Abstract
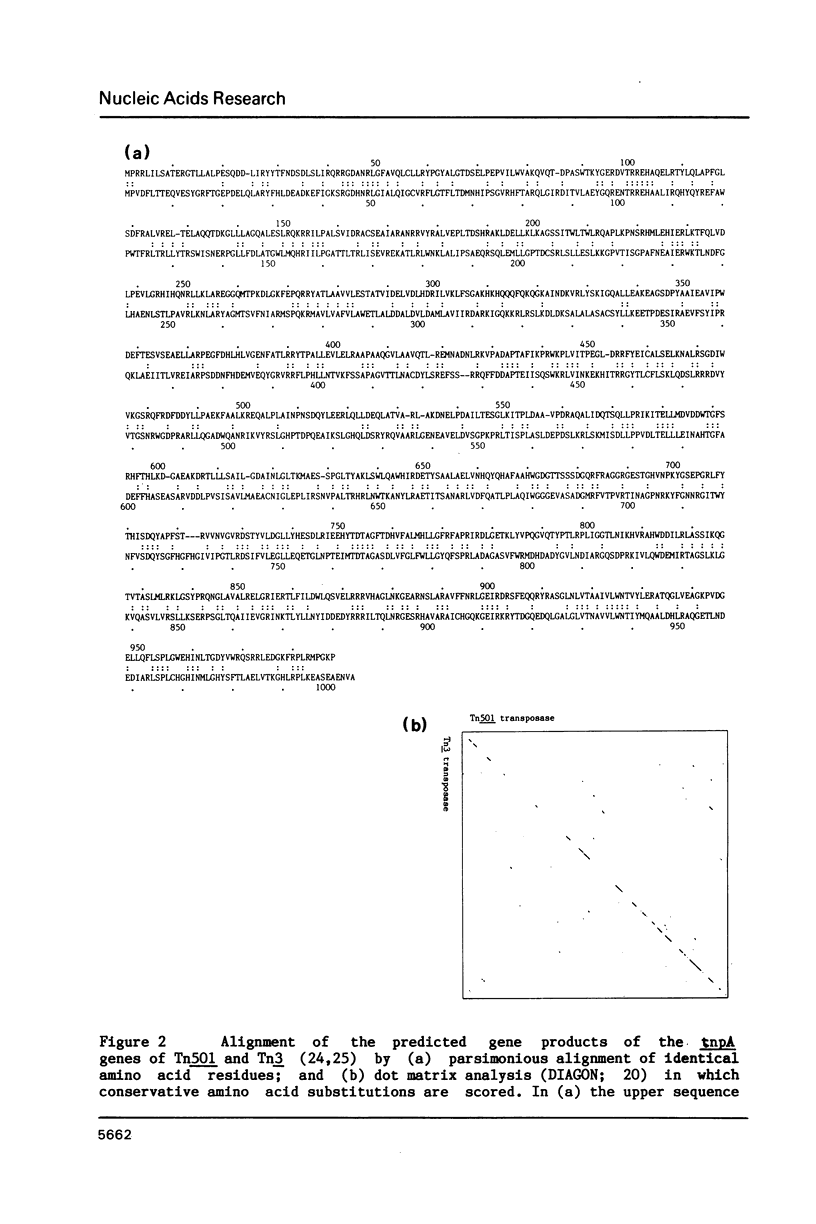
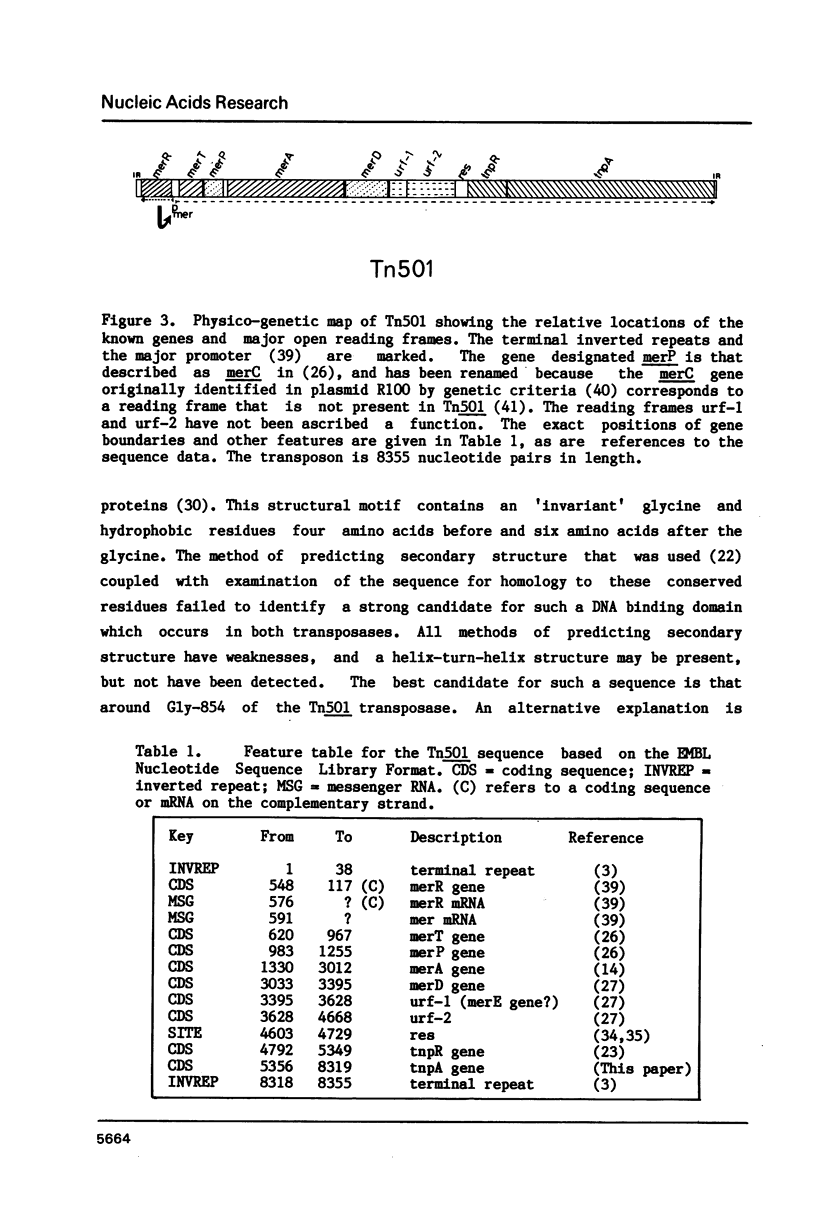
The nucleotide sequence of the gene (tnpA) which codes for the transposase of transposon Tn501 has been determined. It contains an open reading frame for a polypeptide of Mr = 111,500, which terminates within the inverted repeat sequence of the transposon. The reading frame would be transcribed in the same direction as the mercury-resistance genes and the tnpR gene. The amino acid sequence predicted from this reading frame shows 32% identity with that of the transposase of the related transposon Tn3. The C-terminal regions of these two polypeptides show slightly greater homology than the N-terminal regions when conservative amino acid substitutions are considered. With this sequence determination, the nucleotide sequence of Tn501 is fully defined. The main features of the sequence are briefly presented.
Full text
PDF










Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altenbuchner J., Choi C. L., Grinsted J., Schmitt R., Richmond M. H. The transposons Tn501(Hg) and Tn1721(Tc) are related. Genet Res. 1981 Jun;37(3):285–289. doi: 10.1017/s0016672300020280. [DOI] [PubMed] [Google Scholar]
- Altenbuchner J., Schmitt R. Transposon Tn1721: site-specific recombination generates deletions and inversions. Mol Gen Genet. 1983;190(2):300–308. doi: 10.1007/BF00330655. [DOI] [PubMed] [Google Scholar]
- Arthur A., Sherratt D. Dissection of the transposition process: a transposon-encoded site-specific recombination system. Mol Gen Genet. 1979 Oct 1;175(3):267–274. doi: 10.1007/BF00397226. [DOI] [PubMed] [Google Scholar]
- Brown N. L., Choi C. L., Grinsted J., Richmond M. H., Whitehead P. R. Nucleotide sequences at the ends of the mercury resistance transposon, Tn501. Nucleic Acids Res. 1980 May 10;8(9):1933–1945. doi: 10.1093/nar/8.9.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. L., Ford S. J., Pridmore R. D., Fritzinger D. C. Nucleotide sequence of a gene from the Pseudomonas transposon Tn501 encoding mercuric reductase. Biochemistry. 1983 Aug 16;22(17):4089–4095. doi: 10.1021/bi00286a015. [DOI] [PubMed] [Google Scholar]
- Diver W. P., Grinsted J., Fritzinger D. C., Brown N. L., Altenbuchner J., Rogowsky P., Schmitt R. DNA sequences of and complementation by the tnpR genes of Tn21, Tn501 and Tn1721. Mol Gen Genet. 1983;191(2):189–193. doi: 10.1007/BF00334812. [DOI] [PubMed] [Google Scholar]
- Fennewald M. A., Gerrard S. P., Chou J., Casadaban M. J., Cozzarelli N. R. Purification of the Tn3 transposase and analysis of its binding to DNA. J Biol Chem. 1981 May 25;256(10):4687–4690. [PubMed] [Google Scholar]
- Grinsted J., Brown N. L. A Tn21 terminal sequence within Tn501: complementation of tnpA gene function and transposon evolution. Mol Gen Genet. 1984;197(3):497–502. doi: 10.1007/BF00329949. [DOI] [PubMed] [Google Scholar]
- Grinsted J., de la Cruz F., Altenbuchner J., Schmitt R. Complementation of transposition of tnpA mutants of Tn3, Tn21, Tn501, and Tn1721. Plasmid. 1982 Nov;8(3):276–286. doi: 10.1016/0147-619x(82)90065-8. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., McCarthy B. J., Ohtsubo H., Ohtsubo E. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell. 1979 Dec;18(4):1153–1163. doi: 10.1016/0092-8674(79)90228-9. [DOI] [PubMed] [Google Scholar]
- Kitts P. A., Lamond A., Sherratt D. J. Inter-replicon transposition of Tn1/3 occurs in two sequential genetically separable steps. Nature. 1982 Feb 18;295(5850):626–628. doi: 10.1038/295626a0. [DOI] [PubMed] [Google Scholar]
- Kitts P., Symington L., Burke M., Reed R., Sherratt D. Transposon-specified site-specific recombination. Proc Natl Acad Sci U S A. 1982 Jan;79(1):46–50. doi: 10.1073/pnas.79.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Kneale G. G., Kennard O. The EMBL nucleotide sequence data library. Biochem Soc Trans. 1984 Dec;12(6):1011–1014. doi: 10.1042/bst0121011. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Misra T. K., Brown N. L., Fritzinger D. C., Pridmore R. D., Barnes W. M., Haberstroh L., Silver S. Mercuric ion-resistance operons of plasmid R100 and transposon Tn501: the beginning of the operon including the regulatory region and the first two structural genes. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5975–5979. doi: 10.1073/pnas.81.19.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra T. K., Brown N. L., Haberstroh L., Schmidt A., Goddette D., Silver S. Mercuric reductase structural genes from plasmid R100 and transposon Tn501: functional domains of the enzyme. Gene. 1985;34(2-3):253–262. doi: 10.1016/0378-1119(85)90134-9. [DOI] [PubMed] [Google Scholar]
- Ni'Bhriain N. N., Silver S., Foster T. J. Tn5 insertion mutations in the mercuric ion resistance genes derived from plasmid R100. J Bacteriol. 1983 Aug;155(2):690–703. doi: 10.1128/jb.155.2.690-703.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Reed R. R., Shibuya G. I., Steitz J. A. Nucleotide sequence of gamma delta resolvase gene and demonstration that its gene product acts as a repressor of transcription. Nature. 1982 Nov 25;300(5890):381–383. doi: 10.1038/300381a0. [DOI] [PubMed] [Google Scholar]
- Roberts R. J. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 1985;13 (Suppl):r165–r200. doi: 10.1093/nar/13.suppl.r165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogowsky P., Schmitt R. Resolution of a hybrid cointegrate between transposons Tn501 and Tn1721 defines the recombination site. Mol Gen Genet. 1984;193(1):162–166. doi: 10.1007/BF00327431. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Schmidt F., Klopfer-Kaul I. Evolutionary relationship between Tn21-like elements and pBP201, a plasmid from Klebsiella pneumoniae mediating resistance to gentamicin and eight other drugs. Mol Gen Genet. 1984;197(1):109–119. doi: 10.1007/BF00327930. [DOI] [PubMed] [Google Scholar]
- Schöffl F., Arnold W., Pühler A., Altenbuchner J., Schmitt R. The tetracycline resistance transposons Tn1721 and Tn1771 have three 38-base-pair repeats and generate five-base-pair direct repeats. Mol Gen Genet. 1981;181(1):87–94. doi: 10.1007/BF00339010. [DOI] [PubMed] [Google Scholar]
- Staden R. A new computer method for the storage and manipulation of DNA gel reading data. Nucleic Acids Res. 1980 Aug 25;8(16):3673–3694. doi: 10.1093/nar/8.16.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982 May 11;10(9):2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Graphic methods to determine the function of nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):521–538. doi: 10.1093/nar/12.1part2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanisich V. A., Bennett P. M., Richmond M. H. Characterization of a translocation unit encoding resistance to mercuric ions that occurs on a nonconjugative plasmid in Pseudomonas aeruginosa. J Bacteriol. 1977 Mar;129(3):1227–1233. doi: 10.1128/jb.129.3.1227-1233.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Yamamoto T., Sawai T. Evolution of complex resistance transposons from an ancestral mercury transposon. J Bacteriol. 1983 Mar;153(3):1432–1438. doi: 10.1128/jb.153.3.1432-1438.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Yamamoto T., Sawai T. Fine structure of transposition genes on Tn2603 and complementation of its tnpA and tnpR mutations by related transposons. Mol Gen Genet. 1983;191(3):442–450. doi: 10.1007/BF00425761. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart W. L., Broach J. R., Ohtsubo E. ATP-dependent specific binding of Tn3 transposase to Tn3 inverted repeats. Nature. 1985 Apr 11;314(6011):556–558. doi: 10.1038/314556a0. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]



