Abstract
Neurotensin receptor-1 (NTSR-1) is a G-protein coupled receptor (GPCR) that has been recently identified as a mediator of cancer progression. NTSR-1 and its endogenous ligand, neurotensin (NTS), are co-expressed in several breast cancer cell lines and breast cancer tumor samples. Based on our previously published study demonstrating that intact structured membrane microdomains (SMDs) are required for NTSR-1 mitogenic signaling, we hypothesized that regulated receptor palmitoylation is responsible for NTSR-1 localization and signaling within SMDs upon NTS stimulation. Site-directed mutagenesis and pharmacological strategies were utilized to assess NTRS-1 post-translational modifications in an overexpression cell model (HE K293T) as well as a native breast cancer cell model (MDA-MB-231). NTSR-1 palmitoylation was confirmed by multiple chemical and fluororadiographic methodologies. NTSR-1 glycosylation was confirmed by pharmacological (tunicamycin) and chemical (PGNaseF and O-type glycosidase) approaches. Physiological correlates including cell viability (MTS assay), apoptosis (caspase-3/7 assay) and ERK phosphorylation were utilized to assess the consequences of NTRS-1 palmitoylation. The interaction between palmitoylated NTRS-1 and Gαq/11 within SMDS was confirmed with immunopreciptation analysis of detergent-free isolated fractions of caveolin-rich microdomains. We identified dual-palmitoylation at Cys381 and Cys383 of endogenously-expressed NTSR-1 in MDA-MB-231 breast adeno-carcinomas as well as exogenously-expressed NTSR-1 in HEK293T cells (which do not normally express NTSR-1). Pharmacological inhibition of NTSR-1 palmitoylation in MDA-MB-231 cells as well as NTSR-1-expressing HEK293T cells diminished NTS-mediated ERK 1/2 phosphorylation. Additionally, NTSR-1 mutated at Cys381 and Cys383 showed diminished ERK1/2 stimulation and reduced ability to protect HEK293T cells against apoptosis induced by serum starvation. Mechanistically, mutated C381,383S-NTSR-1 showed reduced ability to interact with Gαq/11 and diminished localization to structured membrane microdomains (SMDs), where Gαq/11 preferentially resides. We also demonstrated that only glycosylated isoforms of NTRS-1 localize within SMDs by palmitotylation. Collectively, our data establish palmitoylation as a novel pharmacological target to inhibit NTSR-1 mitogenic signaling in breast cancer cells.
Keywords: neurotensin, palmitoylation, G protein coupled receptor, lipid rafts, glycosylation
Introduction
The overexpression of the G-protein coupled neurotensin receptor, NTSR-1, and its endogenous ligand, NTS, plays a key role in the development and progression of several types of tumors including breast,1 pancreas,2 prostate,3 colon4 and lung5 cancers. In invasive ductal breast carcinomas, NTS expression is regulated by estrogen.1 Moreover, NTSR-1 expression level in breast cancer is correlated with the Scarff-Bloom-Richardson (SBR) grade, tumor size and tendency to metastasize to lymph nodes.1 Additionally, patients with low NTSR-1 expression had better prognosis relative to those with high expression levels.1 Therefore, identification of novel pharmacological approaches to inhibit NTSR-1 mitogenic signaling is needed in breast cancers.
Palmitoylation is the post-translational addition of a 16-carbon fatty acid, palmitate, to a cysteine residue of a protein via a thioester or amide bond.6 Unlike myristoylation and isoprenylation, palmitoylation is a dynamic, reversible process, which allows for regulation of protein lipophilicity during signal transmission.6 Rhodopsin receptor was the first G-protein coupled receptor (GPCR) to be identified as a target for palmitoylation.7 GPCR palmitoylation alters receptor conformation and thereby, regulates the interactions of the receptor with specific downstream effectors.8 A palmitoylation/depalmitoylation cycle upon stimulation was observed for several GPCRs, such as the D1 dopamine,9–11 β2-adrenergic12 and α2A-adrenergic receptors.13 Palmitoylation of chemokine (C-C motif) receptor 5 CCR5 14 and A1 adenosine receptors15 was found to be necessary for receptor delivery to the plasma membrane, while non-palmitoylated receptors were degraded. In addition to receptor palmitoylation, downstream effectors such as heterotrimeric G protein subunits are also subject to several posttranslational modifications through lipidation. For example, Gαq/11 and Gαs palmitoylation is required for membrane anchorage and interaction with GPCRs.16
To date, over 20 putative mammalian palmitoyl acyl transferases (PAT) characterized by the presence of DHHC-cysteine-rich domain have been identified. Recent studies suggest that targeting PATs could lead to the development of novel anti-neoplastic chemotherapeutic agents.17 Although, characterization of the specific PATs that are responsible for GPCR palmitoylation are still not well defined, there are currently several lipid-based and small molecule based-experimental PAT inhibitors that can serve as lead compounds for the development of more specific inhibitors.18
We previously published that intact structured membrane micordomains (SMDs) are required for NTSR-1 signaling and interaction with Gαq/11.19 Specifically, methyl-β-cyclodexrtrin-mediated cholesterol depletion abolished NTSR-1-dependent MAPK activation and the effect was rescued through subsequent cholesterol repletion. These studies provided the rationale to now explore the biophysical and biochemical mechanisms by which SMDs regulate NTSR-1 mitogenic signaling.19 We now report that NTSR-1 is palmitoylated and this post-translational modification is required for optimal receptor signaling within SMDs. In this study, we identify NTSR-1 as a target for PAT and we investigate the functional and mechanistic roles of NTSR-1 palmitoylation in receptor-mediated pro-survival signaling. We validate the potential of utilizing PAT inhibitors to selectively disrupt NTSR-1 mitogenic signaling in breast cancer.
Results
NTSR-1 is palmitoylated in MDA-MB-231 breast cancer cells.
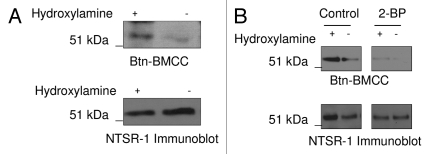
We initially identified Cys 381 and Cys 383 as putative palmitoylation sites based on published sequences. To test the hypothesis that the endogenously expressed NTSR-1 in MDA-MB-231 breast cancer cells is palmitoylated, we utilized a non-radioactive fatty acyl exchange labeling assay. This assay assesses palmitoylation via cleavage of palmitoyl moieties with hydroxyl amine (NH2OH) after blocking all exposed free cysteine residues with N-ethylmaleimide (NEM). To confirm palmitoylation of endogenous NTRS-1, we identified basal-palmitoylation by visualizing NH2OH exposed cysteine(s) conjugation with biotin (Btn-BMCC) (Fig. 1A, top). As an internal negative control, minimal signal was observed for the immunoprecipitated NTSR-1 in the absence of NH2OH. Figure 1A (bottom) demonstrated equal immunoprecipiation of the 54 kD NTSR-1 protein in the presence or absence of NH2OH To further validate the results of this non-radioactive fatty acyl exchange labeling assay that characterized basal palmitoylation of immunoprecipitated proteins, we tested if 2-bromopalmitate (2-BP) pre-treatment would inhibit NTSR-1 palmitoylation. 2-BP is a putative inhibitor of palmitoyl acyl transferases.20 Treatment of MDA-MB-231 cells with 2-BP for 10 min resulted in the disappearance of the band visualized by conjugation with biotin (Fig. 1B, top). Equal protein loading was again confirmed using anti-NTSR-1 antibody (Fig. 1B, bottom). Collectively, these data clearly demonstrate that endogenous NTSR-1 protein is palmitoylated under basal conditions in a breast cancer cell line.
Figure 1.
Endogenously expressed NTSR-1 in breast cancer cells is palmitoylated. (A) MDA-MB-231 cells were seeded in 10 cm plates followed by NTSR-1 immunoprecipitation. After receptor immunoprecipitation, free cysteine residues were protected using Nethylmaleimide (NEM) followed by NH2OH-mediated cleavage of palmitate groups. The free cysteine residues were then labeled with biotin using biotin-conjugated 1-biotin-amido-4-[4-(maleimidomethyl)cyclohexanecarboxamido]butane (BMCC) and immunoprecipitates were analyzed using SDS electrophoresis using an avidin antibody to detect the biotinylated labeled palmitoylated residues. Representative blots from three independent experiments. (B) MDA-MB-231 cells were seeded in 10 cm plates. After 24 h, cells were treated with 10 µM 2-BP followed by analysis using fatty acyl exchange labeling assay as described above. Representative blots from two independent experiments. As described previously in reference 19, under these immunoprecipitation conditions, the high molecular weight NTSR-1 isoform (∼54 kDa) is preferentially precipitated.
Generation and expression of wild type and C381, 383S-mutated NTSR-1 in HEK293T cells.
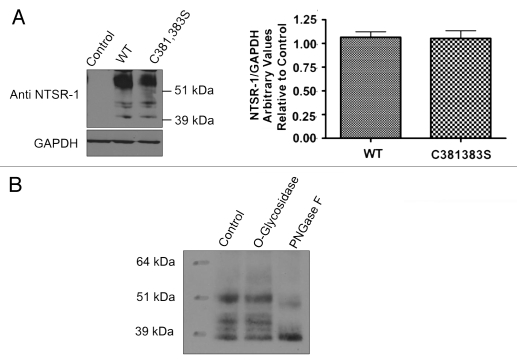
In order to further confirm that NTSR-1 is palmitoylated, we conducted site-directed mutagenesis studies. Cys 381 and Cys 383 were mutated to serine, generating double mutant NTSR-1 constructs. HEK293T cells, which do not normally express the receptor, were transfected with the constructs and grown for 48 h before protein gel blot analysis was performed to measure protein expression. The expression levels of the different constructs were comparable as shown in Figure 2A. No expression was observed with untransfected control lysates. Consistent with our previously published studies in reference 19, fully mature glycosylated receptor isoforms were detected at molecular weight greater than 51 kDa, while non-mature isoforms were detected at molecular weight equal or greater than 39 kDa. To confirm and validate the specificity of glycosylated NTSR-1 species, we assessed N- and O-type glycosylation via PNGaseF enzyme and O-glycosidase, respectively (Fig. 2B). We observed a shift in the high molecular weight isoforms of NTSR-1 for PNGaseF but not O-glycosidase treatment. Taken together, these data suggest that wild type as well as putative palmitoylation-deficient mutant NTSR-1 constructs can be expressed and N-glycosylated in HEK293T cells.
Figure 2.
NTSR-1 proteins can be overexpressed and glycosylated in HEK293T cells. (A) HEK293T cells were seeded in 10 cm plates and allowed to attach before transfection with NTSR-1 constructs using lipofectamine 2000. After 48 h, the cells were lysed using RIPA buffer and samples containing equal total protein were analyzed using SDS gel electrophoresis. The blots were probed with anti-NTSR-1 antibody and quantified using Image-J software Representative plots from three separate experiments. (B) Transfected HEK293T cells were lysed using RIPA lysis buffer and NTSR-1 was immunoprecipitated using anti-NTSR-1 antibody. The protein was subjected to PGNaseF or O-type glycosidase and samples were analyzed using SDS-gel electrophoresis. Similar results were obtained when the immunoprecipitation experiment was performed on endogenously expressed NTSR-1 in MDA-MB-231 cells (data not shown); representative blots from three separate experiments.
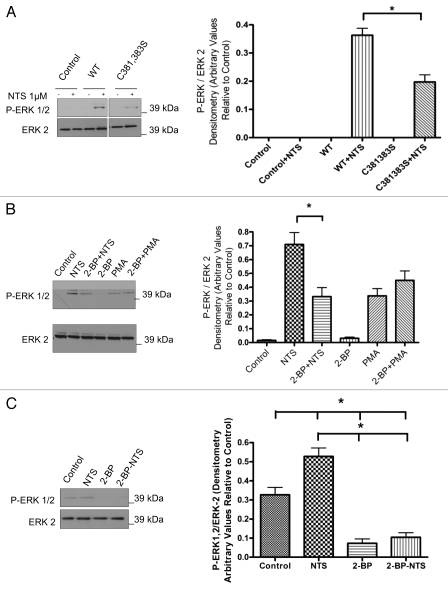
NTS treatment induces palmitoylation of WT-NTSR-1, but not C381,383S-NTSR-1 expressed in HEK293T cells.
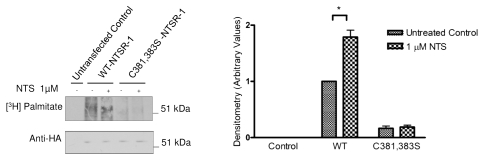
To further confirm if NTSR-1 is palmitoylated at specific cysteine residues, we utilized a metabolic radiolabeling technique using (3H) palmitate. As shown in Figure 3, WT-NTSR-1, but not C381,383S-NTSR-1, is palmitoylated under basal conditions. Additionally, stimulation of the receptor with 1 µM NTS for 15 min resulted in an approximate 50% increase in WT-NTSR-1, but not C381,383S-NTSR-1 receptor palmitoylation, suggesting that this post-translational modification may serve as a regulatory mechanism for NTSR-1.
Figure 3.
Cysteine 381,383 in NTSR-1 are required for optimal palmitoylation. HEK293T cells transfected with HA tagged WT-NTSR-1 and C381,383S NTSR-1 were serum-starved for 24 h before 4 h (3H) palmitate labeling. Cell lysates were subjected to immunoprecipitation using anti-HA antibody. One portion of the samples was analyzed using SDS-gel electrophoresis to determine equal expression level of NTSR-1 constructs. The other portion was analyzed using fluororadiography. The blots were sprayed with ENHANCE and exposed to X-ray film at −80°C for 4–6 weeks. Representative plots from three separate experiments. In this particular experiment, we used a HA tagged construct to increase the efficiency of receptor immunoprecipitation relative to using anti-NTSR-1. Mean ± SE, *p < 0.05.
NTS treatment promotes viability of HEK293T cells expressing WT-NTSR-1 but not C381,383S-NTSR-1.
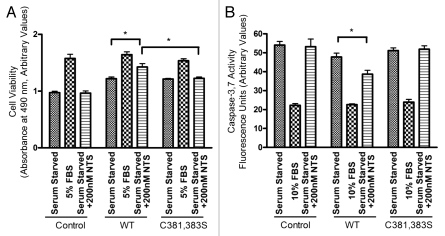
The physiological significance of NTSR-1 mutation at putative palmitoylation sites was evaluated using a MTS cell survival assay. Serum starved HEK293T expressing WT-NTSR-1 that were treated with 200 nM NTS exhibited improved cell viability relative to untransfected HEK293T cells and HEK293T expressing C381,383S-NTSR-1 (Fig. 4A). 10% FBS treatment was used as a positive control in these experiments. These data suggest that these two palmitoylation sites, C381 and C383, play a critical role in receptor pro-survival signaling.
Figure 4.
NTSR-1 palmitoylation is required for NTS-mediated pro-survival activity. (A) Transfected HEK293T cells were seeded in 96 well plates at a density of 4,000 cells/well. Cells were serum-starved for 24 h before stimulation with 5% FBS, or 200 nM NTS every 24 h for 2 d. Cell viability was determined using MTS assay, where absorbance was measured using a plate reader at 495 nm. Results were obtained from n = three separate experiments. Mean ± SE, *p < 0.05. (B) Transfected HEK293T cells were seeded in 96 well plates at density of 400 cells/well. Cells were serum-starved for 24 h before stimulation with 5% FBS, or 200 nM NTS. We used a concentration of 200 nM in these longer term experiments to avoid rapid receptor desensitization. Caspase-3/7 activity was measured using a fluorescence plate reader (560Ex/590Em). Results were obtained from n = three separate experiments. Mean ± SE, *p < 0.05.
NTS treatment protects HEK293T cells expressing WT-NTSR-1 but not C381,383S-NTSR-1 from apoptosis induced by serum starvation.
To further confirm the importance of NTSR-1 palmitoylation for NTS-mediated pro-survival function, we analyzed caspase-3/7 activity after 48 h serum starvation (Fig. 4B). HEK293T cells transfected with WT-NTSR-1 and treated with 200 nM NTS showed less apoptosis relative to untreated cells as well as cells transfected with C381, 383S-NTSR-1. As a negative control, 10% FBS treated cells had minimal caspase-3/7 activity. Collectively, these data suggest that palmitoylation of NTSR-1 at C381 and C383 are necessary to protect against stress-triggered (serum starvation) apoptosis.
WT-NTSR-1, but not C381,383S-NTSR-1 mutant, efficiently mediates NTS-induced ERK 1/2 phosphorylation.
As NTS-induced MAP kinase activity is associated with proliferation and/or survival of breast cancer cells, we next decided to study the importance of palmitoylation for NTSR-1 stimulation of the MAP kinase pathway. After transfecting HEK293T cell with WT-NTSR-1 and C381,383S-NTSR-1, we stimulated the cells using 1 µM NTS for 15 min. As shown in Figure 5A, only WT-NTSR-1 efficiently stimulated ERK phosphorylation compared with an approximate 50% reduction for mutant receptor transfection. These results further support the biochemical importance of NTSR-1 palmitoylation for efficient pro-mitogenic/prosurvival receptor signaling.
Figure 5.
Palmitoylation of glycosylated NTRS-1 isoforms is required for NTS-mediated MAP K mitogenic signaling. (A) HEK293T cells were seeded in 6 well plates and were then transfected with NTSR-1 constructs the next day. After 24 h, the cells were then serum-starved for another 24 h before 1 µM NTS stimulation for 15 min. The cell lysates were then analyzed using SDS-gel electrophoresis and blots were probed using P-ERK1/2 antibody. All blots were generated from the same gel and cropped in order to eliminate lanes irrelevant to this study. Representative blots from three separate experiments. Mean ± SE, *p < 0.05. (B) HEK293T cells were seeded in 6 well plates and were transfected with NTSR-1 constructs the next day. After 24 h, the cells were then serum-starved for another 24 h before 10 µM 2-BP treatment followed by 1 µM NTS stimulation for 15 min. The cell lysates were then analyzed using SDS-gel electrophoresis and blots were probed using a P-ERK1/2 antibody. Representative blots from three separate experiments. Mean ± SE, *p < 0.05. (C) MDA-MB-231 breast cancer cells were seeded in 6 well plates and serum starved for 48 h before 1 µM NTS stimulation for 15 min. The cells were lysed using RIPA buffer and equal total protein samples were analyzed using SDS gel electrophoresis. The blots were probed with P-ERK1/2 antibody and quantified using Image-J software. Representative blots from three separate experiments. Mean ± SE, *p < 0.05.
2-Bbromopalmitate inhibits WT-NTSR-1-mediated, but not PMA-mediated, activation of the MAPK kinase pathway.
To confirm the previous site-directed mutagenesis experimental strategies; we next utilized a pharmacological approach to target palmitoylation-dependent NTSR-1 mitogenic signaling. We utilized 2-BP, which has been shown to be an inhibitor of palmitoyl acyl transferases.20–22 Pretreatment of HEK293T cells transfected with WT-NTSR-1 with 10 µM 2-BP for 30 min significantly reduced NTS-mediated ERK 1/2 phosphorylation (Fig. 5B). In order to ensure the specificity of this effect, we tested 2-BP upon PMA-mediated ERK phosphorylation. 2-BP did not inhibit PMA-induced ERK phosphorylation, suggesting that the 2-BP effect is at the receptor level and not due to non-specific disruption of downstream signaling pathways.
Pharmacological inhibition of NTSR-1 palmitoylation in MDA-MB-231 cells disrupts receptor-mediated mitogenic signaling.
To further define the role of NTSR-1-palmitoylation in a non-overexpression cell system, we examined the pharmacological effect of 2-BP on NTSR-1-mediated-enhancement of the constitutively active MAP kinase signaling pathway in MDA-MB-231 breast cancer cells that endogenously express NTSR-1. Data in Figure 5C demonstrate that 2-BP-pretreatment abrogated both basal and NTS-strengthened P-ERK activity. These results further confirm that the endogenously expressed receptor is indeed palmitoylated and also demonstrate the potential of exploiting PATs inhibitors as a novel pharmacological approach to ameliorate NTSR-1-mediated mitogenic signaling in breast cancer. These 2-BP-treatment conditions were not toxic as observed by data in Figure 5B, where PMA-mediated stimulation of ERK is not inhibited by 2-BP as well as by microscopic examination of cell viability (data not shown).
Palmitoylation of NTSR-1 is required for receptor interaction with Gαq/11.
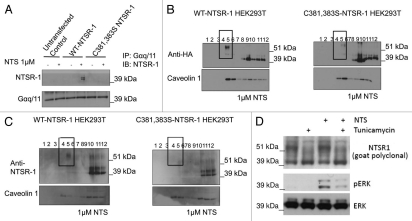
To further identify the biochemical mechanism responsible for regulated NTSR-1 palmitoylation to stimulate the MAP kinase pathway, we next investigated NTSR-1/Gαq/11 protein-protein interaction, which we have shown previously to be essential for efficient induction of ERK phosphorylation by NTS.19 Figure 6A reveals that the palmitoylation-deficient C381, 383S-NTSR-1 mutant failed to interact with Gαq/11 following 15 min stimulation with 1 µM NTS. These co-IP data suggest that palmitoylation is essential for efficient NTSR-1/Gαq/11 coupling. In the next series of experiments, we investigated the biophysical mechanism by which non-palmitoylated NTSR-1 mutants may have impaired ability to co-localize within SMDs, where Gαq/11 mainly resides.19
Figure 6.
NTSR-1 palmitoylation is required for optimal interaction with Gαq/11A within SMDs. (A) HEK293T cells transfected with WT-NTSR-1 and C381,383S NTSR-1 were serum-starved for 24 h before 15 min stimulation with 1 µM NTS. The cells were then lysed using TritonX-100 buffer and immunoprecipitated using anti-Gαq/11 antibody. The immunoprecipitates were immunoblotted with anti-NTSR-1. Representative blots from n = three separate experiments. (B and C) HEK293T cells transfected with HA-tagged (B) or non-HA-tagged (C) WT-NTSR-1 or C381,383S NTSR-1 were serumstarved for 24 h before 15 min stimulation with 1 µM NTS. The lysates were then subjected to detergent free-sucrose gradient fractionation and the fractions were immunoblotted with either anti-HA (B) or anti-NTSR-1 (C). All blots were re-probed with anti-caveolin-1 to assess equal loading as well as to identify caveolin-enriched SMDs. (D) HEK293T cells transfected with WT-NTSR-1 were pretreated with 1 µg/ml tunicamycin for 24 h before stimulation with 1 µM NTS for an additional 15 min. Representative blots from n = three separate experiments.
Palmitoylation of NTSR-1 is required for efficient receptor localization to SMDs.
To test the hypothesis that the reduced NTSR-1/Gαq/11 interaction is due to the diminished ability of the receptor to reside within SMDs for extended intervals, we studied NTSR-1-membrane localization using detergent-free sucrose gradient fractionation. Data in Figure 6B demonstrate that palmitoylation-deficient NTSR-1 localization within caveolinenriched SMDs is diminished upon NTS stimulation, relative to WT-NTSR-1. In particular, we showed reduction in mutated but not wild type NTSR-1 localization within SMDs utilizing HA-tagged (Fig. 6B) constructs. This reduction of glycosylated, but palmitoylation-deficient NTSR-1 within the caveolin enriched domains (fractions 4 and 5) was quantified in n = three separate experiments and localization was reduced from 25 ± 4 to 4 ± 4%. To confirm these experiments, we utilized a non-HA-tagged construct (Fig. 6C) and again visualized decreased localization within caveolin-enriched domains with the double cysteine mutant using an anti-NTSR-1 antibody. Equal loading of the gels for both experiments was confirmed by comparable caveolin levels. Moreover, the data clearly show that only the glycosylated NTSR-1 isoforms localize within SMD fractions 4 and 5. The non-mature, non-glycosylated NSTR-1 isoforms preferentially reside within the non-SMDs fractions 8–12. To further establish that NTSR-1 glycosylation is critical for NTS-mediated signaling (ERK phosphorylation), we pretreated wild type HA-tagged NTSR-1 expressing HEK293T cells with tunicamycin, a well known glycosylation inhibitor. As shown in Figure 6D, 1 µg/ml tunicamycin treatment for 24 h resulted in reduction of the ∼54 kDa band corresponding to glycosylated NTSR-1 isoform accompanied by an increase in the 43 kDa band which represent the non-mature NTSR-1. Additionally, the reduction in NTSR-1 glycosylation was accompanied by significant reduction in NTS-mediated ERK phosphorylation. Taken together, these results suggest that both palmitoylation and glycosylation of NTSR-1 could be necessary for efficient SMDs localization, interaction with Gαq/11 and subsequent mitogenic signaling.
Discussion
The need to identify novel pharmacological approaches to inhibit NTSR-1 mitogenic signaling in cancer is justified by recent studies demonstrating NTSR-1 overexpression correlating with tumor metastatic potential and resistance to anti-neoplastic chemotherapeutic agents.1–5 Our previous findings19 revealed a unique biophysical mechanism that has the potential to yield a more selective targeting approach for NTSR-1 mediated oncogenesis. We specifically demonstrated that NTS-stimulated NTSR-1 preferentially localizes within SMDs, to interact with Gαq/11 subunits. Thus, we hypothesized that agonist-enhanced receptor palmitoylation is the primary signal that targets the receptor to these domains, increasing residence time and facilitating interaction with Gαq/11. Our data suggest that NTS augments basal NTSR-1 palmitoylation which increases the efficiency and potency of MAP kinase signaling through NTSR-1. Palmitoyltation is required for efficient receptor-mediated mitogenic-signaling within SMDs possibly by enhancing the interaction with Gαq/11, which mainly resides within SMDs. Thus, our studies potentially argue for the use of PAT inhibitors as a new therapeutic modality for NTSR-1 expressing cancers. The mechanism(s) by which NTS induces NTSR-1 palmitoylation are not known, however, it is possible that NTS/NTSR-1 binding may fully expose C-terminal palmitoylation sites to interact with PAT(s).6 NTSR-1-palmitoylation could further lead to formation of an additional fourth intra-cellular loop that result in a conformational change, which may serve as a docking site for critical downstream protein/protein interaction.23
We have combined both site-directed mutagenesis and pharmacological approaches to study NTSR-1 palmitoylation. Using a pharmacological approach (2-BP) and a molecular approach (C381,383S mutations), we demonstrated similar effects on ERK phosphorylation in two different cell lines, MDA-MB-231 and HEK-293T cells. We chose 2-BP, a lipid based inhibitor, due to its ability to irreversibly inhibit palmitoyl acytransferase activity of all PATs.20 In order to exclude the possibility of off-target effects that would disrupt signal transduction at downstream levels, we showed that PMA-mediated MAP kinase signaling remains intact, suggesting a direct effect of PAT on NTSR-1 All of our mutants were equally expressed and glycosylated in HEK293T model system. Documentation of physiological dysfunction by both molecular and pharmacological strategies strongly suggests that NTSR-1 palmitoylation is a critical event for efficient NTS-mediated mitogenesis and survival.
Our data suggest that there is potential cross talk between palmitoylation and glycosylation post-translational modifications of NTSR-1. Specifically, we demonstrate that only glycosylated isoforms of NTSR-1 localize within SMDs by palmitoylation. Moreover, inhibition of glycosylation with tunicamycin, as well as, palmitoylation with 2-BP reduced NTS-induced ERK signaling. Our studies further suggest that palmitoylation of glycosylated NTSR-1 can affect downstream G protein signaling via stability and/or membrane micro-localization of the receptor. Although our data suggest that NTSR-1 localization within SMDs is reduced when receptor palmitoylation is impaired, other mechanisms cannot be excluded. For example, palmitoylationdeficient A1 adenosine and CCR5 receptors as well as the palmitoylation-deficient Rous sarcoma virus glycoprotein are more susceptible for proteolysis.24,25 Thus, the diminished NTSR-1 protein level within caveolin-enriched SMD fractions 4 and 5 could also be attributed to enhanced protein degradation.
Enhanced protein palmitoylation is involved in the pathogenesis of many disease conditions such as cancer, Huntington disease, immune diseases and various cardiovascular diseases.26 Therefore, identification of functional roles of protein palmitoylation at the molecular level can further validate palmitoyl acyltransferases as strategic targets for the development of novel pharmacological approaches for disease treatment. Accumulating evidence in the literature suggest that PATs may have substrate specificity.27 For example, Singaraja et al. recently identified DHHC8 as the PAT which regulates ATP-binding cassette transporter (ABC-A1) through palmitoylation.28 They proposed that an increased DHHC8 protein levels could lead to an increase in ABCA-1 mediated lipid efflux. Similarly, our findings could enable selective targeting of the specific PAT(s) which regulate NTSR-1 functions using siRNA approach resulting in the desired selective pharmacological effects. In conclusion, we suggest that NTSR-1 palmitoylation is required for receptor localization and efficient signaling within SMDs. We also provide for the first time a novel pharmacological approach to inhibit NTSR-1 mitogenic signaling in cancer through inhibition of receptor palmitoylation. Our studies demonstrate that NTSR-1 plamitoylation is required for NTSR-1-mediated MAPK signaling and cellular proliferation in breast cancer cells. Mechanistically, we found that palmitoylation is critical for receptor interaction with Gαq/11 within structured membrane microdomains. Pharmacological targeting of PAT could be a viable approach to control breast cancer tumors which overexpress NTSR-1.
Materials and Methods
Materials.
Polyacrylamide gel electrophoresis gradient gels were purchased from Invitrogen (Carlsbad, CA), and enhanced chemiluminescence reagent was purchased from Thermo Fisher Scientific Inc. Human MDA-MB-231 breast adenocarcinoma cells and HEK293T were obtained from American Type Culture Collection (Manassas, VA) and grown at 37°C in RPMI 1640 medium and DMEM, respectively, supplemented with 10% fetal bovine serum (FBS). The MDA-MB-231 cell line is a highly aggressive metastatic, estrogen receptor-negative, epidermal growth factor receptor (EGFR) positive and NTSR-1 positive model of human breast cancer. NTS was purchased from Sigma-Aldrich and was dissolved in 0.05% acetic acid just before use in experiments. All antibodies were purchased from Santa Cruz Corp., CA. Peptide-N-glycosidase F (PNGaseF) and O-glycosidase were obtained from Sigma, St. Louis, MO. Phorbol ester (PMA) was obtained from Promega Inc. (Madison, WI). Human NTSR1 and HA-tagged cDNA was purchased from Missouri S&T cDNA Resource Center, Rolla, MO (Catalogue Number: NTSR100000). MTS [3-(4,5-dimethylthia zol-2-yl) -5- (3-carboxymethoxyphenyl) -2- (4-sulfophenyl)-2H-tetrazolium] cytotoxicity assay and caspase-3/7 apoptosis assay reagents were purchased from Promega. [9,10-3H] palmitate was purchased from American Radiolabeled Chemicals St. Louis, MO.
Site directed mutagenesis.
The coding region for human NTSR-1 was sub cloned into the EcoRI and XbaI restriction sites of the CMV promoter expression plasmids pCDNA3.1(+) and N-terminal HA-tagged pCDNA3.1(+). Cysteine residues 381 and 383 of NTSR-1 were mutated to serine residues by site directed PCR mutagenesis to generate C381,383S-NTSR-1 mutant. All sequences were verified by DNA sequencing (Molecular Genetics Core Facility, Pennsylvania State University, Hershey, PA).
PNGase F and O-glycosidase deglycosylation.
HEK293T cells were seeded in 10-cm plates and allowed to grow until 70% confluency and then transfected with 1 µg DNA using lipofectamine 2000 according to manufacturer recommendation. After 48 h, cells were lysed using modified lysis buffer [50 mmol/L TRIS-HCl [pH 7.5], 150 mmol/L NaCl, 1 mmol/L EGTA, 10 mmol/L MgCl2, 0.5% Triton X-100, protease inhibitor cocktail]. NTSR-1 was immunoprecipitated as previously described in reference 16. Immunoprecipitated protein was subjected to deglycosylation using PNGase F or O-glycosidase according to the manufacturer's recommended protocol. The samples were then analyzed using SDS-PAGE electrophoresis.
Protein gel blot analysis.
Transfected HEK293T cells were seeded at 4.0 × 105 cells/well in 6 well plates and grown overnight prior to 24 h serum starvation. Cells were then stimulated with 1 µM NTS or phorbol 12-myristate 13-acetate (PMA) for 15 min in the presence or absence of 2-bromopalmitate (2-BP0). Cells were washed once with cold PBS followed by the addition of 60 µl of cold lysis buffer (1% Triton X-100, 20 mM Tris, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 2 mM Na4P2O7, 1 mM glycerolphosphate, 1 mM Na3VO4 and 1 µg/ml leupeptin in ddH2O, pH 7.5) on ice. Cells were lysed for 15 min on ice, and cell lysate was harvested and centrifuged at 15,000 g for 15 min. 35 µg of protein were loaded in 4 to 12% precasted SDS-PAGE gradient gels and probed for P-ERK 1/2. Blots were stripped and re-probed for ERK 2 to demonstrate equal loading. Protein bands were visualized using enhanced chemiluminescence and quantified using Image-J software.
Metabolic radiolabeling.
HEK293T cells transiently transfected with HA-tagged WT-NTSR-1 and C381,383S-NTSR-1 constructs were grown in 10 cm plates till they reached 80% confluence then starved over night. 50 µCi/ml [3H] Plamitate was added to the cells for 4 h before stimulation with 1 µM NTS for 15 min. Following stimulation, cells were lysed using radioimmunoprecipitation (RIPA) buffer and NTSR-1 was immunoprecipitated using anti-HA antibody. Immunoprecipitated samples were then split into two parts. One part was analyzed by SDS-gel electrophoreses and the blots were probed with anti-HA antibody to ensure equal loading, while the remaining portions were analyzed using auto-radiography where the blots were sprayed with ENHANCE (PerkinElmer, Waltham MA) and the film was exposed for 6–8 weeks at −81°C.
MTS cell viability assay.
HEK293T cells transfected with WT-NTSR-1 or C381,383S-NTSR-1 were seeded in 96 well plates at a density of 4,000 cell/well. After 48 h, the cells were starved for 24 h and then treated with 200 nM NTS or 5% FBS for additional 24 h. Two hundred nanomolar NTS was added again for another 24 h before the addition of MTS reagent, in the presence of phenazine methosulfate (PMS). Cell viability was quantified according to the manufacturer protocol where absorbance was measured at 495 nm using a plate reader.
Caspase-3/7 assay apoptosis assay.
HEK293T cells transfected with WT-NTSR-1 or C381,383S-NTSR-1 were seeded in 96 well plates at a density of 4,000 cell/well. After 48 h, the cells were starved in the presence or absence of 200 nM NTS for additional 48 h. ApoOne caspase-3/7 assay (Promega, WI) was used to measure caspase-3/7 activity according to the manufacturer instructions.
Acyl exchange palmitoylation assay.29
MDA-MB-231 cells were grown in 10 cm plates till they reached 70–80% confluence then lysed using modified lysis buffer (50 mM TRIS-HCl, pH 7.5; 150 mM NaCl; 1 mM EGTA; 10 mM MgCl2; 0.5% Triton X-100; protease inhibitor cocktail). After measuring protein concentration, cell lysates were solubilized overnight at 4°C. Cell lysates were then centrifuged at 4°C at 15,000 g for 10 min to remove the insoluble material. NTSR-1 was immunoprecipitated over night using anti-NTSR-1 antibody. Gamma bind beads (GE, NJ) were added to the immunoprecipitate mixture and rotated over night at 4°C. The immunoprecipitates were washed with PBS for 15 min, lysis buffer for 1 h then two times with PBS for 15 min. 50 µl N-ethylmaleimide (NEM) (30 mg/ml) added to 700 ul PBS and the samples were rotated at 4°C overnight. The beads were then washed 4–5 times using PBS at a required pH of 7–7.5. After splitting each sample into 2,800 µl 1 M NH2OH (∼pH 7) was added to one sample of each pair while the other served as an internal negative control. The samples were then incubated rotating at 37°C for 3–5 h. The beads were then washed with PBS two to three times. 1-biotinamido-4-[4 (maleimidomethyl)cyclohexanecarboxamido] butane (BMCC) was added to each sample followed by incubation on a rotator overnight at room temperature. The beads were then washed twice with PBS and 1x loading dye was added. After heating at 37°C for 30 min, the samples were analyzed by SDS gel electrophoreses. The blots were probed with avidin antibody and visualized using enhanced chemiluminescence.
Detergent-free fractionation of caveolin-rich domains.17,30
In brief, transfected HEK293T cells were seeded at 4.0 × 105 cells/well in 100-mm plates and allowed to grow till they reached 80% confluency. Cells were then treated with 1 µM NTS (15 min) and then washed with ice-cold PBS twice. Sucrose gradient fractionation was performed according to Song et al. where the cells where lysed using sodium carbonate buffer, pH 10 and lysates were homogenized using a dounce homogenizer (10 strokes), followed by brief sonication. After 20 h centrifugation at 39,000 g, the fractionated samples were collected and analyzed using SDS-PAGE electrophoreses as described in Protein gel blot analysis. Detergent free fractionation strategies of caveolin-enriched domains offer the advantage of less artifactual, more reliable, preparations.31
Statistical analysis.
Differences among treatment groups were statistically analyzed using a two-tailed Student's t-test for statistical analyses. Where appropriate, one-way analysis of variance with Bonferroni multiple comparison post-hoc test and t-test analysis were performed using GraphPad Prism 4.0 software. A statistically significant difference was reported if p < 0.05 or less. Data are reported as mean ± SE from at least n = three separate experiments.
Acknowledgments
The project was funded by a grant to M.K. from the NIH (5R01HL076789-05).
Abbreviations
- NTSR-1
neurotensin receptor-1
- NTS
neurotensin
- GPCR
G protein coupled receptor
- SMD
structured membrane microdomains
- PAT
palmitoyl acyl transferase
References
- 1.Dupouy S, Viardot-Foucault V, Alifano M, Souazé F, Plu-Bureau G, Chaouat M, et al. The neurotensin receptor-1 pathway contributes to human ductal breast cancer progression. PLoS ONE. 2009;4:4223. doi: 10.1371/journal.pone.0004223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L, Friess H, Zhu Z, Graber H, Zimmermann A, Korc M, et al. Neurotensin receptor-1 mRNA analysis in normal pancreas and pancreatic disease. Clin Cancer Res. 2000;6:566–571. [PubMed] [Google Scholar]
- 3.Seethalakshmi L, Mitra SP, Dobner PR, Menon M, Carraway RE. Neurotensin receptor expression in prostate cancer cell line and growth effect of NT at physiological concentrations. Prostate. 1997;31:183–192. doi: 10.1002/(SICI)1097-0045(19970515)31:3<183::AIDPROS7>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 4.Gui X, Guzman G, Dobner PR, Kadkol SS. Increased neurotensin receptor-1 expression during progression of colonic adenocarcinoma. Peptides. 2008;29:1609–1615. doi: 10.1016/j.peptides.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 5.Goedert M, Reeve JG, Emson PC, Bleehen NM. Neurotensin in human small cell lung carcinoma. Br J Cancer. 1984;50:179–183. doi: 10.1038/bjc.1984.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Resh MD. Palmitoylation of ligands, receptors and intracellular signaling molecules. Sci STKE. 2006;2006:14. doi: 10.1126/stke.3592006re14. [DOI] [PubMed] [Google Scholar]
- 7.Karnik SS, Ridge KD, Bhattacharya S, Khorana HG. Palmitoylation of bovine opsin and its cysteine mutants in COS cells. Proc Natl Acad Sci USA. 1993;90:40–44. doi: 10.1073/pnas.90.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chini B, Parenti M. G-protein coupled receptors, cholesterol and palmitoylation: facts about fats. J Mol Endocrinol. 2009;42:371–379. doi: 10.1677/JME-08-0114. [DOI] [PubMed] [Google Scholar]
- 9.Jin H, Zastawny R, George SR, O'Dowd BF. Elimination of palmitoylation sites in the human dopamine D1 receptor does not affect receptor-G protein interaction. Eur J Pharmacol. 1997;324:109–116. doi: 10.1016/S0014-2999(97)00059-9. [DOI] [PubMed] [Google Scholar]
- 10.Jin H, Xie Z, George SR, O'Dowd BF. Palmitoylation occurs at cysteine 347 and cysteine 351 of the dopamine D[1] receptor. Eur J Pharmacol. 1999;386:305–312. doi: 10.1016/S0014-2999(99)00727-X. [DOI] [PubMed] [Google Scholar]
- 11.Ng GY, Mouillac B, George SR, et al. Desensitization, phosphorylation and palmitoylation of the human dopamine D1 receptor. Eur J Pharmacol. 1994;267:7–19. doi: 10.1016/0922-4106(94)90219-4. [DOI] [PubMed] [Google Scholar]
- 12.Loisel TP, Adam L, Hebert TE, Bouvier M. Agonist stimulation increases the turnover rate of beta 2AR-bound palmitate and promotes receptor depalmitoylation. Biochemistry. 1996;35:15923–15932. doi: 10.1021/bi9611321. [DOI] [PubMed] [Google Scholar]
- 13.Kennedy ME, Limbird LE. Palmitoylation of the alpha 2A-adrenergic receptor. Analysis of the sequence requirements for and the dynamic properties of alpha 2A-adrenergic receptor palmitoylation. J Biol Chem. 1994;269:31915–31922. [PubMed] [Google Scholar]
- 14.Blanpain C, Wittamer V, Vanderwinden JM, Boom A, Renneboog B, Lee B, et al. Palmitoylation of CCR5 is critical for receptor trafficking and efficient activation of intracellular signaling pathways. J Biol Chem. 2001;276:23795–23804. doi: 10.1074/jbc.M100583200. [DOI] [PubMed] [Google Scholar]
- 15.Gao Z, Ni Y, Szabo G, Linden J. Palmitoylation of the recombinant human A1 adenosine receptor: enhanced proteolysis of palmitoylation-deficient mutant receptors. Biochem J. 1999;342:387–395. doi: 10.1042/0264-6021:3420387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wedegaertner PB, Chu DH, Wilson PT, Levis MJ, Bourne HR. Palmitoylation is required for signaling functions and membrane attachment of Gqalpha and Gsalpha. J Biol Chem. 1993;268:25001–25008. [PubMed] [Google Scholar]
- 17.Ducker CE, Griffel LK, Smith RA, Keller SN, Zhuang Y, Xia Z, et al. Discovery and characterization of inhibitors of human palmitoyl acyltransferases. Mol Cancer Ther. 2006;5:1647–1659. doi: 10.1158/1535-7163.MCT-06-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Draper JM, Smith CD. Palmitoyl acyltransferase assays and inhibitors [Review] Mol Membr Biol. 2009;26:5–13. doi: 10.1080/09687680802683839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heakal Y, Kester M. Nanoliposomal short-chain ceramide inhibits agonist-dependent translocation of neurotensin receptor 1 to structured membrane microdomains in breast cancer cells. Mol Cancer Res. 2009;7:724–734. doi: 10.1158/1541-7786.MCR-08-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jennings BC, Nadolski MJ, Ling Y, Baker MB, Harrison ML, Deschenes RJ, et al. 2-Bromopalmitate and 2-[2-hydroxy-5-nitro-benzylidene]-benzo[b]thiophen-3-one inhibit DHHC-mediated palmitoylation in vitro. J Lipid Res. 2009;50:233–242. doi: 10.1194/jlr.M800270-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikic I, Planey S, Zhang J, Ceballos C, Seron T, von Massenbach B, et al. A live cell, image-based approach to understanding the enzymology and pharmacology of 2-bromopalmitate and palmitoylation. Methods Enzymol. 2006;414:150–187. doi: 10.1016/S0076-6879(06)14010-0. [DOI] [PubMed] [Google Scholar]
- 22.Webb Y, Hermida-Matsumoto L, Resh MD. Inhibition of protein palmitoylation, raft localization and T cell signaling by 2-bromopalmitate and polyunsaturated fatty acids. J Biol Chem. 2000;275:261–270. doi: 10.1074/jbc.275.1.261. [DOI] [PubMed] [Google Scholar]
- 23.Chini B, Parenti M. G-protein coupled receptors, cholesterol and palmitoylation: facts about fats. J Mol Endocrinol. 2009;42:371–379. doi: 10.1677/JME-08-0114. [DOI] [PubMed] [Google Scholar]
- 24.Ochsenbauer-Jambor C, Miller DC, Roberts CR, Rhee SS, Hunter E. Palmitoylation of the Rous sarcoma virus transmembrane glycoprotein is required for protein stability and virus infectivity. J Virol. 2001;75:11544–11554. doi: 10.1128/JVI.75.23.11544-54.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Percherancier Y, Planchenault T, Valenzuela-Fernandez A, Virelizier JL, Arenzana-Seisdedos F, Bachelerie F, et al. Palmitoylation-dependent control of degradation, life span and membrane expression of the CCR5 receptor. J Biol Chem. 2001;276:31936–31944. doi: 10.1074/jbc.M104013200. [DOI] [PubMed] [Google Scholar]
- 26.Planey SL, Zacharias DA. Palmitoyl acyltransferases, their substrates and novel assays to connect them [Review] Mol Membr Biol. 2009;26:14–31. doi: 10.1080/09687680802646703. [DOI] [PubMed] [Google Scholar]
- 27.Roth AF, Wan J, Bailey AO, et al. Global analysis of protein palmitoylation in yeast. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singaraja RR, Kang MH, Vaid K, Sanders SS, Vilas GL, Arstikaitis P, et al. Palmitoylation of ATP-binding cassette transporter A1 is essential for its trafficking and function. Circ Res. 2009;105:138–147. doi: 10.1161/CIRCRESAHA.108.193011. [DOI] [PubMed] [Google Scholar]
- 29.Drisdel RC, Alexander JK, Sayeed A, Green WN. Assays of protein palmitoylation. Methods. 2006;40:127–134. doi: 10.1016/j.ymeth.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 30.Song KS, Li S, Okamoto T, Quilliam LA, Sargiacomo M, Lisanti MP, et al. Co-purification and direct interaction of Ras with caveolin, an integral membrane protein of caveolae microdomains. Detergent-free purification of caveolae microdomains. J Biol Chem. 1996;271:9690–9697. doi: 10.1074/jbc.271.16.9690. [DOI] [PubMed] [Google Scholar]
- 31.Fox TE, Houck KL, O'Neill SM, Nagarajan M, Stover TC, Pomianowski PT, et al. Ceramide recruits and activates protein kinase C zeta [PKCzeta] within structured membrane microdomains. J Biol Chem. 2007;282:12450–12457. doi: 10.1074/jbc.M700082200. [DOI] [PubMed] [Google Scholar]