Abstract
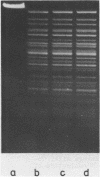
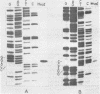
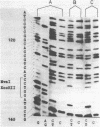
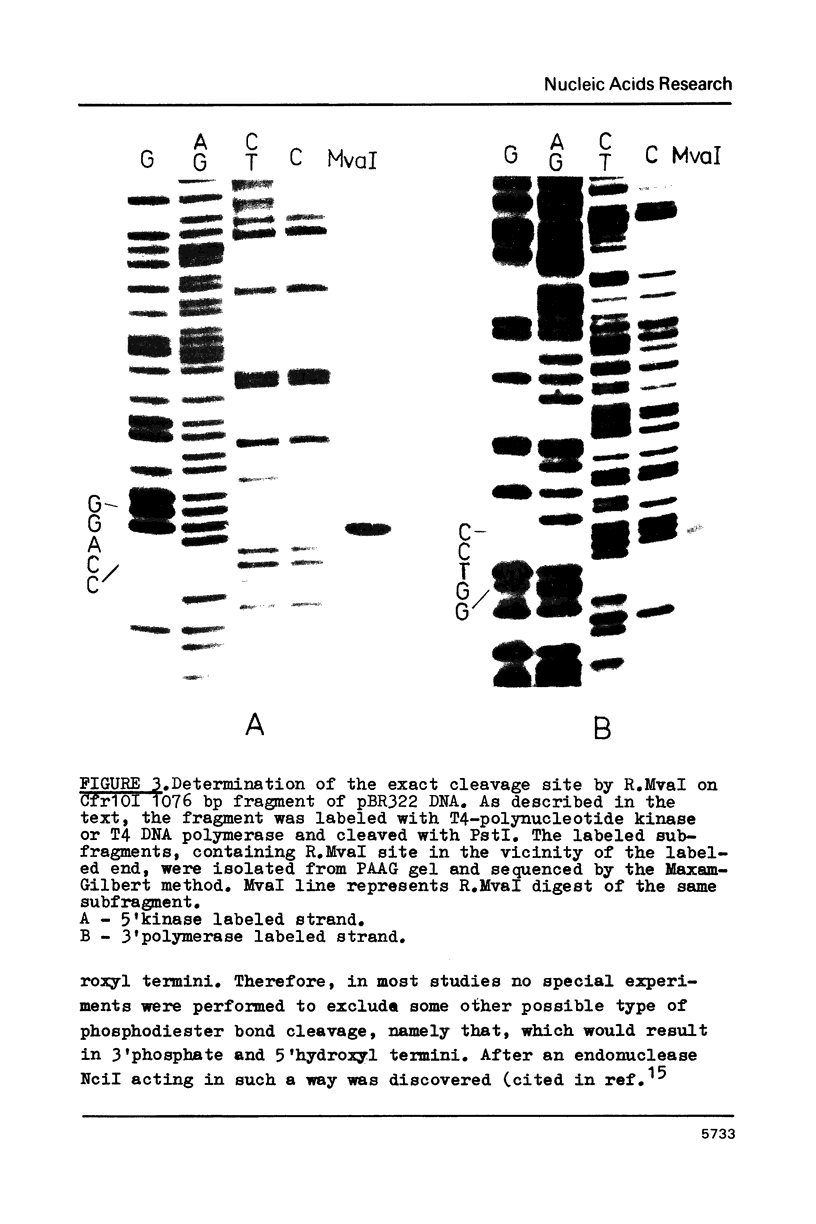
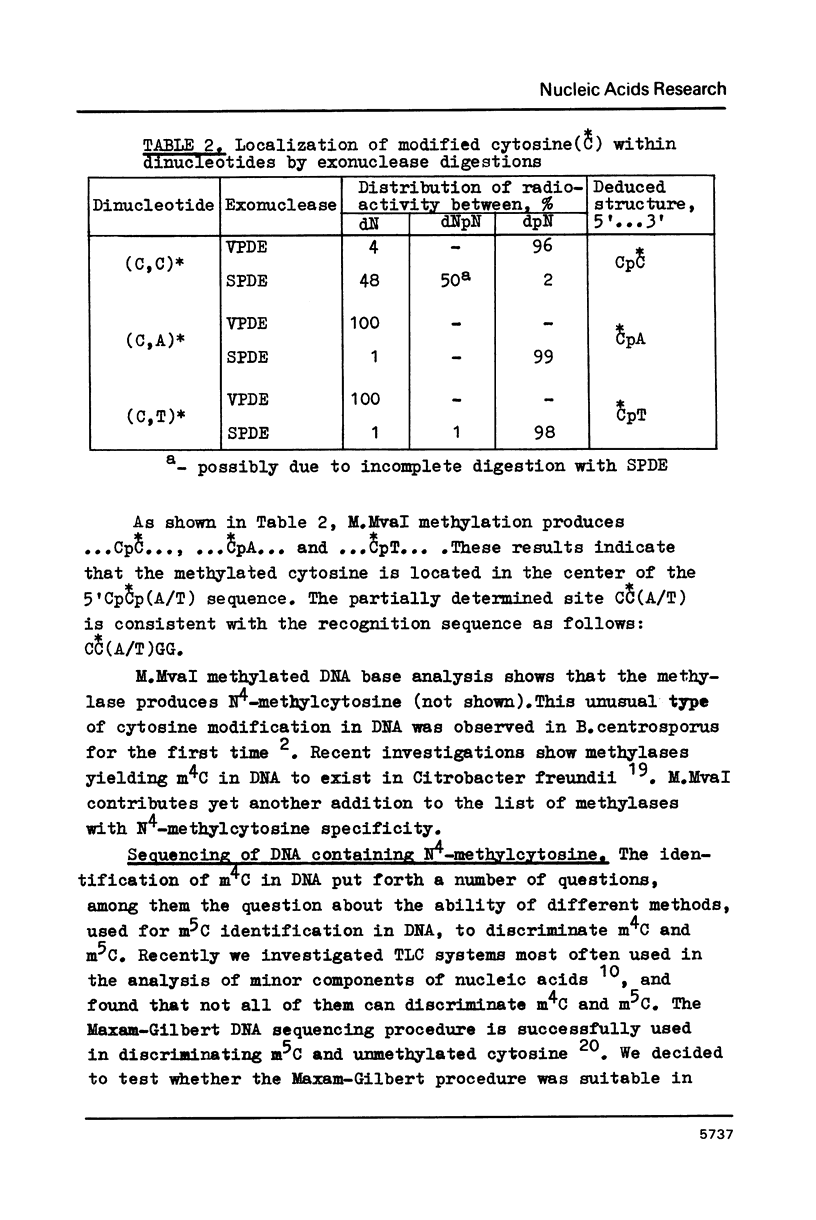
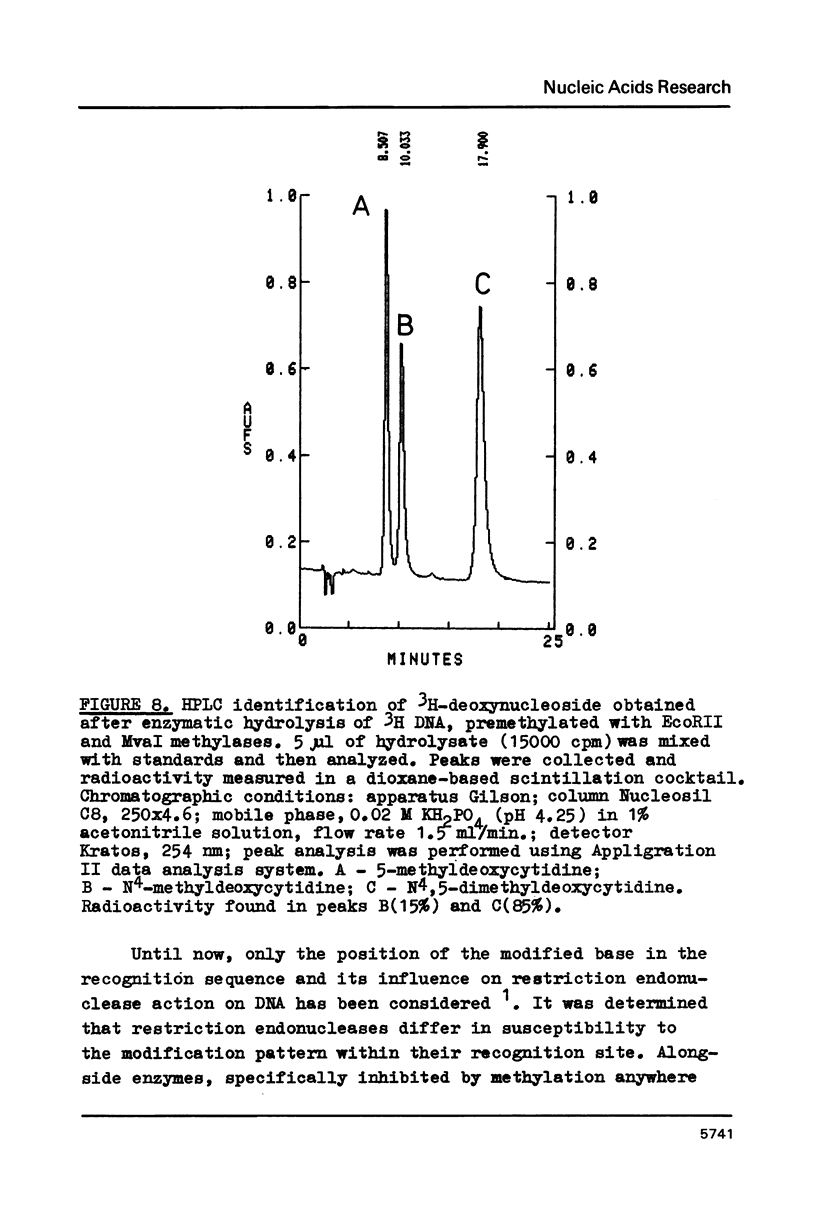
The characterization of MvaI restriction-modification enzymes, isolated from Micrococcus varians RFL19, is reported. Both enzymes recognize the 5'CC decreases (A/T)GG nucleotide sequence. The endonuclease cleaves the sequence at the position indicated by the arrow, whereas the methylase modifies the internal cytosine, yielding N4-methylcytosine. This type of modification protects the substrate from R.MvaI cleavage. 5-Methylcytosine in the same position of the recognition sequence does not protect the substrate from R.MvaI cleavage. R.MvaI proved to be the first example of a restriction endonuclease differentiating the position of the methyl group in the heterocyclic ring of cytosine, located in the same site of the recognition sequence. M.MvaI modifies DNA dcm+ in vitro yielding N4,5-dimethylcytosine. N4-methylcytosine cannot be differentiated from cytosine using the Maxam-Gilbert DNA sequencing procedure.
Full text
PDF
















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Azorin F., Hahn R., Rich A. Restriction endonucleases can be used to study B-Z junctions in supercoiled DNA. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5714–5718. doi: 10.1073/pnas.81.18.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra S. P., Detera S. D., Wilson S. H. Anomalous electrophoretic migration of oligodeoxynucleotides with terminal -OH groups: applications for DNA exonuclease characterization. Anal Biochem. 1983 Feb 15;129(1):200–206. doi: 10.1016/0003-2697(83)90069-6. [DOI] [PubMed] [Google Scholar]
- Behe M., Felsenfeld G. Effects of methylation on a synthetic polynucleotide: the B--Z transition in poly(dG-m5dC).poly(dG-m5dC). Proc Natl Acad Sci U S A. 1981 Mar;78(3):1619–1623. doi: 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Chow L. T., Dugaiczyk A., Hedgpeth J., Goodman H. M. DNA substrate site for the EcoRII restriction endonuclease and modification methylase. Nat New Biol. 1973 Jul 11;244(132):40–43. doi: 10.1038/newbio244040a0. [DOI] [PubMed] [Google Scholar]
- Brown N. L., Smith M. A general method for defining restriction enzyme cleavage and recognition sites. Methods Enzymol. 1980;65(1):391–404. doi: 10.1016/s0076-6879(80)65050-2. [DOI] [PubMed] [Google Scholar]
- Chaconas G., van de Sande J. H. 5'-32P labeling of RNA and DNA restriction fragments. Methods Enzymol. 1980;65(1):75–85. doi: 10.1016/s0076-6879(80)65012-5. [DOI] [PubMed] [Google Scholar]
- Drahovsky D., Boehm T. L. Enzymatic DNA methylation in higher eukaryotes. Int J Biochem. 1980;12(4):523–528. doi: 10.1016/0020-711x(80)90002-6. [DOI] [PubMed] [Google Scholar]
- Dubin D. T., HsuChen C. C. The 3'-terminal region of mosquito mitochondrial small ribosomal subunit RNA: sequence and localization of methylated residues. Plasmid. 1983 May;9(3):307–320. doi: 10.1016/0147-619x(83)90008-2. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Gama-Sosa M. A., Carreira L. H., Ljungdahl L. G., Kuo K. C., Gehrke C. W. DNA methylation in thermophilic bacteria: N4-methylcytosine, 5-methylcytosine, and N6-methyladenine. Nucleic Acids Res. 1985 Feb 25;13(4):1399–1412. doi: 10.1093/nar/13.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M., Wang R. Y. 5-Methylcytosine in eukaryotic DNA. Science. 1981 Jun 19;212(4501):1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- Frederick C. A., Grable J., Melia M., Samudzi C., Jen-Jacobson L., Wang B. C., Greene P., Boyer H. W., Rosenberg J. M. Kinked DNA in crystalline complex with EcoRI endonuclease. Nature. 1984 May 24;309(5966):327–331. doi: 10.1038/309327a0. [DOI] [PubMed] [Google Scholar]
- Garfin D. E., Goodman H. M. Nucleotide sequences at the cleavage sites of two restriction endonucleases from Hemophilus parainfluenzae. Biochem Biophys Res Commun. 1974 Jul 10;59(1):108–116. doi: 10.1016/s0006-291x(74)80181-6. [DOI] [PubMed] [Google Scholar]
- Hanawalt P. C., Cooper P. K., Ganesan A. K., Smith C. A. DNA repair in bacteria and mammalian cells. Annu Rev Biochem. 1979;48:783–836. doi: 10.1146/annurev.bi.48.070179.004031. [DOI] [PubMed] [Google Scholar]
- Janulaitis A., Klimasauskas S., Petrusyte M., Butkus V. Cytosine modification in DNA by BcnI methylase yields N4-methylcytosine. FEBS Lett. 1983 Sep 5;161(1):131–134. doi: 10.1016/0014-5793(83)80745-5. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McClelland M. The effect of site specific methylation on restriction endonuclease cleavage (update). Nucleic Acids Res. 1983 Jan 11;11(1):r169–r173. doi: 10.1093/nar/11.1.235-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller A., Gabriels J. E., Lafer E. M., Nordheim A., Rich A., Stollar B. D. Monoclonal antibodies recognize different parts of Z-DNA. J Biol Chem. 1982 Oct 25;257(20):12081–12085. [PubMed] [Google Scholar]
- Ohmori H., Tomizawa J. I., Maxam A. M. Detection of 5-methylcytosine in DNA sequences. Nucleic Acids Res. 1978 May;5(5):1479–1485. doi: 10.1093/nar/5.5.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Friedman J. DNA methylation and its possible biological roles. Prog Nucleic Acid Res Mol Biol. 1981;25:33–52. doi: 10.1016/s0079-6603(08)60482-1. [DOI] [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Roberts R. J. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 1984;12 (Suppl):r167–r204. doi: 10.1093/nar/12.suppl.r167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976 Mar;73(3):804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stawinski J., Hozumi T., Narang S. A., Bahl C. P., Wu R. Arylsulfonyltetrazoles, new coupling reagents and further improvements in the triester method for the synthesis of deoxyribooligonucleotides. Nucleic Acids Res. 1977 Feb;4(2):353–371. doi: 10.1093/nar/4.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugisaki H. Recognition sequence of a restriction endonuclease from Haemophilus gallinarum. Gene. 1978 Feb;3(1):17–28. doi: 10.1016/0378-1119(78)90004-5. [DOI] [PubMed] [Google Scholar]
- Tran-Dinh S., Taboury J., Neumann J. M., Huynh-Dinh T., Genissel B., Gouyette C., Igolen J. B and Z double helical conformations of d-(m5C-G-C-G-m5C-G) in aqueous solution. FEBS Lett. 1983 Apr 18;154(2):407–410. doi: 10.1016/0014-5793(83)80192-6. [DOI] [PubMed] [Google Scholar]
- Vardimon L., Rich A. In Z-DNA the sequence G-C-G-C is neither methylated by Hha I methyltransferase nor cleaved by Hha I restriction endonuclease. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3268–3272. doi: 10.1073/pnas.81.11.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarling D. A., Arndt-Jovin D. J., Robert-Nicoud M., McIntosh L. P., Thomae R., Jovin T. M. Immunoglobulin recognition of synthetic and natural left-handed Z DNA conformations and sequences. J Mol Biol. 1984 Jul 5;176(3):369–415. doi: 10.1016/0022-2836(84)90495-9. [DOI] [PubMed] [Google Scholar]