Abstract
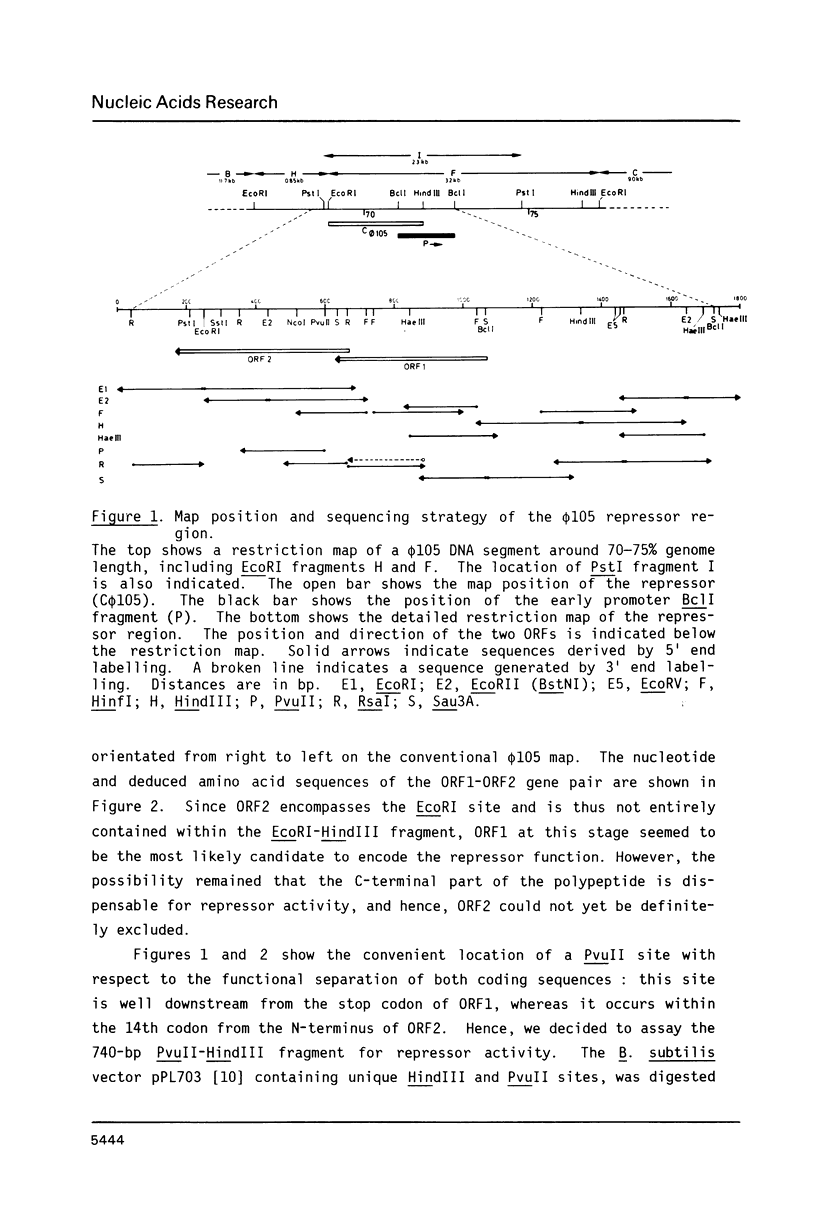
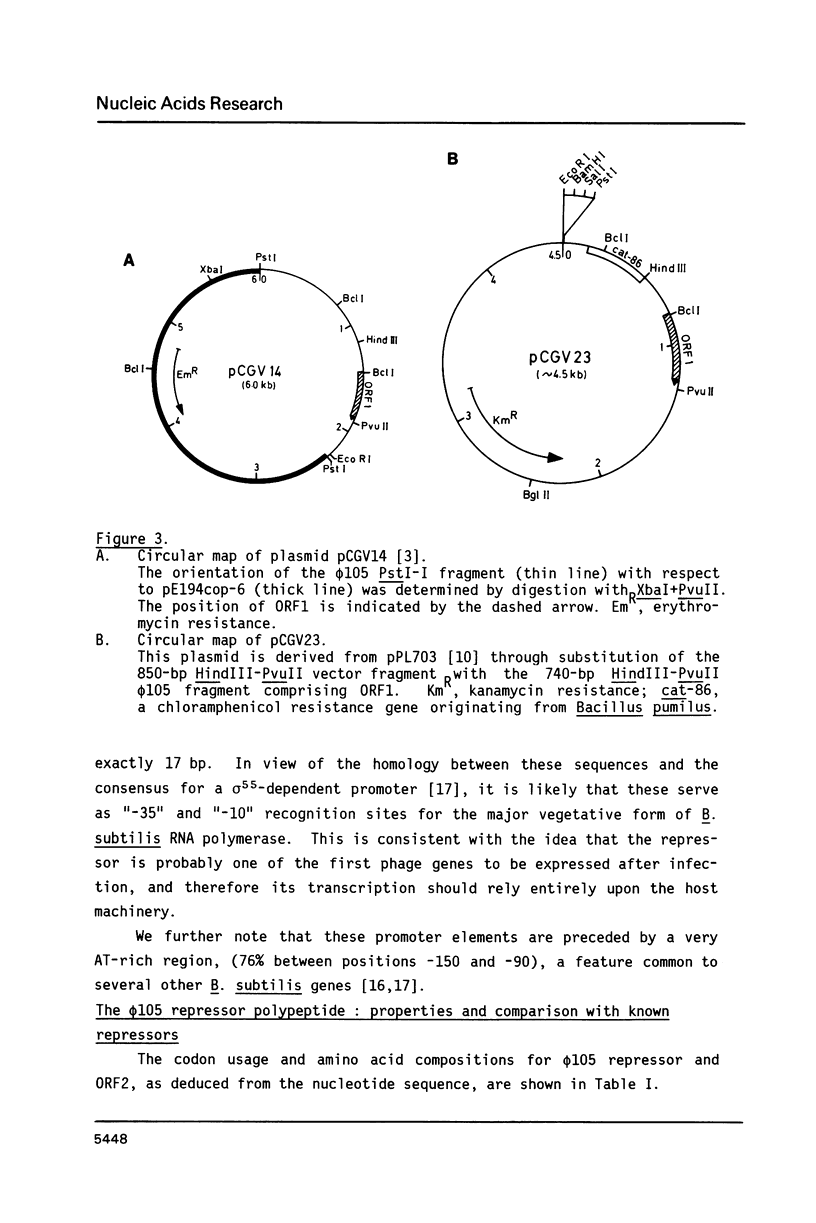
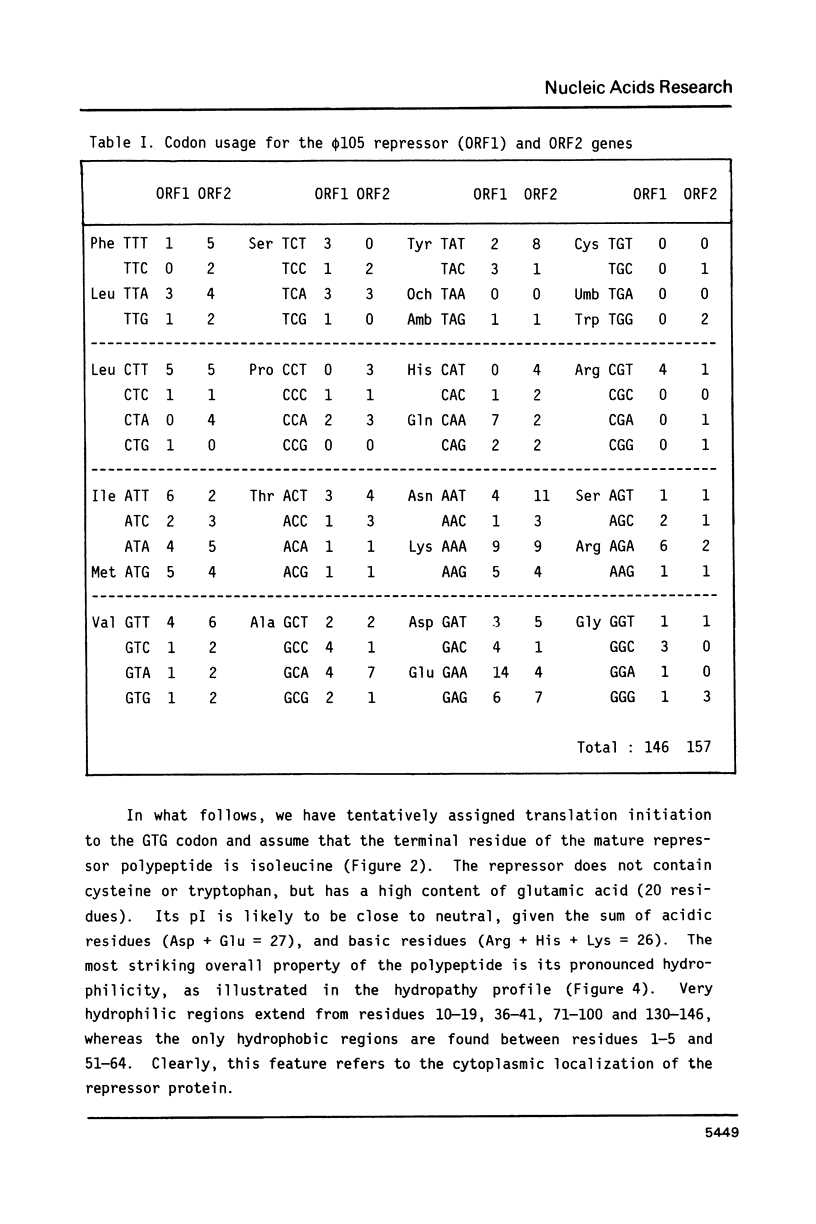
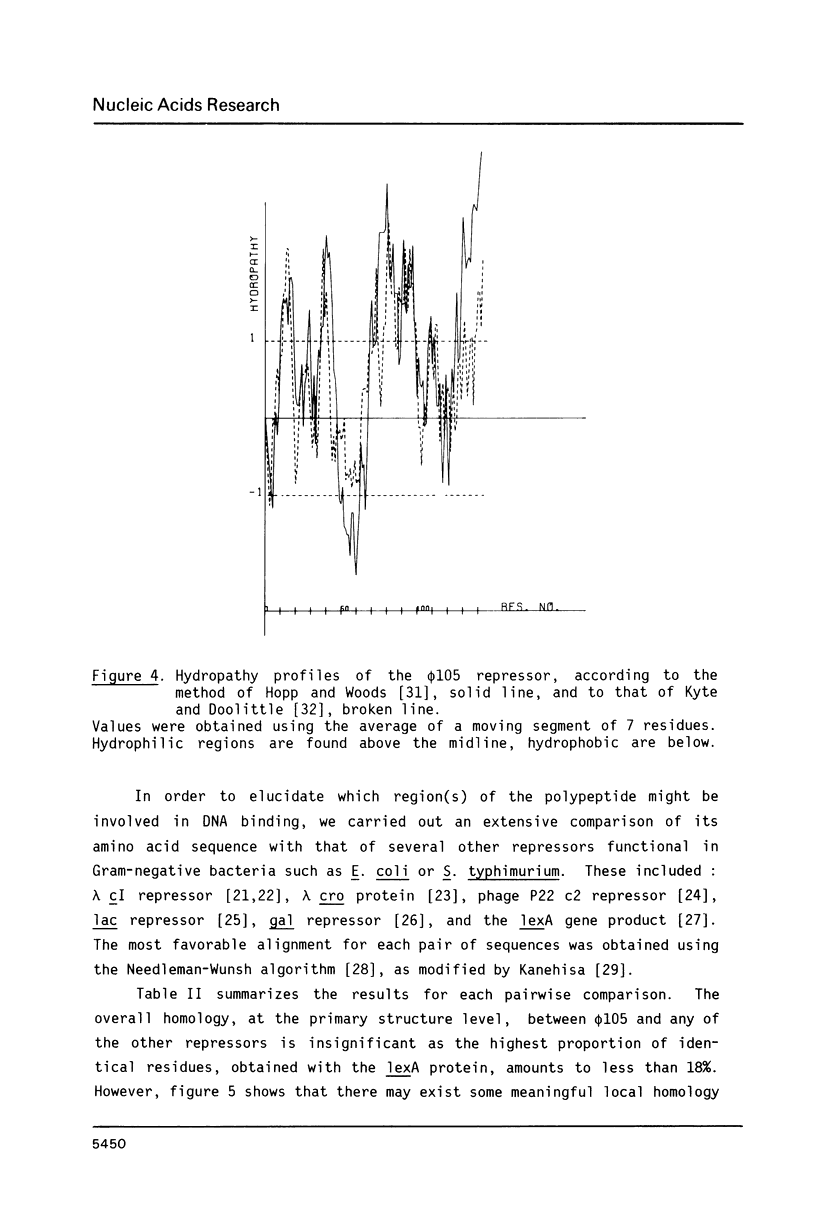
We have identified and sequenced a bacteriophage phi 105 gene encoding an immunity repressor, the first to be characterized from a temperate phage infecting a Gram-positive host. Using superinfection immunity as an assay for repressor function, the phi 105 repressor gene was located within a 740-bp PvuII-HindIII subfragment near the left end of the phi 105 EcoRI-F fragment. We show that the repressor is specified by the 5'-proximal coding sequence of a translationally overlapping gene pair, transcribed from right to left on the conventional phi 105 map. Comparison of its amino acid sequence (146 residues) with that of a large number of Gram-negative bacterial and phage repressors revealed a putative DNA-binding region between positions 20 and 39. The coding region is preceded by a strong Shine-Dalgarno sequence 5' AAAGGAG 3'. Deletion analysis of the 5'-flanking DNA allowed to identify transcriptional control elements. Their structure, 5' TTGTAT 3' at -35 and 5' TATAAT 3' at -10, strongly suggests that the phi 105 repressor gene is transcribed by the major vegetative form of B. subtilis RNA polymerase, as would be expected for an early phage gene.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bugaichuk U. D., Deadman M., Errington J., Savva D. Restriction enzyme analysis of Bacillus subtilis bacteriophage phi 105 DNA. J Gen Microbiol. 1984 Aug;130(8):2165–2167. doi: 10.1099/00221287-130-8-2165. [DOI] [PubMed] [Google Scholar]
- Cully D. F., Garro A. J. Expression of superinfection immunity to bacteriophage phi 105 by Bacillus subtilis cells carrying a plasmic chimera of pUB110 and EcoRI fragment F of phi 105 DNA. J Virol. 1980 Jun;34(3):789–791. doi: 10.1128/jvi.34.3.789-791.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaese P., Hussey C., Van Montagu M. Thermo-inducible gene expression in Bacillus subtilis using transcriptional regulatory elements from temperate phage phi 105. Gene. 1984 Dec;32(1-2):181–194. doi: 10.1016/0378-1119(84)90046-5. [DOI] [PubMed] [Google Scholar]
- Doi R. H. Genetic engineering in Bacillus subtilis. Biotechnol Genet Eng Rev. 1984;2:121–155. doi: 10.1080/02648725.1984.10647797. [DOI] [PubMed] [Google Scholar]
- Flock J. I. Deletion mutants of temperate Bacillus subtilis bacteriophage phi105. Mol Gen Genet. 1977 Oct 24;155(3):241–247. doi: 10.1007/BF00272803. [DOI] [PubMed] [Google Scholar]
- Hopp T. P., Woods K. R. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii T., Ogawa T., Ogawa H. Nucleotide sequence of the lexA gene of E. coli. Cell. 1981 Mar;23(3):689–697. doi: 10.1016/0092-8674(81)90432-3. [DOI] [PubMed] [Google Scholar]
- Hsiang M. W., Cole R. D., Takeda Y., Echols H. Amino acid sequence of Cro regulatory protein of bacteriophage lambda. Nature. 1977 Nov 17;270(5634):275–277. doi: 10.1038/270275a0. [DOI] [PubMed] [Google Scholar]
- Kanehisa M. I. Los Alamos sequence analysis package for nucleic acids and proteins. Nucleic Acids Res. 1982 Jan 11;10(1):183–196. doi: 10.1093/nar/10.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastelein R. A., Remaut E., Fiers W., van Duin J. Lysis gene expression of RNA phage MS2 depends on a frameshift during translation of the overlapping coat protein gene. Nature. 1982 Jan 7;295(5844):35–41. doi: 10.1038/295035a0. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Legerski R. J., Hodnett J. L., Gray H. B., Jr Extracellular nucleases of pseudomonas BAL 31. III. Use of the double-strand deoxyriboexonuclease activity as the basis of a convenient method for the mapping of fragments of DNA produced by cleavage with restriction enzymes. Nucleic Acids Res. 1978 May;5(5):1445–1464. doi: 10.1093/nar/5.5.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mongkolsuk S., Chiang Y. W., Reynolds R. B., Lovett P. S. Restriction fragments that exert promoter activity during postexponential growth of Bacillus subtilis. J Bacteriol. 1983 Sep;155(3):1399–1406. doi: 10.1128/jb.155.3.1399-1406.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., LeGrice S. F., Lee G., Stephens M., Sonenshein A. L., Pero J., Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186(3):339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Murray C. L., Rabinowitz J. C. Nucleotide sequences of transcription and translation initiation regions in Bacillus phage phi 29 early genes. J Biol Chem. 1982 Jan 25;257(2):1053–1062. [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Normark S., Bergström S., Edlund T., Grundström T., Jaurin B., Lindberg F. P., Olsson O. Overlapping genes. Annu Rev Genet. 1983;17:499–525. doi: 10.1146/annurev.ge.17.120183.002435. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Poteete A. R., Ptashne M. Control of transcription by the bacteriophage P22 repressor. J Mol Biol. 1982 May 5;157(1):21–48. doi: 10.1016/0022-2836(82)90511-3. [DOI] [PubMed] [Google Scholar]
- Ptashne M., Backman K., Humayun M. Z., Jeffrey A., Maurer R., Meyer B., Sauer R. T. Autoregulation and function of a repressor in bacteriophage lambda. Science. 1976 Oct 8;194(4261):156–161. doi: 10.1126/science.959843. [DOI] [PubMed] [Google Scholar]
- Reeve J. N. Bacteriophage infection of minicells: a general method for identification of "in vivo" bacteriophage directed polypeptide biosynthesis. Mol Gen Genet. 1977 Dec 14;158(1):73–79. doi: 10.1007/BF00455121. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Anderegg R. Primary structure of the lambda repressor. Biochemistry. 1978 Mar 21;17(6):1092–1100. doi: 10.1021/bi00599a024. [DOI] [PubMed] [Google Scholar]
- Sauer R. T. DNA sequence of the bacteriophage gama cI gene. Nature. 1978 Nov 16;276(5685):301–302. doi: 10.1038/276301a0. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Pan J., Hopper P., Hehir K., Brown J., Poteete A. R. Primary structure of the phage P22 repressor and its gene c2. Biochemistry. 1981 Jun 9;20(12):3591–3598. doi: 10.1021/bi00515a044. [DOI] [PubMed] [Google Scholar]
- Uhlén M., Flock J. I., Philipson L. RecE independent deletions of recombinant plasmids in Bacillus subtilis. Plasmid. 1981 Mar;5(2):161–169. doi: 10.1016/0147-619x(81)90017-2. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Weisblum B., Graham M. Y., Gryczan T., Dubnau D. Plasmid copy number control: isolation and characterization of high-copy-number mutants of plasmid pE194. J Bacteriol. 1979 Jan;137(1):635–643. doi: 10.1128/jb.137.1.635-643.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wilcken-Bergmann B., Müller-Hill B. Sequence of galR gene indicates a common evolutionary origin of lac and gal repressor in Escherichia coli. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2427–2431. doi: 10.1073/pnas.79.8.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]



