Abstract
Background:
Germ-cell nuclear factor (GCNF, NR6AI) is an orphan nuclear receptor. Its expression pattern suggests it functions during embryogenesis, in the placenta and in germ-cell development. Mouse GCNF cDNA codes for a protein of 495 amino acids, whereas the four reported human cDNA variants code for proteins of 454 to 480 amino acids. Apart from this size difference, there is sequence conservation of up to 98.7%. To elucidate the genomic structure that gives rise to the different human GCNF mRNAs, the sequence information of the human GCNF locus is compared to the previously reported structure of the mouse locus.
Results:
The genomic structures of the mouse and human GCNF genes are highly conserved. The comparison reveals that the shorter human protein results from skipping the 45 base-pair third exon. Three different human isoforms - GCNF-1, GCNF-2a and GCNF-2b - are generated by differential usage of alternative splice acceptor sites of the fourth and the seventh exon.
Conclusion:
By homology with the mouse gene, 11 GCNF coding exons can be defined on human chromosome 9. All human GCNF cDNAs identified so far are, however, derived from mRNAs generated by splicing the fourth to the second exon. Although the genomic sequence is highly conserved, the analysis suggests that alternative splicing generates a higher complexity of human GCNF isoforms compared with the situation in the mouse.
Background
The nuclear receptors comprise a family of transcriptional regulators involved in a wide variety of biological processes such as embryonic development, differentiation and homeostasis [1, 2]. The family includes ligand-dependent zinc-finger transcription factors for steroid hormones, estrogens, thyroid hormones, retinoids, vitamin D and other hydrophobic molecules. In addition, several family members are 'orphan receptors' for which ligands have yet to be identified. Nuclear receptors have been assigned to six subfamilies on the basis of evolutionary studies [3]. As the first member of the sixth subfamily, GCNF is also known by its systematic name NR6A1 [4]. On the basis of homology and expression profile, the receptor has been given the alternative name of retinoic acid receptor-related testis-associated receptor (RTR) [5]. GCNF lacks known ligands and is therefore referred to as an orphan receptor. The gene has been mapped to chromosome 9q33-q34.1 [6]. Transfection experiments reveal that GCNF can act as a constitutive repressor when binding as a homodimer to promoters containing a direct repeat DNA element 5'-AGGTCAAGGTCA-3' (DRo) [7, 8, 9, 10]. Gene targeting in the mouse shows that GCNF has essential functions during embryogenesis [11]. The mouse receptor (mGCNF) is highly expressed in the developing embryonic nervous system and the labyrinthine layer of the placenta [12, 13]. In the adult, high transcript levels are restricted to the developing germ cells [5, 14, 15, 16]. Northern analysis reveals a transcript of 7.5 kilobases (kb) in somatic cells and an additional message of approximately 2.4 kb in male germ cells. This size difference is at least partially due to different polyadenylation sites [16], and it is therefore assumed that both transcripts code for identical proteins of 495 amino acids. The protein sequence is encoded by 11 exons [17]. When differentiation of P19 embryonal carcinoma cells is triggered by retinoic acid, the transcript and the protein are temporarily upregulated and then downregulated [18].
Isolation of a human cDNA coding for a protein (hGCNF) with an identity to the mouse protein of 98.7%, similar regulation in mouse P19 cells and in the human embryonal carcinoma cell line NT2/D1, together with the presence of two mRNAs of approximately 7.5 and 2.2 kb in human testis, suggested similar functions for mouse and human GCNF [18, 19, 20]. The cloning of human cDNAs that give rise to different hGCNF isoforms, however, suggests a higher complexity in humans. Currently, four different hGCNF cDNAs have been isolated that code for isoforms ranging in size from 454 to 480 amino acids [21].
We have investigated the genomic structure of mammalian GCNF to determine how the different GCNF isoforms are generated. Here we compare the exon/intron structure of the previously characterized mouse gene with the human ortholog [17]. Our study shows that alternative splicing generates at least three of the different GCNF isoforms.
Results and discussion
Structure of the first coding exon
To understand how the different human GCNF mRNA isoforms are generated, we have identified all human protein-coding exons. The alignment of the full-length human GCNF cDNA (GenBank accession number S83009) with the genome sequencing data at the NCBI localized the first protein-coding exon on chromosome-9-derived working draft sequence element NT_008491. The genomic sequence was aligned with the previously identified mouse exon 1 containing the putative translational start site. The comparison was extended up to position -100 with respect to the mouse cDNA reaching farthest in the furthest 5' direction. In addition, 100 base-pairs (bp) of the first identified intron were included (Figure 1). While the transcriptional start sites of GCNF are still elusive, the 5' ends of the first protein-coding exons cannot be defined. With respect to the sequence in Figure 1, the furthest 5'-reaching human cDNA (S83309) starts with nucleotide 171. The putative translational start codons are in positions 346 and 350 of the mouse and human sequences, respectively. These start codons are present in all full-length mammalian GCNF cDNAs characterized so far, suggesting a common amino terminus for the different GCNF isoforms. The first splice donor site is conserved. The comparison of the mouse 5'-untranslated sequence with the human genomic DNA reveals high conservation with identical sequence elements of up to 50 nucleotides. The presence of 18 CG dinucleotides conserved between human and mouse is suggestive of a regulatory function of the untranslated sequence. Five different human cDNAs with alternative 5' ends have, however, been reported to GenBank (S83309, U80802, AF004291, NM001489/U64876, X99975). A comparison with the genomic sequence (NT_008491) shows the sequence variation (Figure 2). Single-nucleotide polymorphism among cDNAs isolated from different human libraries may reflect variants in the human population. Two cDNAs (U64876/NM001489, X99975) differ in their untranslated region with respect to the genomic sequence. The genomic sequence shows no obvious splice signals in this region. Therefore, it cannot be ruled out that these cDNA ends may have been generated during the cloning process. In addition, one of the cloned cDNAs, coding for hGCNF-3 (AF004291), has a deletion in the coding region of the first exon, giving rise to an open reading frame of 454 amino acids. The 5' part of hGCNF-3 has been isolated by the polymerase chain reaction, suggesting that this deletion may have been generated during the synthesis. The isolation of additional cDNAs may give a clue as to which variants are true GCNF isoforms. The functional significance of the different isoforms is, at present, unknown but may lead to different transcriptional properties of GCNF isoforms.
Figure 1.
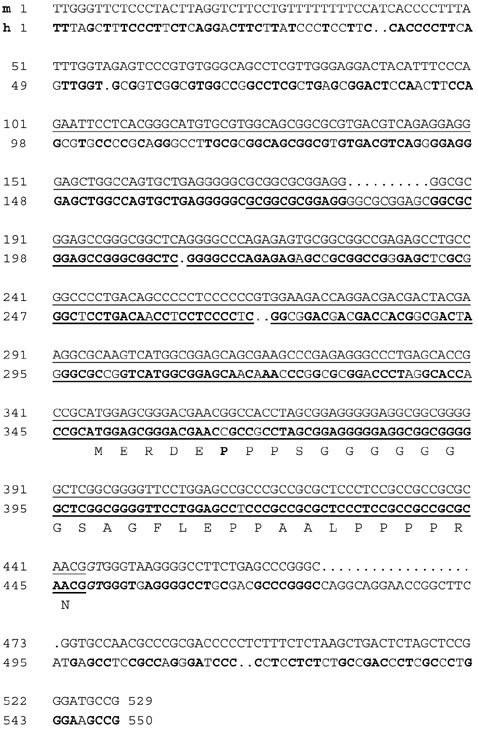
Sequence comparison of the first protein-coding exon of GCNF. The mouse-derived (m) DNA sequence (upper line, GenBank accession number AF254575) of the first protein-coding exon and flanking sequences are compared with the human-derived (h) sequence (lower line, S83309 for the coding, and NT_008491 for the flanking sequences). Identical nucleotides are highlighted in the human sequence by bold letters, gaps in the alignment are shown as dots. The deduced amino-acid sequence of the human protein is shown in the single-letter code. The bold P (proline) marks the position of an arginine in the mouse protein. Sequences represented in mouse and human cDNAs are underlined. The conserved GT-motif of the splice acceptor site is shown in italics.
Figure 2.
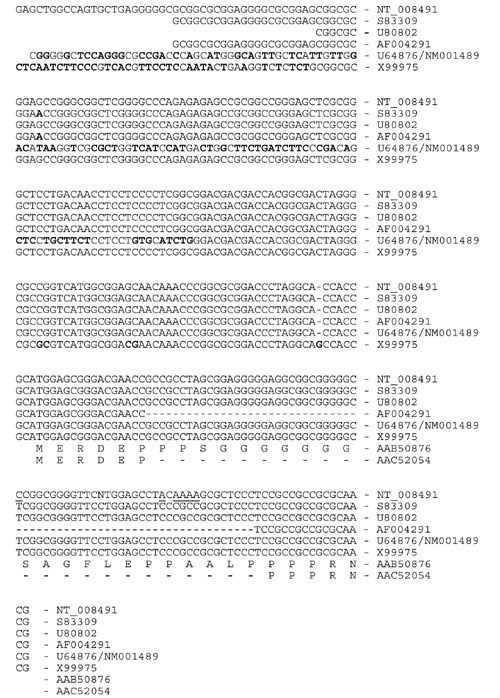
Sequence comparison of the 5' ends of human GCNF cDNAs coding for a full-length protein with the genomic DNA sequence. The genomic sequence NT_008491 is shown in the upper lines as indicated. Human cDNA sequences have been aligned omitting the cloning sites at the 5' ends. Accession numbers are shown. One cytosine and five adenosines in the genomic sequence not found in any of the cDNAs are underlined. Nucleotides in the cDNAs that differ in additional positions are highlighted in bold letters. The deduced amino-acid sequences (AAB50876, AAC52054) are given in the single-letter code.
Conserved structure of exons 2 to 11
The comparison of the genomic sequences of exons 2 to 11 was extended by 100 bp of intronic sequence in both directions (Figure 3). During the preparation of this manuscript all sequence information was made available by the International Human Genome Project collaborators at the NCBI database and included in the contig NT_008491. Sequences of the 5'-untranslated region and of exon 7 obtained with a genome walking approach did not diverge from the sequence at the NCBI.
Figure 3.

Comparison of the exons of the mouse and human GCNF genes. The upper line of nucleotide sequence shows the murine protein-coding exons and their flanking sequences (AF254815-AF254821). Protein-coding nucleotides are underlined. The lower line of nucleotide sequence shows the corresponding human sequence. Identical nucleotides are highlighted by bold letters. The AG/GT splice signals are shown in italics. mGCNF indicates the deduced mouse protein sequence; hGCNF indicates the deduced human protein sequences; GCNF indicates identical protein sequences. (a) The second exon and its flanking regions. Forty-one out of 42 nucleotides are identical and the flanking splicing signals are conserved. (b) The third exon and its flanking regions. A homologous sequence coding for identical amino acids was found in the human genomic sequence. No human isoform containing this sequence has been reported. The splice donor site shows the typical pyrimidine-rich sequence followed by the sequence 5'-NCAG in both sequences, but the comparison reveals several base transitions. (c) A single splice donor site in the fourth exon coding for the DNA-binding domain is used in all mouse-derived cDNAs described so far. For the human isoform GCNF-2, the corresponding splice site is used, giving rise to a protein containing the sequence ISVSDD instead of the VSVPDD in mouse. Usage of an alternative splice site located 12 bp further downstream gives rise to the shorter isoform GCNF-1. An asparagine (N) is underlined because one of the human cDNA clones codes for a lysine in this position (U64876/NM_001489). (d) Sequences of the fifth exon coding for the carboxy-terminal extension of the DNA-binding domain are highly conserved. (e) The DNA sequence of the sixth exon is highly conserved. An arginine in hGCNF-2b instead of serine results from alternative splice donor sites of the seventh exon. (f) The comparison of the seventh exon reveals three positions where the mouse and the human isoforms diverge. Isoform hGCNF-2b is generated by using a splice donor site located three nucleotides further downstream. The exons coding for the putative α-helices 3 to 6 (g), 7 and 8 (h), 9 and 10 (i), 11 and 12 (j) in the ligand-binding domain are highly conserved. The comparison of the last coding exon in (j) was extended up to the end of the human cDNA sequence of S88309.
Two short exons of 42 bp and 45 bp, respectively, follow the first protein-coding exon in the mouse [17]. Short exons are relatively rare in mammalian genomes. The structure of the second protein-coding exon is conserved (Figure 3a). Splice donor and acceptor sites are identical in both species. Interestingly, on the basis of the genomic cDNA, the third exon is highly conserved as well (Figure 3b). The human splicing apparatus preferentially, or exclusively, skips this putative exon, however. As splicing is highly regulated, a splice enhancer present in the mouse genome may not be present in the human genome. Consequently, all known human GCNF isoforms lack the amino acids encoded by the putative third exon.
Of the 243 bp exon 4 that encodes the core of the DNA-binding domain, 225 bp are identical in both species (Figure 3c). One of the reported sequences (U64876/NM_001489) has a C to A transversion, however, which changes a codon for asparagine to one for lysine. Splicing of exon 2 to exon 4 at the position characterized in the mouse results in isoform hGCNF-2. In addition to this splice acceptor position, a splice acceptor site located 12 nucleotides further downstream is used to generate hGCNF-1. Exons 5 and 6 are highly conserved (Figure 3d,e). Two hGCNF-2 variants, hGCNF-2A and hGCNF-2B, which differ by a single amino acid, have been isolated. As speculated, alternative splicing generates the isoform 2B with a deletion of a serine residue. Splicing to an acceptor site of exon 7 located three nucleotides further downstream gives rise to this shorter isoform (Figure 3e,f). The sequence and structure of exons 8 to 11 are also highly conserved (Figure 3g,h,i,j). The comparison of the 11th exon was extended up to the end of the human cDNA sequence of S88309. Highly conserved sequence elements of up to 91 identical nucleotides indicate a regulatory function of the 3'-untranslated sequence following the translational stop codon.
All intron-exon boundaries obeyed the GT/AG rule [22]. The AceView analysis at the NCBI based on the draft sequence and a Blast search with S83309 of the Celera Genomics freely accessible whole-genome sequence data gave mostly similar intron sizes. Both analyses revealed a large first intron of 37,652 bp in the public sequence data, and 37,157 bp in the private data. The size of the second intron separating exon 2 and exon 4 was only available in the NCBI database (14,869 bp). According to the NCBI and Celera databases, introns 3 to 9 have sizes of 10,486 (NCBI) (10,471, Celera) bp, 3629 (3615) bp, 190,321 (1708) bp, 1963 (1960) bp, 2716 (9019) bp, 1905 (1912) bp, and 1927 (1928) bp, respectively. The comparison of both analyses shows that the deduced sizes of two of the human introns differs greatly. It seems likely that these inconsistencies will be corrected in the final assembly of the human genome.
Conclusion
In summary, our analysis reveals a conserved structure for GCNF, allows the verification and systematic analysis of splice variants, and may be the basis of a better understanding of GCNF. The human GCNF gene consists of at least 10 exons. The conservation of the intron-exon boundaries is consistent with the extremely high degree of amino-acid conservation between the human and the mouse proteins. The generation of the proteins hGCNF-1, hGCNF-2a and hGCNF-2b can be explained by alternative splicing of the RNA. The sequence of the third coding mouse exon, including the splice sites, is highly conserved; however, at present no human cDNA has been isolated containing this putative exon. Alternative splicing provides a plausible means for generating diversity and may contribute to a higher instructive complexity in human GCNF.
Materials and methods
Database search
Exons of GCNF were identified by a Blast [23] search with the human GCNF cDNA sequence (S83009) in the "unfinished high throughput genomic sequences" and in the Homo sapiens genomic contig sequences at the NCBI [24, 25]. Intron sizes given by the AceView analysis [26] were compared with the numbers obtained by a Blast search of Celera's assembled sequence of the human genome [27]. The putative human GCNF exon 3 was identified by a Blast search with the sequence of the third mouse exon.
Sequence analysis
Sequences were aligned using the Wisconsin Package Version 10.0 of the Genetics Computer Group (GCG), Madison, Wisconsin.
References
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giguère V. Orphan nuclear receptors: from gene to function. Endocr Rev. 1999;20:689–725. doi: 10.1210/edrv.20.5.0378. [DOI] [PubMed] [Google Scholar]
- Laudet V. Evolution of the nuclear receptor superfamily: early diversification from an ancestral orphan receptor. J Mol Endocrinol. 1997;19:207–226. doi: 10.1677/jme.0.0190207. [DOI] [PubMed] [Google Scholar]
- The Nuclear Receptor Committee A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161–163. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- Hirose T, O'Brien DA, Jetten AM. RTR: a new member of the nuclear receptor superfamily that is highly expressed in murine testis. Gene. 1995;152:247–251. doi: 10.1016/0378-1119(94)00656-d. [DOI] [PubMed] [Google Scholar]
- Agoulnik IY, Cho Y, Niederberger C, Kieback DG, Cooney AJ. Cloning, expression analysis and chromosomal localization of the human nuclear receptor gene GCNF. FEBS Lett. 1998;424:73–78. doi: 10.1016/s0014-5793(98)00142-2. [DOI] [PubMed] [Google Scholar]
- Bauer U-M, Schneider-Hirsch S, Reinhardt S, Pauly T, Maus A, Wang F, Heiermann R, Rentrop M, Maelicke A. Neuronal cell nuclear factor-a nuclear receptor possibly involved in the control of neurogenesis and neuronal differentiation. Eur J Biochem. 1997;249:826–837. doi: 10.1111/j.1432-1033.1997.t01-1-00826.x. [DOI] [PubMed] [Google Scholar]
- Cooney AJ, Hummelke GC, Herman T, Chen F, Jackson KJ. Germ cell nuclear factor is a response element-specific repressor of transcription. Biochem Biophys Res Commun. 1998;245:94–100. doi: 10.1006/bbrc.1998.8391. [DOI] [PubMed] [Google Scholar]
- Greschik H, Wurtz J-M, Hublitz P, Köhler F, Moras D, Schüle R. Characterization of the DNA-binding and dimerization properties of the nuclear orphan receptor germ cell nuclear factor. Mol Cell Biol. 1999;19:690–703. doi: 10.1128/mcb.19.1.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Jetten AM. Characterization of the repressor function of the nuclear orphan receptor retinoid receptor-related testis-associated receptor/germ cell nuclear factor. J Biol Chem. 2000;275:10565–10572. doi: 10.1074/jbc.M005566200. [DOI] [PubMed] [Google Scholar]
- Chung AC-K, Katz D, Pereira FA, Jackson KJ, DeMayo FJ, Cooney AJ, O'Malley BW. Loss of orphan receptor germ cell nuclear factor function results in ectopic development of the tail bud and a novel posterior truncation. Mol Cell Biol. 2001;21:663–677. doi: 10.1128/MCB.21.2.663-677.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Süsens U, Aguiluz JB, Evans RM, Borgmeyer U. The germ cell nuclear factor mGCNF is expressed in the developing nervous system. Dev Neurosci. 1997;19:410–420. doi: 10.1159/000111238. [DOI] [PubMed] [Google Scholar]
- Morasso MI, Grinberg A, Robinson G, Sargent TD, Mahon KA. Placental failure in mice lacking the homeobox gene Dlx3. Proc Natl Acad Sci USA. 1999;96:162–167. doi: 10.1073/pnas.96.1.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Cooney AJ, Wang Y, Law SW, O'Malley BW. Cloning of a novel orphan receptor (GCNF) expressed during germ cell development. Mol Endocrinol. 1994;8:1434–1444. doi: 10.1210/mend.8.10.7854358. [DOI] [PubMed] [Google Scholar]
- Katz D, Niederberger C, Slaughter GR, Cooney AJ. Characterization of germ cell-specific expression of the orphan nuclear receptor, germ cell nuclear factor. Endocrinology. 1997;138:4364–4372. doi: 10.1210/endo.138.10.5444. [DOI] [PubMed] [Google Scholar]
- Zhang YL, Akmal KM, Tsuruta JK, Shang Q, Hirose T, Jetten AM, Kim KH, O'Brien DA. Expression of germ cell nuclear factor (GCNF/RTR) during spermatogenesis. Mol Reprod Dev. 1998;50:93–102. doi: 10.1002/(SICI)1098-2795(199805)50:1<93::AID-MRD12>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Süsens U, Borgmeyer U. Genomic structure of the mouse germ cell nuclear factor (GCNF) gene. Genome Biol. 2000;1:research0006.1–0006.3. doi: 10.1186/gb-2000-1-3-research0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzer C, Süsens U, Schmitz TP, Borgmeyer U. Retinoids induce differential expression and DNA binding of the mouse germ cell nuclear factor in P19 embryonal carcinoma cells. Biol Chem. 1998;379:349–359. doi: 10.1515/bchm.1998.379.3.349. [DOI] [PubMed] [Google Scholar]
- Süsens U, Borgmeyer U. Characterization of the human germ cell nuclear factor gene. Biochim Biophys Acta. 1996;1309:179–182. doi: 10.1016/s0167-4781(96)00157-1. [DOI] [PubMed] [Google Scholar]
- Schmitz TP, Süsens U, Borgmeyer U. DNA binding, protein interaction and differential expression of the human germ cell nuclear factor. Biochim Biophys Acta. 1999;1446:173–180. doi: 10.1016/s0167-4781(99)00079-2. [DOI] [PubMed] [Google Scholar]
- Greschik H, Schüle R. Germ cell nuclear factor: an orphan receptor with unexpected properties. J Mol Med. 1998;76:800–810. doi: 10.1007/s001090050284. [DOI] [PubMed] [Google Scholar]
- Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- NCBI: Basic BLAST http://www.ncbi.nlm.nih.gov/blast/blast.cgi
- NCBI: Genome Sequencing - BLAST the Human Genome http://www.ncbi.nlm.nih.gov/genome/seq/HsBlast.html
- The AceView at the NCBI http://www.ncbi.nlm.nih.gov/AceView/
- CELERA: Consensus Human Genome http://public.celera.com


