Abstract
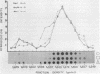
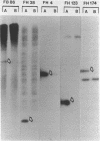
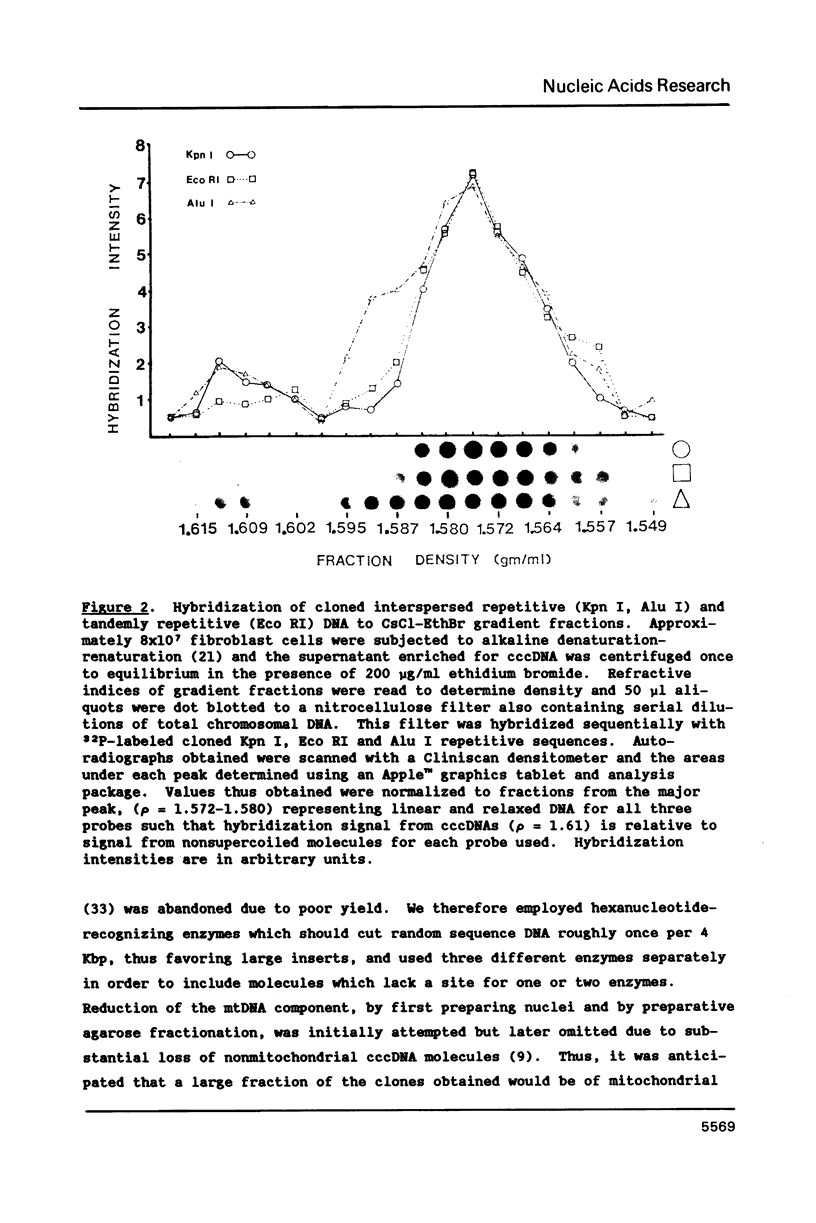
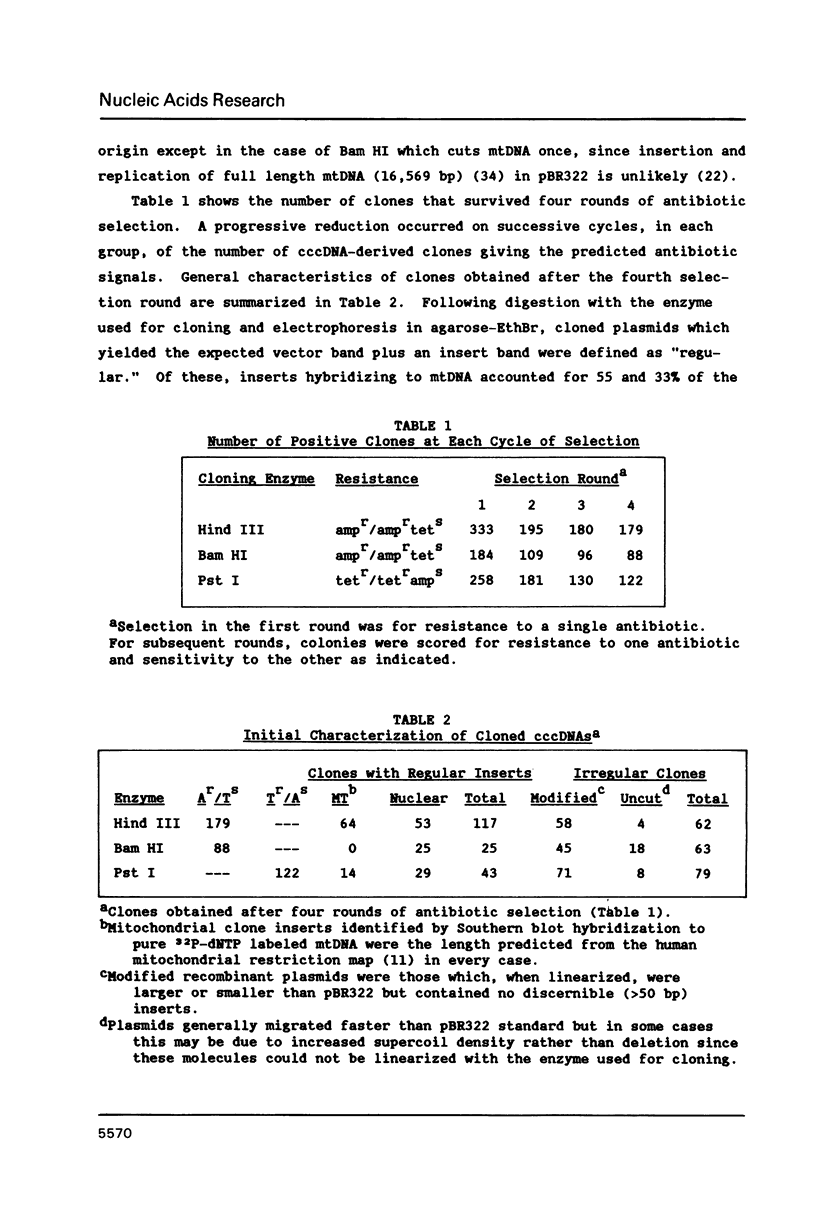
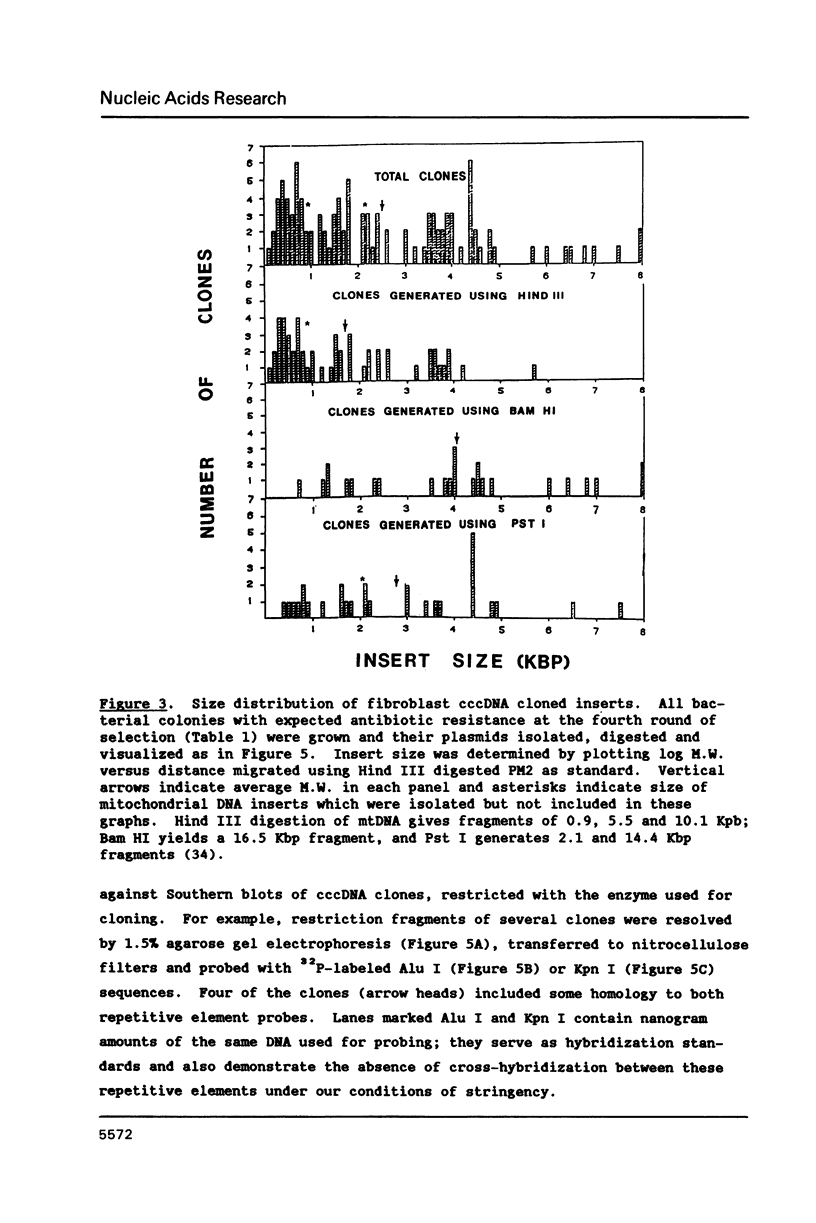
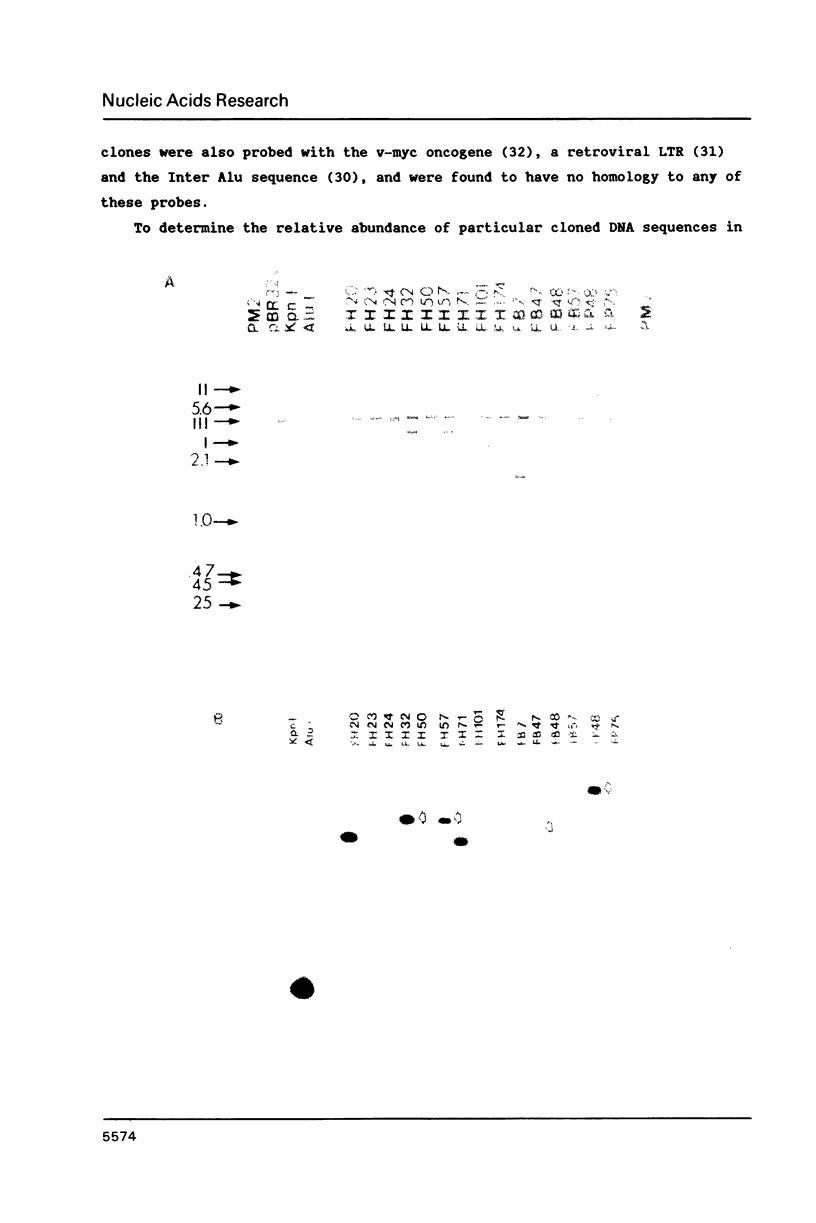
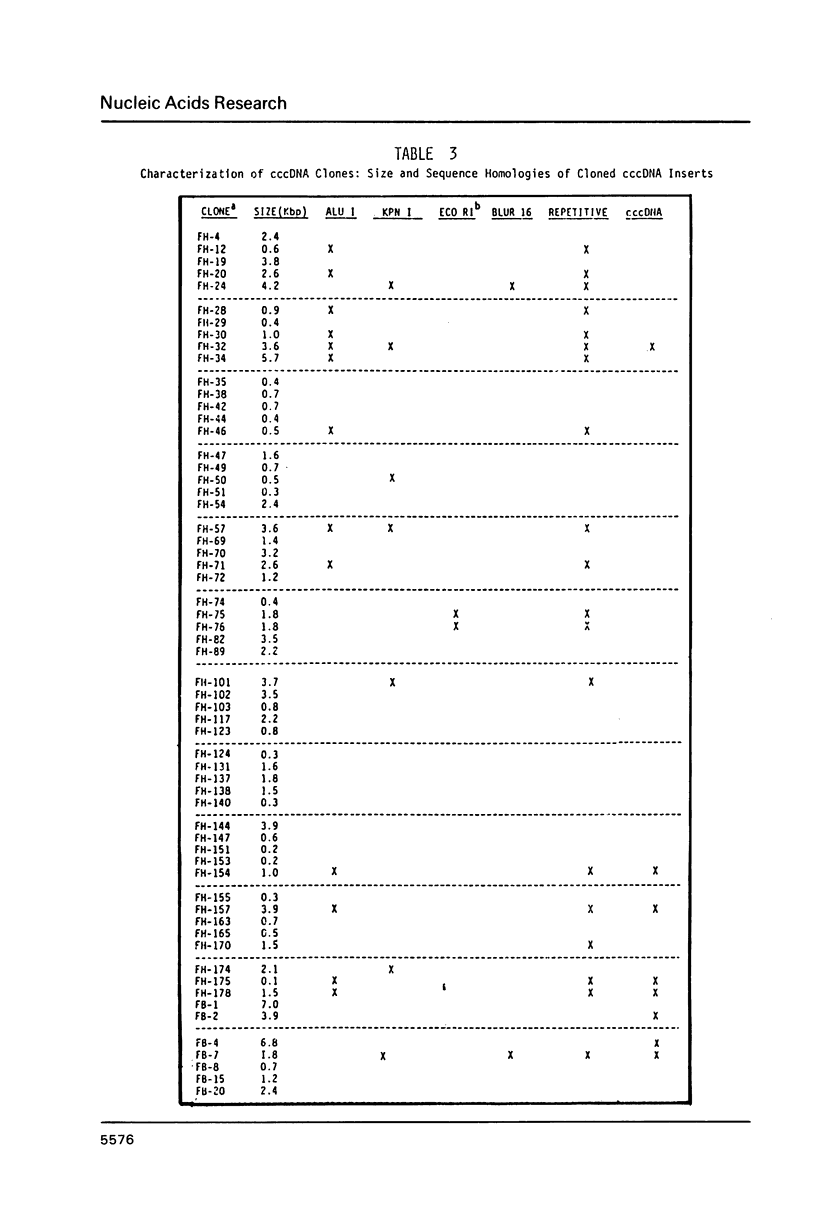
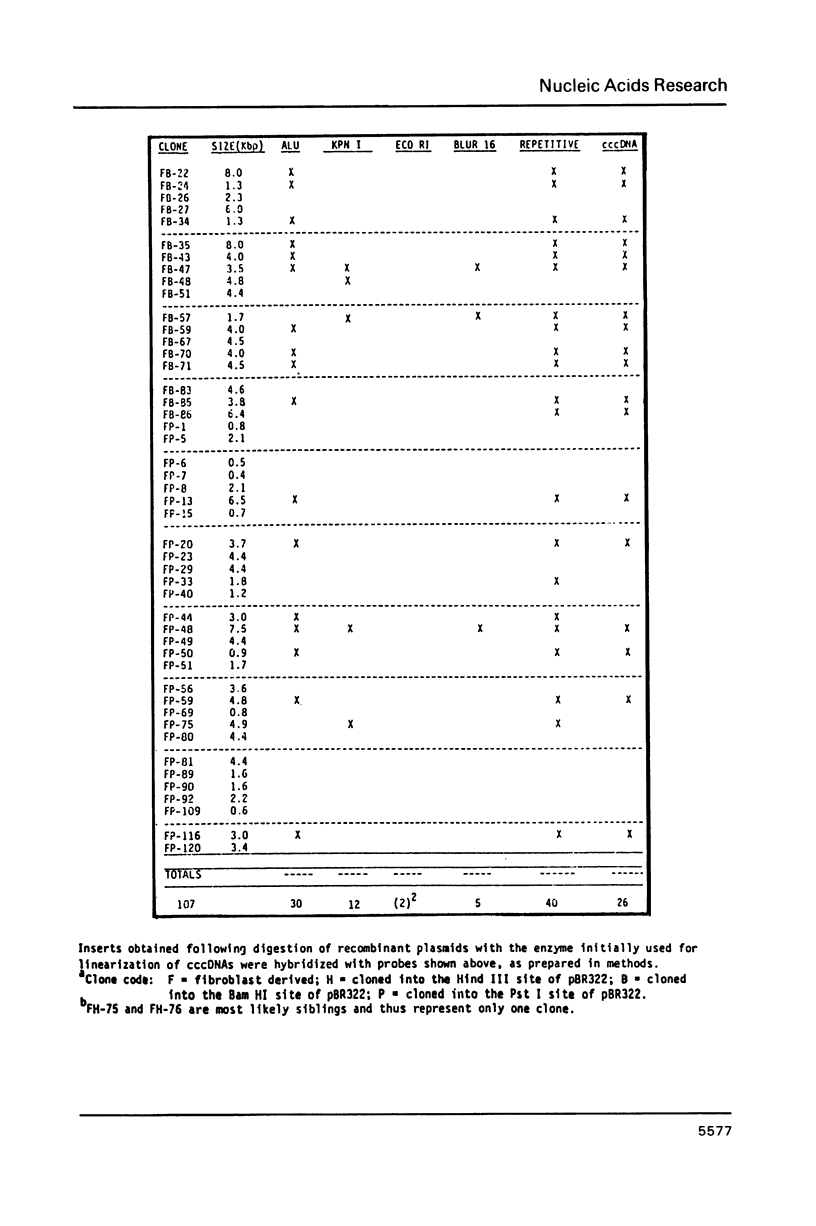
Extrachromosomal covalently closed circular DNA (cccDNA) was isolated from human diploid fibroblasts by alkaline denaturation/renaturation and CsCl-ethidium bromide isopycnic centrifugation. Probing across these gradient fractions showed a higher proportion of cccDNA sequences homologous to the interspersed highly repetitive Alu I and Kpn I sequences than to the human tandemly-repetitive Eco RI (alphoid) DNA. Cloning of these cccDNAs was then carried out following digestion with restriction endonucleases Hind III, Bam HI or Pst I, and ligation into plasmid pBR322. Many isolated recombinant clones were unstable as seen by a high rate of loss over four cycles of antibiotic selection, and frequent plasmid modifications including deletions adjoining the site of insertion. Of 107 cloned sequences which appeared relatively stable, i.e., survived four cycles of antibiotic selection without incurring detectable deletions, 28% and 11% showed homology to Alu I and Kpn I families, respectively, while 4% contained sequences homologous to both. In contrast, less than one percent hybridized to probes for tandemly-repetitive sequences, Eco RI and Satellite III. The average insert size of cloned cccDNA derived from human fibroblasts, 2.52 Kbp, was larger than previously reported for similar clones derived from genetically less stable permanent lines, which may reflect differences in the process of cccDNA generation.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benz E. J., Jr, Murnane M. J., Tonkonow B. L., Berman B. W., Mazur E. M., Cavallesco C., Jenko T., Snyder E. L., Forget B. G., Hoffman R. Embryonic-fetal erythroid characteristics of a human leukemic cell line. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3509–3513. doi: 10.1073/pnas.77.6.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelsen A. H., Humayun M. Z., Karfopoulos S. G., Rush M. G. Molecular characterization of small polydisperse circular deoxyribonucleic acid from an African green monkey cell line. Biochemistry. 1982 Apr 27;21(9):2076–2085. doi: 10.1021/bi00538a015. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabretta B., Robberson D. L., Barrera-Saldaña H. A., Lambrou T. P., Saunders G. F. Genome instability in a region of human DNA enriched in Alu repeat sequences. Nature. 1982 Mar 18;296(5854):219–225. doi: 10.1038/296219a0. [DOI] [PubMed] [Google Scholar]
- DeLap R. J., Rush M. G. Change in quantity and size distribution of small circular DNAs during development of chicken bursa. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5855–5859. doi: 10.1073/pnas.75.12.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLap R. J., Rush M. G., Zouzias D., Khan S. Isolation and preliminary characterization of the small circular DNA present in African green monkey kidney (BSC-1) cells. Plasmid. 1978 Sep;1(4):508–521. doi: 10.1016/0147-619x(78)90008-2. [DOI] [PubMed] [Google Scholar]
- DiGiovanni L., Haynes S. R., Misra R., Jelinek W. R. Kpn I family of long-dispersed repeated DNA sequences of man: evidence for entry into genomic DNA of DNA copies of poly(A)-terminated Kpn I RNAs. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6533–6537. doi: 10.1073/pnas.80.21.6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin J. Cloning of human mitochondrial DNA in Escherichia coli. J Mol Biol. 1980 Jun 15;140(1):15–34. doi: 10.1016/0022-2836(80)90354-x. [DOI] [PubMed] [Google Scholar]
- Gillespie D., Adams J. W., Costanzi C., Caranfa M. J. New orientations of ancestral, "long interspersed repeated sequences" (LINES) in human DNA. Gene. 1982 Dec;20(3):409–414. doi: 10.1016/0378-1119(82)90209-8. [DOI] [PubMed] [Google Scholar]
- Grimaldi G., Singer M. F. A monkey Alu sequence is flanked by 13-base pair direct repeats by an interrupted alpha-satellite DNA sequence. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1497–1500. doi: 10.1073/pnas.79.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi G., Singer M. F. Members of the KpnI family of long interspersed repeated sequences join and interrupt alpha-satellite in the monkey genome. Nucleic Acids Res. 1983 Jan 25;11(2):321–338. doi: 10.1093/nar/11.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi G., Skowronski J., Singer M. F. Defining the beginning and end of KpnI family segments. EMBO J. 1984 Aug;3(8):1753–1759. doi: 10.1002/j.1460-2075.1984.tb02042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C. B., Goldstein S. Cultured human fibroblasts: distribution of cell generations and a critical limit. J Cell Physiol. 1978 Dec;97(3 Pt 2 Suppl 1):509–516. doi: 10.1002/jcp.1040970326. [DOI] [PubMed] [Google Scholar]
- Houck C. M., Rinehart F. P., Schmid C. W. A ubiquitous family of repeated DNA sequences in the human genome. J Mol Biol. 1979 Aug 15;132(3):289–306. doi: 10.1016/0022-2836(79)90261-4. [DOI] [PubMed] [Google Scholar]
- Jelinek W. R., Toomey T. P., Leinwand L., Duncan C. H., Biro P. A., Choudary P. V., Weissman S. M., Rubin C. M., Houck C. M., Deininger P. L. Ubiquitous, interspersed repeated sequences in mammalian genomes. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1398–1402. doi: 10.1073/pnas.77.3.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krolewski J. J., Bertelsen A. H., Humayun M. Z., Rush M. G. Members of the Alu family of interspersed, repetitive DNA sequences are in the small circular DNA population of monkey cells grown in culture. J Mol Biol. 1982 Jan 25;154(3):399–415. doi: 10.1016/s0022-2836(82)80003-x. [DOI] [PubMed] [Google Scholar]
- Krolewski J. J., Schindler C. W., Rush M. G. Structure of extrachromosomal circular DNAs containing both the Alu family of dispersed repetitive sequences and other regions of chromosomal DNA. J Mol Biol. 1984 Mar 25;174(1):41–54. doi: 10.1016/0022-2836(84)90364-4. [DOI] [PubMed] [Google Scholar]
- Mariani B. D., Schimke R. T. Gene amplification in a single cell cycle in Chinese hamster ovary cells. J Biol Chem. 1984 Feb 10;259(3):1901–1910. [PubMed] [Google Scholar]
- Martin R. G., Oppenheim A. Initiation points for DNA replication in nontransformed and simian virus 40-transformed Chinese hamster lung cells. Cell. 1977 Aug;11(4):859–869. doi: 10.1016/0092-8674(77)90297-5. [DOI] [PubMed] [Google Scholar]
- McClements W. L., Enquist L. W., Oskarsson M., Sullivan M., Vande Woude G. F. Frequent site-specific deletion of coliphage lambda murine sarcoma virus recombinants and its use in the identification of a retrovirus integration site. J Virol. 1980 Aug;35(2):488–497. doi: 10.1128/jvi.35.2.488-497.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore G. P., Moore A. R. The average spacing of restriction enzyme recognition sites in DNA. J Theor Biol. 1982 Sep 7;98(1):165–169. doi: 10.1016/0022-5193(82)90064-9. [DOI] [PubMed] [Google Scholar]
- Osterlund M., Luthman H., Nilsson S. V., Magnusson G. Ethidium-bromide-inhibited restriction endonucleases cleave one strand of circular DNA. Gene. 1982 Nov;20(1):121–125. doi: 10.1016/0378-1119(82)90093-2. [DOI] [PubMed] [Google Scholar]
- Potter S. S. Rearranged sequences of a human Kpn I element. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1012–1016. doi: 10.1073/pnas.81.4.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rush M. G., Eason R., Vinograd J. Identification and properties of complex forms of SV40 DNA isolated from SV40-infected African Green monkey (BSC-1) cells. Biochim Biophys Acta. 1971 Feb 11;228(3):585–594. doi: 10.1016/0005-2787(71)90723-4. [DOI] [PubMed] [Google Scholar]
- Shafit-Zagardo B., Maio J. J., Brown F. L. KpnI families of long, interspersed repetitive DNAs in human and other primate genomes. Nucleic Acids Res. 1982 May 25;10(10):3175–3193. doi: 10.1093/nar/10.10.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M. F. Highly repeated sequences in mammalian genomes. Int Rev Cytol. 1982;76:67–112. doi: 10.1016/s0074-7696(08)61789-1. [DOI] [PubMed] [Google Scholar]
- Stanfield S. W., Lengyel J. A. Small circular DNA of Drosophila melanogaster: chromosomal homology and kinetic complexity. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6142–6146. doi: 10.1073/pnas.76.12.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield S., Helinski D. R. Small circular DNA in Drosophila melanogaster. Cell. 1976 Oct;9(2):333–345. doi: 10.1016/0092-8674(76)90123-9. [DOI] [PubMed] [Google Scholar]
- Sun L., Paulson K. E., Schmid C. W., Kadyk L., Leinwand L. Non-Alu family interspersed repeats in human DNA and their transcriptional activity. Nucleic Acids Res. 1984 Mar 26;12(6):2669–2690. doi: 10.1093/nar/12.6.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudzynski P., Esser K. Chromosomal and extrachromosomal control of senescence in the ascomycete Podospora anserina. Mol Gen Genet. 1979 May 23;173(1):71–84. doi: 10.1007/BF00267692. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. On the possibility of metabolic control of replicon "misfiring": relationship to emergence of malignant phenotypes in mammalian cell lineages. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3673–3677. doi: 10.1073/pnas.78.6.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. C., Manuelidis L. Sequence definition and organization of a human repeated DNA. J Mol Biol. 1980 Sep 25;142(3):363–386. doi: 10.1016/0022-2836(80)90277-6. [DOI] [PubMed] [Google Scholar]
- Yamagishi H., Kunisada T., Tsuda T. Small circular DNA complexes in eucaryotic cells. Plasmid. 1982 Nov;8(3):299–306. doi: 10.1016/0147-619x(82)90067-1. [DOI] [PubMed] [Google Scholar]