownstein BH, Fay A, MacMillan S, Laramie J, Xiao W, Moldawer LL, Cobb JP, Laudanski K, Miller-Graziano CL, Maier RV, Schoenfeld D, Davis RW, Tompkins RG. Whole blood and leukocyte RNA isolation for gene expression analyses. Physiol Genomics. 2004;19:247–254. doi: 10.1152/physiolgenomics.00020.2004. [DOI] [PubMed] [Google Scholar]
- Shen-Orr SS, Tibshirani R, Khatri P, Bodian DL, Staedtler F, Perry NM, Hastie T, Sarwal MM, Davis MM, Butte AJ. Cell type-specific gene expression differences in complex tissues. Nat Methods. 2010;7:287–289. doi: 10.1038/nmeth.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas AR, Wolslegel K, Seshasayee D, Modrusan Z, Clark HF. Deconvolution of blood microarray data identifies cellular activation patterns in systemic lupus erythematosus. PLoS One. 2009;4:e6098. doi: 10.1371/journal.pone.0006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repsilber D, Kern S, Telaar A, Walzl G, Black GF, Selbig J, Parida SK, Kaufmann SH, Jacobsen M. Biomarker discovery in heterogeneous tissue samples -taking the in-silico deconfounding approach. BMC Bioinformatics. 2010;11:27. doi: 10.1186/1471-2105-11-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki P, Lachmann N, Cai J. Summary of the effect of de novo HLA antibodies on chronic kidney graft failure. Clin Transpl. 2006. pp. 455–462. [PubMed]
- Israni A, Leduc R, Holmes J, Jacobson PA, Lamba V, Guan W, Schladt D, Chen J, Matas AJ, Oetting WS. Single-nucleotide polymorphisms, acute rejection, and severity of tubulitis in kidney transplantation, accounting for center-to-center variation. Transplantation. 2010;90:1401–1408. doi: 10.1097/TP.0b013e3182000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger B, Banas MC, Walberer A, Boger CA, Farkas S, Hoffmann U, Fischereder M, Banas B, Kramer BK. A comprehensive genotype-phenotype interaction of different Toll-like receptor variations in a renal transplant cohort. Clin Sci (Lond) 2010;119:535–544. doi: 10.1042/CS20100190. [DOI] [PubMed] [Google Scholar]
- Dhillon N, Walsh L, Kruger B, Ward SC, Godbold JH, Radwan M, Schiano T, Murphy BT, Schroppel B. A single nucleotide polymorphism of Toll-like receptor 4 identifies the risk of developing graft failure after liver transplantation. J Hepatol. 2010;53:67–72. doi: 10.1016/j.jhep.2009.12.044. [DOI] [PubMed] [Google Scholar]
- Ducloux D, Deschamps M, Yannaraki M, Ferrand C, Bamoulid J, Saas P, Kazory A, Chalopin JM, Tiberghien P. Relevance of Toll-like receptor-4 polymorphisms in renal transplantation. Kidney Int. 2005;67:2454–2461. doi: 10.1111/j.1523-1755.2005.00354.x. [DOI] [PubMed] [Google Scholar]
- Eid AJ, Brown RA, Paya CV, Razonable RR. Association between toll-like receptor polymorphisms and the outcome of liver transplantation for chronic hepatitis C virus. Transplantation. 2007;84:511–516. doi: 10.1097/01.tp.0000276960.35313.bf. [DOI] [PubMed] [Google Scholar]
- Hwang YH, Ro H, Choi I, Kim H, Oh KH, Hwang JI, Park MH, Kim S, Yang J, Ahn C. Impact of polymorphisms of TLR4/CD14 and TLR3 on acute rejection in kidney transplantation. Transplantation. 2009;88:699–705. doi: 10.1097/TP.0b013e3181b2f34a. [DOI] [PubMed] [Google Scholar]
- Kastelijn EA, van Moorsel CH, Rijkers GT, Ruven HJ, Karthaus V, Kwakkel-van Erp JM, van de Graaf EA, Zanen P, van Kessel DA, Grutters JC, van den Bosch JM. Polymorphisms in innate immunity genes associated with development of bronchiolitis obliterans after lung transplantation. J Heart Lung Transplant. 2010;29:665–671. doi: 10.1016/j.healun.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Naesens M, Butte AJ, Sarwal MM. C3 polymorphisms and outcomes of renal allografts. N Engl J Med. 2009;360:2478–2479. [PubMed] [Google Scholar]
- Naesens M, Li L, Ying L, Sansanwal P, Sigdel TK, Hsieh SC, Kambham N, Lerut E, Salvatierra O, Butte AJ, Sarwal MM. Expression of complement components differs between kidney allografts from living and deceased donors. J Am Soc Nephrol. 2009;20:1839–1851. doi: 10.1681/ASN.2008111145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland SM, Li L, Sigdel TK, Wadia PP, Miklos DB, Butte AJ, Sarwal MM. Protein microarrays identify antibodies to protein kinase Czeta that are associated with a greater risk of allograft loss in pediatric renal transplant recipients. Kidney Int. 2009;76:1277–1283. doi: 10.1038/ki.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Chen A, Chaudhuri A, Kambham N, Sigdel T, Chen R, Sarwal MM. Compartmental localization and clinical relevance of MICA antibodies after renal transplantation. Transplantation. 2010;89:312–319. doi: 10.1097/TP.0b013e3181bbbe4c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Stastny P, Susal C, Dohler B, Opelz G. Antibodies against MICA antigens and kidney-transplant rejection. N Engl J Med. 2007;357:1293–1300. doi: 10.1056/NEJMoa067160. [DOI] [PubMed] [Google Scholar]
- Sarwal M, Chua MS, Kambham N, Hsieh SC, Satterwhite T, Masek M, Salvatierra O Jr. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med. 2003;349:125–138. doi: 10.1056/NEJMoa035588. [DOI] [PubMed] [Google Scholar]
- Hertz MI, Aurora P, Christie JD, Dobbels F, Edwards LB, Kirk R, Kucheryavaya AY, Rahmel AO, Rowe AW, Stehlik J, Taylor DO. Scientific Registry of the International Society for Heart and Lung Transplantation: introduction to the 2009 Annual Reports. J Heart Lung Transplant. 2009;28:989–992. doi: 10.1016/j.healun.2009.08.005. [DOI] [PubMed] [Google Scholar]
- National Institutes of Health. U.S. Renal Data System, USRDS 2010 Annual data report: atlas of chronic kidney disease and end-stage renal disease in the United States. http://www.usrds.org/adr.htm
- Choy JC. Granzymes and perforin in solid organ transplant rejection. Cell Death Differ. 2010;17:567–576. doi: 10.1038/cdd.2009.161. [DOI] [PubMed] [Google Scholar]
- Sarwal MM, Jani A, Chang S, Huie P, Wang Z, Salvatierra O Jr, Clayberger C, Sibley R, Krensky AM, Pavlakis M. Granulysin expression is a marker for acute rejection and steroid resistance in human renal transplantation. Hum Immunol. 2001;62:21–31. doi: 10.1016/S0198-8859(00)00228-7. [DOI] [PubMed] [Google Scholar]
- Li B, Hartono C, Ding R, Sharma VK, Ramaswamy R, Qian B, Serur D, Mouradian J, Schwartz JE, Suthanthiran M. Noninvasive diagnosis of renal-allograft rejection by measurement of messenger RNA for perforin and granzyme B in urine. N Engl J Med. 2001;344:947–954. doi: 10.1056/NEJM200103293441301. [DOI] [PubMed] [Google Scholar]
- Kotsch K, Mashreghi MF, Bold G, Tretow P, Beyer J, Matz M, Hoerstrup J, Pratschke J, Ding R, Suthanthiran M, Volk HD, Reinke P. Enhanced granulysin mRNA expression in urinary sediment in early and delayed acute renal allograft rejection. Transplantation. 2004;77:1866–1875. doi: 10.1097/01.TP.0000131157.19937.3F. [DOI] [PubMed] [Google Scholar]
- Galante NZ, Camara NO, Kallas EG, Salomao R, Pacheco-Silva A, Medina-Pestana JO. Noninvasive immune monitoring assessed by flow cytometry and real time RT-PCR in urine of renal transplantation recipients. Transpl Immunol. 2006;16:73–80. doi: 10.1016/j.trim.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Krukemeyer MG, Moeller J, Morawietz L, Rudolph B, Neumann U, Theruvath T, Neuhaus P, Krenn V. Description of B lymphocytes and plasma cells, complement, and chemokines/receptors in acute liver allograft rejection. Transplantation. 2004;78:65–70. doi: 10.1097/01.tp.0000132324.14207.8b. [DOI] [PubMed] [Google Scholar]
- Hauser IA, Spiegler S, Kiss E, Gauer S, Sichler O, Scheuermann EH, Ackermann H, Pfeilschifter JM, Geiger H, Grone HJ, Radeke HH. Prediction of acute renal allograft rejection by urinary monokine induced by IFN-gamma (MIG). J Am Soc Nephrol. 2005;16:1849–1858. doi: 10.1681/ASN.2004100836. [DOI] [PubMed] [Google Scholar]
- Schaub S, Nickerson P, Rush D, Mayr M, Hess C, Golian M, Stefura W, Hayglass K. Urinary CXCL9 and CXCL10 levels correlate with the extent of subclinical tubulitis. Am J Transplant. 2009;9:1347–1353. doi: 10.1111/j.1600-6143.2009.02645.x. [DOI] [PubMed] [Google Scholar]
- Belperio JA, Keane MP, Burdick MD, Lynch JP, Zisman DA, Xue YY, Li K, Ardehali A, Ross DJ, Strieter RM. Role of CXCL9/CXCR3 chemokine biology during pathogenesis of acute lung allograft rejection. J Immunol. 2003;171:4844–4852. doi: 10.4049/jimmunol.171.9.4844. [DOI] [PubMed] [Google Scholar]
- Hancock WW, Gao W, Csizmadia V, Faia KL, Shemmeri N, Luster AD. Donor-derived IP-10 initiates development of acute allograft rejection. J Exp Med. 2001;193:975–980. doi: 10.1084/jem.193.8.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu AB, Wilkinson MF, van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008;27:471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anglicheau D, Sharma VK, Ding R, Hummel A, Snopkowski C, Dadhania D, Seshan SV, Suthanthiran M. MicroRNA expression profiles predictive of human renal allograft status. Proc Natl Acad Sci USA. 2009;106:5330–5335. doi: 10.1073/pnas.0813121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A, Krams SM, Martinez OM. MicroRNAs as immune regulators: implications for transplantation. Am J Transplant. 2010;10:713–719. doi: 10.1111/j.1600-6143.2010.03032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall A, Hariharan S. BK virus nephritis after renal transplantation. Clin J Am Soc Nephrol. 2008;3(Suppl 2):S68–S75. doi: 10.2215/CJN.02770707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschner S, Gerke P, Geyer M, Wilpert J, Krumme B, Benzing T, Walz G. Leflunomide therapy for polyomavirus-induced allograft nephropathy: efficient BK virus elimination without increased risk of rejection. Transplant Proc. 2009;41:2533–2538. doi: 10.1016/j.transproceed.2009.06.099. [DOI] [PubMed] [Google Scholar]
- Sis B, Jhangri GS, Bunnag S, Allanach K, Kaplan B, Halloran PF. Endothelial gene expression in kidney transplants with alloantibody indicates antibody-mediated damage despite lack of C4d staining. Am J Transplant. 2009;9:2312–2323. doi: 10.1111/j.1600-6143.2009.02761.x. [DOI] [PubMed] [Google Scholar]
- Zarkhin V, Kambham N, Li L, Kwok S, Hsieh SC, Salvatierra O, Sarwal MM. Characterization of intra-graft B cells during renal allograft rejection. Kidney Int. 2008;74:664–673. doi: 10.1038/ki.2008.249. [DOI] [PubMed] [Google Scholar]
- Dragun D, Müller DN, Bräsen JH, Fritsche L, Nieminen-Kelhä M, Dechend R, Kintscher U, Rudolph B, Hoebeke J, Eckert D, Mazak I, Plehm R, Schönemann C, Unger T, Budde K, Neumayer HH, Luft FC, Wallukat G. Angiotensin II type 1-receptor activating antibodies in renal-allograft rejection. N Engl J Med. 2005;352:558–569. doi: 10.1056/NEJMoa035717. [DOI] [PubMed] [Google Scholar]
- Terasaki PI, Ozawa M, Castro R. Four-year follow-up of a prospective trial of HLA and MICA antibodies on kidney graft survival. Am J Transplant. 2007;7:408–415. doi: 10.1111/j.1600-6143.2006.01644.x. [DOI] [PubMed] [Google Scholar]
- Lerut E, Van Damme B, Noizat-Pirenne F, Emonds MP, Rouger P, Vanrenterghem Y, Pirenne J, Ansart-Pirenne H. Duffy and Kidd blood group antigens: minor histocompatibility antigens involved in renal allograft rejection? Transfusion. 2007;47:28–40. doi: 10.1111/j.1537-2995.2007.01060.x. [DOI] [PubMed] [Google Scholar]
- Joosten SA, Sijpkens YW, van Ham V, Trouw LA, van der Vlag J, van den Heuvel B, van Kooten C, Paul LC. Antibody response against the glomerular basement membrane protein agrin in patients with transplant glomerulopathy. Am J Transplant. 2005;5:383–393. doi: 10.1111/j.1600-6143.2005.00690.x. [DOI] [PubMed] [Google Scholar]
- Chen R, Sigdel TK, Li L, Kambham N, Dudley JT, Hsieh SC, Klassen RB, Chen A, Caohuu T, Morgan AA, Valantine HA, Khush KK, Sarwal MM, Butte AJ. Differentially expressed RNA from public microarray data identifies serum protein biomarkers for cross-organ transplant rejection and other conditions. PLoS Comput Biol. 2010;6:e1000940. doi: 10.1371/journal.pcbi.1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton-Chess J, Giral M, Mengel M, Renaudin K, Foucher Y, Gwinner W, Braud C, Dugast E, Quillard T, Thebault P, Chiffoleau E, Braudeau C, Charreau B, Soulillou JP, Brouard S. Tribbles-1 as a novel biomarker of chronic antibody-mediated rejection. J Am Soc Nephrol. 2008;19:1116–1127. doi: 10.1681/ASN.2007101056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihn H. Pathogenesis of fibrosis: role of TGF-beta and CTGF. Curr Opin Rheumatol. 2002;14:681–685. doi: 10.1097/00002281-200211000-00009. [DOI] [PubMed] [Google Scholar]
- Booth AJ, Csencsits-Smith K, Wood SC, Lu G, Lipson KE, Bishop DK. Connective tissue growth factor promotes fibrosis downstream of TGFbeta and IL-6 in chronic cardiac allograft rejection. Am J Transplant. 2010;10:220–230. doi: 10.1111/j.1600-6143.2009.02826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Tu Z, Bao J, Sun H, Wang W, Luo G, Li S, Li Y, Bu H. Urinary connective tissue growth factor increases far earlier than histopathological damage and functional deterioration in early chronic renal allograft injury. Scand J Urol Nephrol. 2009;43:390–399. doi: 10.3109/00365520903142244. [DOI] [PubMed] [Google Scholar]
- Luo GH, Lu YP, Song J, Yang L, Shi YJ, Li YP. Inhibition of connective tissue growth factor by small interfering RNA prevents renal fibrosis in rats undergoing chronic allograft nephropathy. Transplant Proc. 2008;40:2365–2369. doi: 10.1016/j.transproceed.2008.07.100. [DOI] [PubMed] [Google Scholar]
- Ho J, Rush DN, Gibson IW, Karpinski M, Storsley L, Bestland J, Stefura W, HayGlass KT, Nickerson PW. Early urinary CCL2 is associated with the later development of interstitial fibrosis and tubular atrophy in renal allografts. Transplantation. 2010;90:394–400. doi: 10.1097/TP.0b013e3181e6424d. [DOI] [PubMed] [Google Scholar]
- van Timmeren MM, van den Heuvel MC, Bailly V, Bakker SJ, van Goor H, Stegeman CA. Tubular kidney injury molecule-1 (KIM-1) in human renal disease. J Pathol. 2007;212:209–217. doi: 10.1002/path.2175. [DOI] [PubMed] [Google Scholar]
- Han WK, Bailly V, Abichandani R, Thadhani R, Bonventre JV. Kidney Injury Molecule-1 (KIM-1): a novel biomarker for human renal proximal tubule injury. Kidney Int. 2002;62:237–244. doi: 10.1046/j.1523-1755.2002.00433.x. [DOI] [PubMed] [Google Scholar]
- Malyszko J, Koc-Zorawska E, Malyszko JS, Mysliwiec M. Kidney injury molecule-1 correlates with kidney function in renal allograft recipients. Transplant Proc. 2010;42:3957–3959. doi: 10.1016/j.transproceed.2010.10.005. [DOI] [PubMed] [Google Scholar]
- van Timmeren MM, Vaidya VS, van Ree RM, Oterdoom LH, de Vries AP, Gans RO, van Goor H, Stegeman CA, Bonventre JV, Bakker SJ. High urinary excretion of kidney injury molecule-1 is an independent predictor of graft loss in renal transplant recipients. Transplantation. 2007;84:1625–1630. doi: 10.1097/01.tp.0000295982.78039.ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Vaidya VS, Brown RP, Zhang J, Rosenzweig BA, Thompson KL, Miller TJ, Bonventre JV, Goering PL. Comparison of kidney injury molecule-1 and other nephrotoxicity biomarkers in urine and kidney following acute exposure to gentamicin, mercury, and chromium. Toxicol Sci. 2008;101:159–170. doi: 10.1093/toxsci/kfm260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando G, Hematti P, Stratta RJ, Burke GW, Cocco PD, Pisani F, Soker S, Wood K. Clinical operational tolerance after renal transplantation: current status and future challenges. Ann Surg. 2010;252:915–928. doi: 10.1097/SLA.0b013e3181f3efb0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Llordella M, Lozano JJ, Puig-Pey I, Orlando G, Tisone G, Lerut J, Benítez C, Pons JA, Parrilla P, Ramírez P, Bruguera M, Rimola A, Sánchez-Fueyo A. Using transcriptional profiling to develop a diagnostic test of operational tolerance in liver transplant recipients. J Clin Invest. 2008;118:2845–2857. doi: 10.1172/JCI35342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell KA, Asare A, Kirk AD, Gisler TD, Bourcier K, Suthanthiran M, Burlingham WJ, Marks WH, Sanz I, Lechler RI, Hernandez-Fuentes MP, Turka LA, Seyfert-Margolis VL. Immune Tolerance Network ST507 Study Group. Identification of a B cell signature associated with renal transplant tolerance in humans. J Clin Invest. 2010;120:1836–1847. doi: 10.1172/JCI39933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagoo P, Perucha E, Sawitzki B, Tomiuk S, Stephens DA, Miqueu P, Chapman S, Craciun L, Sergeant R, Brouard S, Rovis F, Jimenez E, Ballow A, Giral M, Rebollo-Mesa I, Le Moine A, Braudeau C, Hilton R, Gerstmayer B, Bourcier K, Sharif A, Krajewska M, Lord GM, Roberts I, Goldman M, Wood KJ, Newell K, Seyfert-Margolis V, Warrens AN, Janssen U. et al. Development of a cross-platform biomarker signature to detect renal transplant tolerance in humans. J Clin Invest. 2010;120:1848–1861. doi: 10.1172/JCI39922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng MC, Eisen HJ, Mehra MR, Billingham M, Marboe CC, Berry G, Kobashigawa J, Johnson FL, Starling RC, Murali S, Pauly DF, Baron H, Wohlgemuth JG, Woodward RN, Klingler TM, Walther D, Lal PG, Rosenberg S, Hunt S. Noninvasive discrimination of rejection in cardiac allograft recipients using gene expression profiling. Am J Transplant. 2006;6:150–160. doi: 10.1111/j.1600-6143.2005.01175.x. [DOI] [PubMed] [Google Scholar]
- Bernstein D, Williams GE, Eisen H, Mital S, Wohlgemuth JG, Klingler TM, Fang KC, Deng MC, Kobashigawa J. Gene expression profiling distinguishes a molecular signature for grade 1B mild acute cellular rejection in cardiac allograft recipients. J Heart Lung Transplant. 2007;26:1270–1280. doi: 10.1016/j.healun.2007.09.017. [DOI] [PubMed] [Google Scholar]
- Perez-Flores I, Sanchez-Fructuoso A, Santiago JL, Fernandez-Arquero M, Calvo N, de la Concha EG, Barrientos A. Intracellular ATP levels in CD4+ lymphocytes are a risk marker of rejection and infection in renal graft recipients. Transplant Proc. 2009;41:2106–2108. doi: 10.1016/j.transproceed.2009.06.136. [DOI] [PubMed] [Google Scholar]
- Reinsmoen NL, Cornett KM, Kloehn R, Burnette AD, McHugh L, Flewellen BK, Matas A, Savik K. Pretransplant donor-specific and non-specific immune parameters associated with early acute rejection. Transplantation. 2008;85:462–470. doi: 10.1097/TP.0b013e3181612ead. [DOI] [PubMed] [Google Scholar]
- Sanchez-Velasco P, Rodrigo E, Valero R, Ruiz JC, Fernandez-Fresnedo G, Lopez-Hoyos M, Pinera C, Palomar R, Leyva-Cobian F, Arias M. Intracellular ATP concentrations of CD4 cells in kidney transplant patients with and without infection. Clin Transplant. 2008;22:55–60. doi: 10.1111/j.1399-0012.2007.00744.x. [DOI] [PubMed] [Google Scholar]
- Cabrera R, Ararat M, Soldevila-Pico C, Dixon L, Pan JJ, Firpi R, Machicao V, Levy C, Nelson D, Morelli G. Using an immune functional assay to differentiate acute cellular rejection from recurrent hepatitis C in liver transplant patients. Liver Transpl. 2009;15:216–222. doi: 10.1002/lt.21666. [DOI] [PubMed] [Google Scholar]
- Rossano JW, Denfield SW, Kim JJ, Price JF, Jefferies JL, Decker JA, Smith EO, Clunie SK, Towbin JA, Dreyer WJ. Assessment of the Cylex ImmuKnow cell function assay in pediatric heart transplant patients. J Heart Lung Transplant. 2009;28:26–31. doi: 10.1016/j.healun.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Bhorade SM, Janata K, Vigneswaran WT, Alex CG, Garrity ER. Cylex ImmuKnow assay levels are lower in lung transplant recipients with infection. J Heart Lung Transplant. 2008;27:990–994. doi: 10.1016/j.healun.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Cadillo-Chavez R, de Echegaray S, Santiago-Delpin EA, Rodriguez-Trinidad AT, Camacho-Carrazo B, Alfaro T, Saavedra-Pozo M, Carrasquillo L, Gonzalez-Caraballo ZA, Morales-Otero LA. Assessing the risk of infection and rejection in Hispanic renal transplant recipients by means of an adenosine triphosphate release assay. Transplant Proc. 2006;38:918–920. doi: 10.1016/j.transproceed.2006.02.051. [DOI] [PubMed] [Google Scholar]