Abstract
Introduction
The C allele of the nonsynonymous Arg265His (rs3733591) variant of SLC2A9 confers risk for gout in Han Chinese, Solomon Island and Japanese samples, with a stronger role in tophaceous gout. There is no evidence for an association with gout in Caucasian populations. In the present study, we tested rs3733591 for association with gout in New Zealand (NZ) Māori, Pacific Island and Caucasian samples.
Methods
Rs3733591 was genotyped across gout patients (n = 229, 232 and 327 NZ Māori, Pacific Island and Caucasian samples, respectively) and non-gout controls (n = 343, 174 and 638 Māori, Pacific Island and Caucasian samples, respectively). Further Caucasian sample sets consisting of 67 cases and 4,712 controls as well as 153 cases and 6,969 controls were obtained from the Framingham Heart Study and the Atherosclerosis Risk in Communities study, respectively. The Polynesian samples were analyzed according to Eastern and Western Polynesian ancestry.
Results
No evidence for risk conferred by the C allele of rs3733591 with gout was found in the sample sets of NZ Māori (odd ratio (OR) = 0.98, P = 0.86), Eastern Polynesians (OR = 0.99, P = 0.92), Western Polynesians (OR = 1.16, P = 0.36) or combined Caucasians (OR = 1.15, P = 0.13). The C allele was significantly overrepresented in Māori tophaceous cases compared to cases without tophi (OR = 2.21, P = 0.008), but not in the other ancestral groupings.
Conclusions
Noting that our study's power was limited for detecting weak genetic effects, we were unable to replicate associations of rs3733591 with gout in Eastern Polynesian, Western Polynesian and Caucasian samples. However, consistent with a previous study of Han Chinese and Solomon Island populations, our data suggest that rs3733591 could be a marker of severe gout in some populations. Our results also suggest that the effect of this variant is population-specific, further confirming population heterogeneity regarding the association of SLC2A9 with gout.
Introduction
Gout is a common form of inflammatory arthritis predominantly affecting men, with hyperuricemia being an essential predeterminant. As urate concentrations reach saturation in the blood, monosodium urate (MSU) crystals are deposited in the joints and tissues. An acute self-limiting inflammatory reaction to these MSU crystals leads to severe pain and debilitation (gout). Without resolution, the MSU crystals and subsequent inflammation can lead to chronic tophaceous gout, bony erosions and permanent disability. In New Zealand (NZ; Aotearoa), gout is common in Māori and Pacific Island men, with the prevalence estimated to range from 9.3% to 13.9% and 14.9%, respectively [1,2]. Renal underexcretion of uric acid has been determined to be an underlying characteristic of gout and is more pronounced in people of Māori and Pacific Island descent, and even more so in patients with hyperuricemia and/or gout [3,4].
Genomewide association studies in Caucasian cohorts have shown that intronic variants (rs7442295 and surrogate marker rs11942223) within the solute carrier family 2, member 9/facilitated glucose transporter 9 (SLC2A9/GLUT9) gene are associated with high serum urate concentrations and gout [5-10]. The intronic SLC2A9 variant rs11942223 best explains the strong role that SLC2A9 played in the development of gout in NZ Māori, Pacific Island and Caucasian sample sets [11]. Interestingly, this variant is very rare in Chinese, Japanese and Solomon Island people and does not play a genetic role in the development of gout in these populations [12,13]. The intronic SLC2A9 variants have gender-specific effects on serum urate, with the effect being stronger in women [14].
SLC2A9 has been confirmed to be a renal urate transporter [9,15,16]. It is a urate reuptake molecule that has two isoforms, with the long-form being expressed on the basal side and the short-form being expressed on the apical side of the proximal renal tubule [17]. Mice with overexpression of the long-form of SLC2A9 (also known as hURATv1) on the basolateral surface have a much greater reuptake of urate from the lumen into the blood and reduced urinary urate excretion [15]. In contrast, overexpression of the URAT1 renal urate transporter does not enhance urate reabsorption, indicating that SLC2A9 is the rate-limiting step in urate reuptake in mice [17].
An additional SLC2A9 variant, R265H (rs3733591), contributes significantly to the development of elevated urate concentrations and gout in Han Chinese, Solomon Island and Japanese sample sets [12,13], but not in a Caucasian sample set [8]. Han Chinese and Solomon Island gout patients with the risk C allele had a higher risk for tophi [12]. There are currently no data indicating how R265H may influence SLC2A9 function. Importantly, the effect of R265H in these populations is independent of the previously gout-associated intronic rs7442295/rs11942223 variants. The measure of linkage disequilibrium, r2, is < 0.05 between rs3733591 and rs11942223 in HapMap Caucasian, Chinese and Japanese samples. Given the existing evidence for population heterogeneity in association of SLC2A9 variants (intronic and R265H) with gout [8,11-13], we investigated a possible role for R265H (rs3733591) in gout in NZ Māori, Pacific Island and Caucasian case-control sample sets in a study adequately powered to detect an effect equivalent to that observed in other populations (odds ratio (OR) > 1.4).
Material and methods
Study participants
Genotyping of NZ sample sets of Māori (229 cases and 343 controls), Pacific Island (232 cases and 174 controls) and Caucasian (327 cases and 638 controls) was done (Table 1). All cases were recruited from rheumatology outpatient clinics, and their gout diagnoses were confirmed by a rheumatologist according to the American College of Rheumatology preliminary diagnostic criteria for acute gout [18]. The controls had no history of arthritis and were recruited from the wider community. Our recruitment of gout patients was approved by the NZ Multi-region Ethics Committee (MREC 05/10/130), and the recruitment of the controls was approved by the Lower South and Multi-region Ethics Committees (OTA/99/11/098 and MREC 05/10/130). All participants provided written informed consent for the collection of samples and subsequent analysis.
Table 1.
Demographic and clinical characteristics of gout cases and controlsa
| Gout | Controls | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristics | Māorib | EPb | WP | NZ Caucasian | FHS | ARIC | Māori | NZ EP | NZ WP | NZ Caucasian | FHS | ARIC |
| Number of cases/controls | 229 | 261 | 187 | 327 | 67 | 153 | 343 | 348 | 142 | 638 | 47,12 | 6,969 |
| Males, % | 78 | 76 | 95 | 85 | 94 | 73 | 31 | 33 | 57 | 42 | 45 | 45 |
| Mean ± 1 SD grandparents of stated ancestry | 2.87 ± 1.15 | 3.10 ± 1.07 | 3.72 ± 0.59 | 4.00 ± 0.00 | - | - | 2.33 ± 1.20 | 2.46 ± 1.22 | 3.49 ± 0.86 | 4.00 ± 0.00 | - | - |
| Mean serum urate at recruitment, mmol/L (range) | 0.45 (0.21 to 0.98) |
0.47 (0.19 to 0.98) |
0.49 (0.17 to 0.72) |
0.41 (0.18 to 1.07) |
0.42 (0.21 to 0.58) |
0.42 (0.11-0.70) |
- | - | - | - | 0.29 (0.07 to 0.63) |
0.34 (0.03 to 0.72) |
| Confirmed tophi, % | 38 | 39 | 56 | 30 | - | - | - | - | - | - | - | - |
| Mean age at onset, years (range) | 39 (15 to 74) |
39 (15 to 74) |
34 (15 to 75) |
47 (15 to 83) |
- | - | - | - | - | - | - | - |
| Mean age at recruitment, years (range) | - | - | - | - | 38 (18 to 55) |
54 (45-65) |
44 (17 to 80) |
44 (17 to 85) |
38 (17 to 86) |
52 (17 to 95) |
37 (5 to 72) |
54 (44 to 66) |
| Mean gout attacks in past year, n (range) | 11 (0 to > 1/week) |
11 (0 to > 1/week) |
16 (0 to > 1/week) |
7.5 (0 to > 1/week) |
- | - | - | - | - | - | - | - |
| First-degree relative with gout, % | 63 | 64 | 53 | 42 | - | - | 30 | 31 | 23 | 14 | - | - |
| Allopurinol treatment, % | 83 | 82 | 87 | 78 | - | - | - | - | - | - | - | - |
| Probenecid treatment, % | 6 | 6 | 13 | 6 | - | - | - | - | - | - | - | - |
| Mean BMI (range) | 35 (22 to 62) |
35 (22 to 66) |
38 (22 to 93) |
30 (19 to 62) |
29 (20 to 37) |
28 (21-41) |
32 (20 to 77) |
32 (20 to 77) |
35 (21 to 63) |
28 (19 to 56) | 27 (15 to 61) |
27 (15 to 56) |
| Type 2 DM, % | 28 | 30 | 18 | 13 | 15 | 7 | 10 | 9 | 11 | 7 | 4 | 7 |
| Hypertension, % | 63 | 63 | 46 | 49 | - | - | 16 | 16 | 17 | 14 | - | - |
| Dyslipidemia, % | 51 | 52 | 53 | 48 | - | - | 12 | 12 | 11 | 16 | - | - |
| Cardiovascular disease, % | 40 | 38 | 20 | 37 | - | - | 3 | 3 | 1 | 6 | - | - |
| Renal disease, % | 30 | 30 | 22 | 22 | 21 | - | 2 | 2 | 1 | 2 | 6 | - |
aARIC = Atherosclerosis Risk in Communities study; BMI = body mass index; DM = diabetes mellitus; EP = Eastern Polynesian; FHS = Framingham Heart Study; NZ = New Zealand; SD = standard deviation; WP = Western Polynesian. bFull clinical data were available in 76% of Māori and 63% of EP gout cases (> 90% in other sample sets).
Given data from previous work investigating the ABCG2 rs2231142 variant in the NZ sample sets [19], along with knowledge of ancestral Polynesian and Māori migration [20-22], the groups analyzed were Māori and Eastern Polynesian (Māori and Cook Island), Western Polynesian (Tonga, Samoa, Niue and Tokelau) and Caucasian. A total of 223 cases and 327 controls overlapped between the Māori and Eastern Polynesian sample sets. People of mixed Eastern and Western Polynesian ancestry were excluded from the Eastern and Western Polynesian sample sets.
Fifty-five gout cases were obtained from the Framingham Heart Study (FHS) Offspring data set and combined with 17 gout cases from the Generation 3 data. Self-reported gout cases from the Offspring data set were included if the participants reported having gout on two or more survey occasions or reported having gout on one survey occasion and were also taking antigout medication. Self-reported gout cases from the Generation 3 data set were included if the participants had also answered "no" to taking medication for hypertension or high blood pressure. Control participants included those who were Caucasian and were unrelated to the gout patients. Genotypes were available for 67 gout cases and 4,712 control samples. One hundred fifty-three self-reported Caucasian gout patients who were not taking hypertensive medication were obtained from the Atherosclerosis Risk in Communities (ARIC) study and were compared to 6,969 unrelated controls.
Genotyping
The rs3733591 variant of SLC2A9 was genotyped across the NZ Caucasian, Māori and Pacific Island sample sets using the TaqMan Allelic Discrimination Assay Kit (probe ID C__25803684_10; Applied Biosystems, Foster City, CA, USA) and a LightCycler 480 Real-Time Polymerase Chain Reaction System (Roche, Indianapolis, IN, USA).
Statistical analysis
A priori calculations were performed to test the power of the NZ Polynesian sample sets to detect association of rs3733591 with gout on the basis of previous data [12,13]. Statistical power in the Māori, Eastern Polynesian and Western Polynesian sample sets was 79%, 82% and 58%, respectively (OR = 1.41, minor allele frequency = 0.346).
Allelic and genotypic frequencies were compared between case and control samples, and ORs and adherence to the Hardy-Weinberg equilibrium were calculated using the SHEsis package [23]. The genotype frequencies for rs3733591 were in Hardy-Weinberg equilibrium (P > 0.01) for all case and control sample sets.
Twenty-five biallelic markers were used as genomic controls to account for differing levels of non-Māori and non-Eastern Polynesian and non-Western Polynesian ancestry between the case and control samples. The stratification markers used were as follows: rs2075876 (AIRE), rs1816532 (ERBB4), rs13419122 (GFPT1), rs12401573 (SEMA4A), rs6945435 (MGC87315), rs743777 (IL2RB), rs10511216 (Intergenic), rs12745968 (FAM69A), rs1539438 (AP4B1), rs729749 (NCF4), rs3738919 (ITGAV), rs1130214 (AKT1), rs755622 (MIF), rs7901695 (TCF7L2), rs7578597 (THADA), rs2043211 (CARD8), rs10733113 (NLRP3), rs900865 (SOX6), rs2059606 (PGDS), rs4129148 (PseudoY), rs831628 (CD59), rs7725 (GFPT), rs573816 (upstream of ALDOB), rs1929480 (ALDOB) and rs12917707 (UMOD). There was an average allele frequency difference of 0.22 (0.03 to 0.61) between a subset of 469 Māori cases and controls and 505 Caucasian controls and a difference of 0.22 (allele frequency range 0.03 to 0.59) and 0.29 (0.04 to 0.67) between subsets of 417 Eastern Polynesian and 215 Western Polynesian cases and controls and 505 Caucasian controls, respectively. The genotype frequencies for the stratification markers all exhibited Hardy-Weinberg equilibrium P values > 0.003 for all case and control sample sets. STRUCTURE [24] was used to assign Māori, Eastern Polynesian and Western Polynesian individuals into non-Caucasian populations (parameters: number of populations was assumed to be two, 30,000 burn-in period and 1 million Markov chain Monte Carlo replications after burn-in). The 505 Caucasian control individuals were included in the STRUCTURE procedure to aid in population clustering as being representative of the ancestral Caucasian population. After running STRUCTURE on the Māori samples, the proportions of samples in the inferred Caucasian cluster were 0.95 for the 505 Caucasian controls and 0.06 for the total 572 Māori samples, 0.95 and 0.06 for the 608 Eastern Polynesian samples and 0.98 and 0.05 for the 330 Western Polynesian samples. The STRUCTURE output was used to run STRAT [24] to test for association (PSTRAT) of the variant with disease in the presence of admixture. The phenotype of the 505 Caucasian individuals was set as unknown.
Gender and gender-genotype interaction analysis was performed using Stata software (StataCorp, College Station, TX, USA). Meta-analysis was performed to combine data from independent data sets using rmeta software (http://cran.r-project.org/web/packages/rmeta/index.html) (within Stata) to calculate the combined Mantel-Haenszel OR using a fixed effects model and the Breslow-Day test for heterogeneity between studies. Imputation of rs3733591 genotypes in the FHS and ARIC samples was done with IMPUTE2 using HapMap3 CEU (NCBI Build 36, db126b) as reference data and a quality threshold of 0.9.
Results
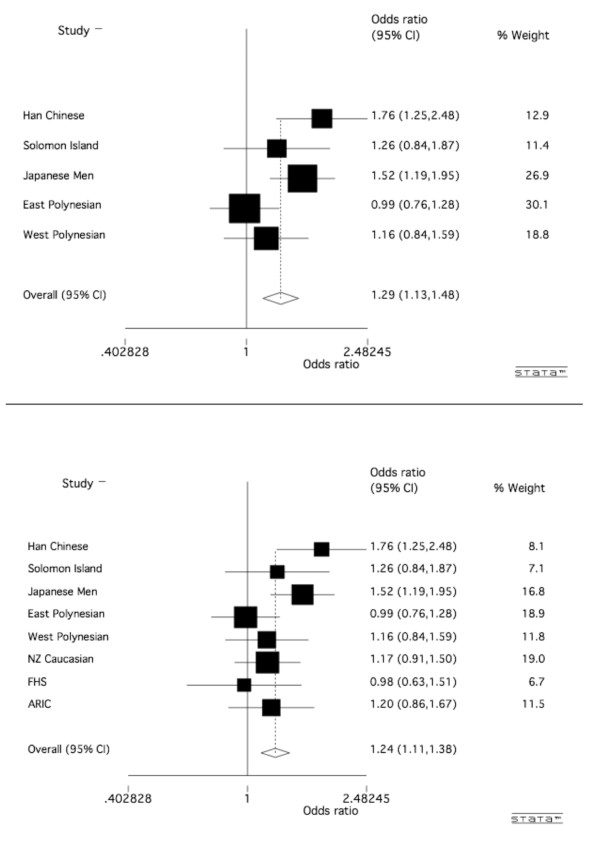
The demographic and clinical characteristics of study participants are presented in Table 1 and genotype and allele distributions of the rs3733591 variant are shown in Table 2. There was no evidence for an association of the risk C allele of rs3733591 with gout in any of the Māori, Eastern Polynesian, Western Polynesian or Caucasian analyses (OR = 0.98, PSTRAT = 0.93; OR = 0.99, PSTRAT = 0.80; OR = 1.16, PSTRAT = 0.65; and OR = 1.15, PMeta = 0.13, PBreslow-Day = 0.84, respectively). The r2 values (measure of linkage disequilibrium) between the previously associated SLC2A9 variant, rs11942223 [11], and the variant tested here, rs3733591, were 0.04, 0.03, 0.04 and 0.05 for the Māori, Eastern Polynesian, Western Polynesian and Caucasian sample sets, respectively. Similarly, there was no evidence for an association on the basis of meta-analysis of the Eastern and Western Polynesian sample sets (OR = 1.05 (95% CI: 0.86 to 1.29); P = 0.62 and PBreslow-Day = 0.44, respectively). Meta-analysis of data from Polynesian, Han Chinese, Solomon Islands and Japanese sample sets (Table 2) [12,13] indicated heterogeneity (PBreslow-Day = 0.05); however, there was strong evidence for an association of rs3733591 in these combined Asian-Pacific populations (Figure 1) (OR = 1.29 (95% CI: 1.13 to 1.48, P = 1.6 × 10-4). Addition of Caucasian data to the meta-analysis weakened the overall effect; however, a strong association was maintained (Figure 1) (OR = 1.24 (95% CI: 1.13 to 1.48); PBreslow-Day = 0.13 and P = 1.1 × 10-4).
Table 2.
Association analysis of rs3733591 with gout in NZ Māori, EP, WP and Caucasian sample sets and in the FHS cohorta
| Case genotypesb, n (OR) | Control genotypes, n (OR) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample set | CC | CT | TT | C-allele frequency | CC | CT | TT | C-allele frequency | Genotypic P | Allelic P | P STRAT | Allelic OR, C allele (95% CI) |
| Māori | 101 (0.500) | 83 (0.411) | 18 (0.089) | 0.705 | 164 (0.500) | 138 (0.421) | 26 (0.079) | 0.710 | 0.92 | 0.86 | 0.93 | 0.98 (0.74 to 1.28) |
| EP | 119 (0.513) | 92 (0.397) | 21 (0.091) | 0.711 | 169 (0.506) | 139 (0.416) | 26 (0.078) | 0.714 | 0.82 | 0.92 | 0.80 | 0.99 (0.76 to 1.28) |
| WP | 39 (0.227) | 88 (0.512) | 45 (0.262) | 0.483 | 33 (0.239) | 57 (0.413) | 48 (0.348) | 0.446 | 0.17 | 0.36 | 0.65 | 1.16 (0.84 to 1.59) |
| NZ Caucasian | 217 (0.693) | 87 (0.278) | 9 (0.029) | 0.832 | 417 (0.656) | 196 (0.308) | 23 (0.036) | 0.810 | 0.49 | 0.23 | - | 1.17 (0.91 to 1.50) |
| FHS | 44 (0.657) | 21 (0.313) | 2 (0.030) | 0.813 | 3158 (0.670) | 1,385 (0.294) | 169 (0.036) | 0.817 | 0.92 | 0.91 | - | 0.98 (0.63 to 1.51) |
| ARIC | 117 (0.765) | 30 (0.196) | 6 (0.039) | 0.863 | 4902 (0.704) | 1,898 (0.272) | 168 (0.024) | 0.840 | 0.07 | 0.28 | - | 1.20 (0.86 to 1.67) |
aARIC = Atherosclerosis Risk in Communities study; CI = confidence interval; FHS = Framingham Heart Study; OR = odds ratio; EP = Eastern Polynesian; NZ = New Zealand; WP = Western Polynesian PSTRAT = STRAT test for association. bThere was successful genotyping of 202 Māori cases (88%) and 328 Māori controls (96%), 232 EP cases (89%) and 334 EP controls (96%), 172 WP cases (92%) and 138 WP controls (97%) and 313 NZ Caucasian cases (96%) and 636 NZ Caucasian controls (100%).
Figure 1.
Meta-analysis of association of rs3733591 with gout in Asian-Pacific ancestral groups without (top) and with (bottom) New Zealand (NZ) Caucasians. ARIC = Atherosclerosis Risk in Communities study; CI = confidence interval; FHS = Framingham Heart Study.
A stronger effect of R265H with tophaceous gout has been reported [12]; therefore, we tested whether the effect of this variant is stronger in patients with tophi than in patients without tophi. We found significant overrepresentation of the C allele of rs3733591 in the group with tophaceous gout in the Māori analysis (OR = 2.21, PSTRAT = 0.01), but not in the analyses of Eastern Polynesian (OR = 1.53, PSTRAT = 0.17), Western Polynesian (OR = 0.97, PSTRAT = 0.54) or NZ Caucasian (OR = 0.80, P = 0.33) population samples (Table 3).
Table 3.
Rs3733591 and tophaceous gout in the Māori, EP, WP and NZ Caucasian casesa
| Tophaceous cases, n (OR) | Nontophaceous cases, n (OR) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample set | CC | CT | TT | C-allele frequency | CC | CT | TT | C-allele frequency | Genotypic P | Allelic P | P STRAT | Allelic OR, C allele (95% CI) |
| Māori | 36 (0.692) | 13 (0.250) | 3 (0.058) | 0.817 | 37 (0.446) | 37 (0.446) | 9 (0.108) | 0.669 | 0.02 | 0.008 | 0.01 | 2.21 (1.22 to 4.01) |
| EP | 43 (0.652) | 17 (0.258) | 6 (0.091) | 0.780 | 48 (0.490) | 41 (0.418) | 9 (0.092) | 0.699 | 0.09 | 0.10 | 0.17 | 1.53 (0.92 to 2.55) |
| WP | 22 (0.234) | 46 (0.489) | 26 (0.277) | 0.479 | 16 (0.213) | 41 (0.547) | 18 (0.240) | 0.487 | 0.76 | 0.88 | 0.54 | 0.97 (0.63 to 1.48) |
| NZ Caucasian | 63 (0.656) | 30 (0.313) | 3 (0.031) | 0.812 | 158 (0.715) | 57 (0.258) | 6 (0.027) | 0.844 | 0.58 | 0.33 | - | 0.88 (0.51 to 1.24) |
aCI, confidence interval; OR, odds ratio; CI, confidence interval; EP, Eastern Polynesian; NZ, New Zealand; WP, Western Polynesian; PSTRAT = STRAT test for association.
Although the intronic SLC2A9 polymorphisms (rs7442295/rs11942223) have shown gender-specific effects on serum urate [14], there was no evidence for rs3733591 exerting a gender influence in the gout sample sets comprising NZ Māori (C/C, C/T, T/T genotypes in men and women were 78, 58 and 13 in men and 19, 18 and 3 in women; P = 0.78), Eastern Polynesians (C/C, C/T, T/T genotypes were 91, 61 and 15 in men and 24, 23 and 4 in women; P = 0.55) or NZ Caucasians (C/C, C/T, T/T genotypes in men and women were 187, 74 and 8 in men and 31, 13 and 1 in women; P = 0.95) (Western Polynesian samples were not stratified according to gender, as there were too few women (n = 6)). On the basis of logistic regression models, there was no evidence of an interaction between rs3733591 genotype and gender for the Māori, Eastern Polynesian or Caucasian sample sets (P = 0.77, P = 0.46 and P = 0.49, respectively). There was also no evidence for an association of rs3733591 with gout in the Māori, Eastern Polynesian or Caucasian sample sets when only males were analyzed (P = 0.59, P = 0.37 and P = 0.19, respectively).
Discussion
The R265H nonsynonymous SLC2A9 variant has previously been demonstrated to be associated with gout and tophaceous gout in Han Chinese and Solomon Islanders [12], and it is associated with the development of gout in Japanese males [13]. We found no evidence for an association of this variant with gout in Māori, Eastern Polynesian, Western Polynesian or Caucasian sample sets (Table 2). However, consistent with the data from Han Chinese and Solomon Island sample sets [12], we found that the C allele conferred an increased risk of tophaceous gout in the Māori samples (OR = 2.21, PSTRAT = 0.01), but not in the other sample sets (Table 3). At this stage, the simplest conclusion is that the C allele of rs3733591 has a weaker effect on gout per se in Caucasian and Polynesian populations than in the Asian and Melanesian populations studied thus far. However, it is very important to note that our sample sets had insufficient power to detect an association at OR < 1.4, although a priori power calculations indicated that they were sufficiently powered to detect an effect of OR > 1.4, which is equivalent to the risk observed in the sample sets described by Tu et al. [12] and Urano et al. [13]. Note that post priori power calculations for Eastern and Western Polynesian sample sets using the actual risk C-allele frequencies revealed similar power for OR = 1.4 (78% and 57%, respectively).
Despite not finding any evidence for an association of rs3733591 with gout in the Māori sample set (OR = 0.98), there was an effect conferred by the C allele when the Māori cases were stratified for the presence or absence of tophi (OR = 2.21). This supports the previous findings of Tu et al. [12], who concluded that rs3733591 might be a genetic checkpoint for tophaceous gout. Whether this is true will require further study in larger sample sets drawn from diverse ancestral groups. In the Asia-Pacific region, an effect for rs3733591 in tophaceous gout has been observed in the Han Chinese, Solomon Island and Polynesian NZ Māori populations, but not in Western Polynesian populations (Samoa, Tonga, Niue). Why this is the case is unclear; however, it is worth pointing out that unexpected heterogeneity in genetic association with gout in the Asia-Pacific region has also been documented at ABCG2 [19]. Furthermore, given that the C-allele frequency was similar between Māori and Caucasian sample sets (0.71 and 0.81, respectively, in controls), the disparate ORs for tophaceous gout are notable (OR = 2.21 and OR = 0.88, respectively, for tophaceous gout compared to nontophaceous gout). The reason for this is unclear; it could be a consequence of moderate sample size (power) or it could reflect differences in disease pathogenesis between the different population groups.
It is of interest that tophus formation is not consistently present in all patients with long-standing hyperuricemia and gout, suggesting that additional factors may regulate the development of these lesions. It is possible that genetic variation in SLC2A9 is one of these factors. SLC2A9 is expressed in the chondrocytes of human articular cartilage [25]. The minor allele of R265H (rs3733591, or one in linkage disequilibrium) may influence the activity of SLC2A9 in articular chondrocytes and increase the risk for deposition of MSU crystals and formation of tophi in joint structures. A further issue is that host factors other than hyperuricemia may contribute to the development of tophi; quantitative analysis of tophus histology indicates that, in contrast to the innate immune responses that are activated in acute gout, both innate and adaptive immunity are implicated in the development of the tophus [26]. Thus different immune responses to MSU crystals may lead to different manifestations of disease.
Interestingly, the C-allele risk variant of rs3733591 is the minor allele in control sample sets of Han Chinese (controls = 0.32) [12] Solomon Islanders (controls = 0.40) [12], Japanese (controls = 0.32) [13] and Western Polynesians (controls = 0.43), yet it is the major allele in sample sets taken from the FHS (controls = 0.82), NZ Caucasians (controls = 0.81) and Eastern Polynesians (controls = 0.71) (Table 2). There is a large shift in the minor allele frequency between Eastern and Western Polynesian sample sets, and such a change in allele frequency is consistent with that seen at another gout locus, ABCG2 (rs2231142) (the risk allele frequency is 0.29 in Western Polynesians, 0.13 in Caucasians and 0.10 in Eastern Polynesians) [19]. It can be observed that, as the frequency of the rs3733591 C allele increases, the risk conferred by rs3733591 for gout decreases, suggesting that population allele frequency is important at R265H to detect an effect on the risk of gout. It is also notable that the situation is reversed for the SLC2A9 intronic variants (rs11942223 and surrogates), with a strong association in Caucasian [5-7,9,10] and Polynesian populations (driven by protective haplotypes) [11], but not in Melanesian and Asian populations, studied thus far [12,13].
In Caucasians, in addition to R265H, there are other nonsynonymous genetic variants within SLC2A9, including A17T (rs6820230), G25R (rs2276961), V253I (rs16890979) and P321L (rs2280205) [7]. Only rs16890979 is associated with gout in Caucasians [7,11], possibly because of linkage disequilibrium with the more strongly associated intronic variants that include rs6855911 and rs11942223 [7,11]. Rs16890979 is also associated with gout in Polynesians; however, this is also likely to be secondary to an association with the intronic variants [11]. The minor allele of rs16890979 is rare in Han Chinese and Japanese [12,13]. Of the other variants, only P321L has been tested for association with gout in an Asia-Pacific region population (Japanese), in whom there was no evidence for association [13]. The impact of these nonsynonymous variants on the function of SLC2A9 has not been reported to date.
Given the heterogeneity evident in risk conferred for gout at SLC2A9 and ABCG2 in the Asian and Austronesian populations studied so far, further investigation of these genes in samples derived from Asian and Austronesian people is warranted. These populations have shared geographical history, although it is increasingly evident that considerable genetic heterogeneity exists within Asians and Austronesians, at least in gout and possibly in other complex diseases.
Conclusions
We were unable to replicate association of the rs3733591 variant with gout in NZ Eastern Polynesian, Western Polynesian or Caucasian sample sets. It is possible that rs3733591 has a weak effect in these populations; however, our study's power was limited to detecting an effect less than that previously reported (OR = 1.4). However, consistent with a previous study in Han Chinese and Solomon Islanders, our data suggest that rs3733591 could be a marker of severe gout in some populations.
Abbreviations
ABCG2: ATP-binding cassette-dependent transporter G2; ARIC: Atherosclerosis Risk in Communities; FHS: Framingham Heart Study; MSU: monosodium urate; NHLBI: National Heart, Lung and Blood Institute; NZ: New Zealand; OR: odds ratio; P: probability value; SHARe: SNP Health Association Resource; SLC2A9: solute carrier family 2, member 9; SNP: single-nucleotide polymorphism; URAT1: urate transporter 1.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JEHM and TRM helped to design the study, oversaw its execution and prepared the manuscript. PJG, AAH, JH, PBBJ, LKS and ND helped to provide clinical recruitment and prepared the manuscript. All authors read and approved the final manuscript.
Contributor Information
Jade E Hollis-Moffatt, Email: jade.hollis-moffatt@otago.ac.nz.
Peter J Gow, Email: PGow@Middlemore.co.nz.
Andrew A Harrison, Email: andrew.harrison@otago.ac.nz.
John Highton, Email: john.highton@otago.ac.nz.
Peter BB Jones, Email: p.jones@auckland.ac.nz.
Lisa K Stamp, Email: lisa.stamp@cdhb.govt.nz.
Nicola Dalbeth, Email: n.dalbeth@auckland.ac.nz.
Tony R Merriman, Email: tony.merriman@otago.ac.nz.
Acknowledgements
This work was supported by the Health Research Council of New Zealand and Arthritis New Zealand. JEHM was supported by a New Zealand National Heart Foundation Research Fellowship. Amanda Phipps-Green, Marilyn Merriman and Ruth Topless are thanked for expert technical work. Gael Hewett, Jill Drake, Roddi Laurence, Karen Lindsay, Maria Lobo, Karen Pui and Gabrielle Sexton are thanked for assistance in clinical recruitment. Mik Black is thanked for his assistance with statistical analysis. The Framingham Heart Study and the Framingham SHARe project are conducted and supported by the National Heart, Lung and Blood Institute (NHLBI) in collaboration with Boston University. The Framingham SHARe data used for the analyses described herein were obtained through dbGaP. This article was not prepared in collaboration with investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, Boston University or the NHLBI. The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by NHLBI contracts N01-HC-55015, N01 HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, N01-HC-55022, R01 HL087641, R01 HL59367 and R01 HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health (NIH) contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by grant UL1RR025005, a component of the NIH and the NIH Roadmap for Medical Research.
References
- Klemp P, Stansfield SA, Castle B, Robertson MC. Gout is on the increase in New Zealand. Ann Rheum Dis. 1997;56:22–26. doi: 10.1136/ard.56.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnard D, Kake T, Gow P, Barratt-Boyes C, Harris V, Hall DA, Mason H, Merriman T, Dalbeth N. Maaori Gout Action Group in Counties Manukau District Health Board. Debunking the myths to provide 21st century management of gout. N Z Med J. 2008;121:79–85. [PubMed] [Google Scholar]
- Simmonds HA, McBride MB, Hatfield PJ, Graham R, McCaskey J, Jackson M. Polynesian women are also at risk for hyperuricaemia and gout because of a genetic defect in renal urate handling. Br J Rheumatol. 1994;33:932–937. doi: 10.1093/rheumatology/33.10.932. [DOI] [PubMed] [Google Scholar]
- Gibson T, Waterworth R, Hatfield P, Robinson G, Bremner K. Hyperuricaemia, gout and kidney function in New Zealand Māori men. Br J Rheumatol. 1984;23:276–282. doi: 10.1093/rheumatology/23.4.276. [DOI] [PubMed] [Google Scholar]
- Yang Q, Köttgen A, Dehghan A, Smith AV, Glazer NL, Chen MH, Chasman DI, Aspelund T, Eiriksdottir G, Harris TB, Launer L, Nalls M, Hernandez D, Arking DE, Boerwinkle E, Grove ML, Li M, Linda Kao WH, Chonchol M, Haritunians T, Li G, Lumley T, Psaty BM, Shlipak M, Hwang SJ, Larson MG, O'Donnell CJ, Upadhyay A, van Duijn CM, Hofman A. et al. Multiple genetic loci influence serum urate levels and their relationship with gout and cardiovascular disease risk factors. Circ Cardiovasc Genet. 2010;3:523–530. doi: 10.1161/CIRCGENETICS.109.934455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring A, Gieger C, Mehta D, Gohlke H, Prokisch H, Coassin S, Fischer G, Henke K, Klopp N, Kronenberg F, Paulweber B, Pfeufer A, Rosskopf D, Völzke H, Illig T, Meitinger T, Wichmann HE, Meisinger C. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat Genet. 2008;40:430–436. doi: 10.1038/ng.107. [DOI] [PubMed] [Google Scholar]
- Li S, Sanna S, Maschio A, Busonero F, Usala G, Mulas A, Lai S, Dei M, Orrù M, Albai G, Bandinelli S, Schlessinger D, Lakatta E, Scuteri A, Najjar SS, Guralnik J, Naitza S, Crisponi L, Cao A, Abecasis G, Ferrucci L, Uda M, Chen WM, Nagaraja R. The GLUT9 gene is associated with serum uric acid levels in Sardinia and Chianti cohorts. PLoS Genet. 2007;3:e194. doi: 10.1371/journal.pgen.0030194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArdle PF, Parsa A, Chang YP, Weir MR, O'Connell JR, Mitchell BD, Shuldiner AR. Association of a common nonsynonymous variant in GLUT9 with serum uric acid levels in Old Order Amish. Arthritis Rheum. 2008;58:2874–2881. doi: 10.1002/art.23752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitart V, Rudan I, Hayward C, Gray NK, Floyd J, Palmer CN, Knott SA, Kolcic I, Polasek O, Graessler J, Wilson JF, Marinaki A, Riches PL, Shu X, Janicijevic B, Smolej-Narancic N, Gorgoni B, Morgan J, Campbell S, Biloglav Z, Barac-Lauc L, Pericic M, Klaric IM, Zgaga L, Skaric-Juric T, Wild SH, Richardson WA, Hohenstein P, Kimber CH, Tenesa A. et al. SLC2A9 is a newly identified urate transporter influencing serum urate concentration, urate excretion and gout. Nat Genet. 2008;40:437–442. doi: 10.1038/ng.106. [DOI] [PubMed] [Google Scholar]
- Wallace C, Newhouse SJ, Braund P, Zhang F, Tobin M, Falchi M, Ahmadi K, Dobson RJ, Marçano AC, Hajat C, Burton P, Deloukas P, Brown M, Connell JM, Dominiczak A, Lathrop GM, Webster J, Farrall M, Spector T, Samani NJ, Caulfield MJ, Munroe PB. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am J Hum Genet. 2008;82:139–149. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis-Moffatt JE, Xu X, Dalbeth N, Merriman ME, Topless R, Waddell C, Gow PJ, Harrison AA, Highton J, Jones PB, Stamp LK, Merriman TR. Role of the urate transporter SLC2A9 gene in susceptibility to gout in New Zealand Māori, Pacific Island, and Caucasian case-control sample sets. Arthritis Rheum. 2009;60:3485–3492. doi: 10.1002/art.24938. [DOI] [PubMed] [Google Scholar]
- Tu HP, Chen CJ, Tovosia S, Ko AM, Lee CH, Ou TT, Lin GT, Chang SJ, Chiang SL, Chiang HC, Chen PH, Wang SJ, Lai HM, Ko YC. Associations of a non-synonymous variant in SLC2A9 with gouty arthritis and uric acid levels in Han Chinese subjects and Solomon Islanders. Ann Rheum Dis. 2010;69:887–890. doi: 10.1136/ard.2009.113357. [DOI] [PubMed] [Google Scholar]
- Urano W, Taniguchi A, Anzai N, Inoue E, Sekita C, Endou H, Kamatani N, Yamanaka H. Association between GLUT9 and gout in Japanese men. Ann Rheum Dis. 2010;69:932–933. doi: 10.1136/ard.2009.111096. [DOI] [PubMed] [Google Scholar]
- Dehghan A, Köttgen A, Yang Q, Hwang SJ, Kao WL, Rivadeneira F, Boerwinkle E, Levy D, Hofman A, Astor BC, Benjamin EJ, van Duijn CM, Witteman JC, Coresh J, Fox CS. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 2008;372:1953–1961. doi: 10.1016/S0140-6736(08)61343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzai N, Ichida K, Jutabha P, Kimura T, Babu E, Jin CJ, Srivastava S, Kitamura K, Hisatome I, Endou H, Sakurai H. Plasma urate level is directly regulated by a voltage-driven urate efflux transporter URATv1 (SLC2A9) in humans. J Biol Chem. 2008;283:26834–26838. doi: 10.1074/jbc.C800156200. [DOI] [PubMed] [Google Scholar]
- Caulfield MJ, Munroe PB, O'Neill D, Witkowska K, Charchar FJ, Doblado M, Evans S, Eyheramendy S, Onipinla A, Howard P, Shaw-Hawkins S, Dobson RJ, Wallace C, Newhouse SJ, Brown M, Connell JM, Dominiczak A, Farrall M, Lathrop GM, Samani NJ, Kumari M, Marmot M, Brunner E, Chambers J, Elliott P, Kooner J, Laan M, Org E, Veldre G, Viigimaa M. et al. SLC2A9 is a high-capacity urate transporter in humans. PLoS Med. 2008;5:e197. doi: 10.1371/journal.pmed.0050197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzai N, Jutabha P, Kimura T, Fukutomi T. Urate transport: regulators of serum urate levels in humans. Curr Rheumatol Rev. 2011;7:123–131. doi: 10.2174/157339711795305059. [DOI] [Google Scholar]
- Wallace SL, Robinson H, Masi AT, Decker JL, McCarty DJ, Yu TF. Preliminary criteria for the classification of the acute arthritis of primary gout. Arthritis Rheum. 1977;20:895–900. doi: 10.1002/art.1780200320. [DOI] [PubMed] [Google Scholar]
- Phipps-Green AJ, Hollis-Moffatt JE, Dalbeth N, Merriman ME, Topless R, Gow PJ, Harrison AA, Highton J, Jones PB, Stamp LK, Merriman TR. A strong role for the ABCG2 gene in susceptibility to gout in New Zealand Pacific Island and Caucasian, but not Māori, case and control sample sets. Hum Mol Genet. 2010;19:4813–4819. doi: 10.1093/hmg/ddq412. [DOI] [PubMed] [Google Scholar]
- Friedlaender JS, Friedlaender FR, Reed FA, Kidd KK, Kidd JR, Chambers GK, Lea RA, Loo JH, Koki G, Hodgson JA, Merriwether DA, Weber JL. The genetic structure of Pacific Islanders. PLoS Genet. 2008;4:e19. doi: 10.1371/journal.pgen.0040019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Brauer S, Cordaux R, Casto A, Lao O, Zhivotovsky LA, Moyse-Faurie C, Rutledge RB, Schiefenhoevel W, Gil D, Lin AA, Underhill PA, Oefner PJ, Trent RJ, Stoneking M. Melanesian and Asian origins of Polynesians: mtDNA and Y chromosome gradients across the Pacific. Mol Biol Evol. 2006;23:2234–2244. doi: 10.1093/molbev/msl093. [DOI] [PubMed] [Google Scholar]
- Trent RJ, Mickleson KN, Yakas J, Hertzberg M. Population genetics of the globin genes in Polynesians. Hemoglobin. 1988;12:533–537. doi: 10.3109/03630268808991642. [DOI] [PubMed] [Google Scholar]
- Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- Software for Genetic Analysis. http://pritch.bsd.uchicago.edu/software.html
- Mobasheri A, Neama G, Bell S, Richardson S, Carter SD. Human articular chondrocytes express three facilitative glucose transporter isoforms: GLUT1, GLUT3 and GLUT9. Cell Biol Int. 2002;26:297–300. doi: 10.1006/cbir.2001.0850. [DOI] [PubMed] [Google Scholar]
- Dalbeth N, Pool B, Gamble GD, Smith T, Callon KE, McQueen FM, Cornish J. Cellular characterization of the gouty tophus: a quantitative analysis. Arthritis Rheum. 2010;62:1549–1556. doi: 10.1002/art.27356. [DOI] [PubMed] [Google Scholar]