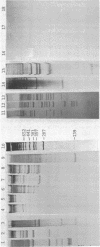
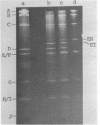
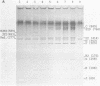
Abstract
Restriction endonucleases (13 out of 18 species used for the test) were certified to cleave single-stranded(ss)DNA. Such enzymes as AvaII, HaeII, DdeI, AluI, Sau3AI, AccII,TthHB8I and HapII were newly reported to cleave ssDNA. A model to account for the cleavage of ssDNA by restriction enzymes was proposed with supportive data. The essential part of the model was that restriction enzymes preferentially cleave transiently formed secondary structures (called canonical structures) in ssDNA composed of two recognition sequences with two fold rotational symmetry. This means that a restriction enzyme can cleave ssDNAs in general so far as the DNAs have the sequences of restriction sites for the enzyme, and that the rate of cleavage depends on the stabilities of canonical structures.
Full text
PDF



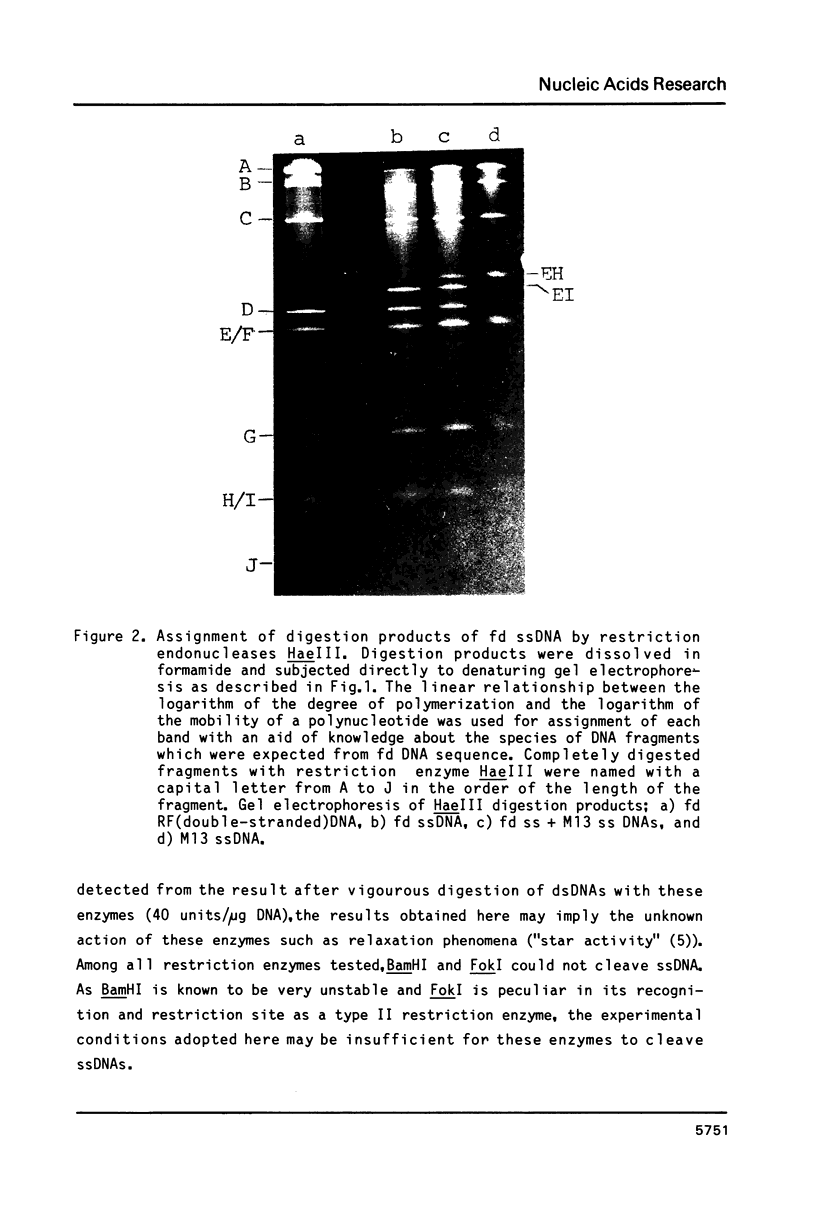

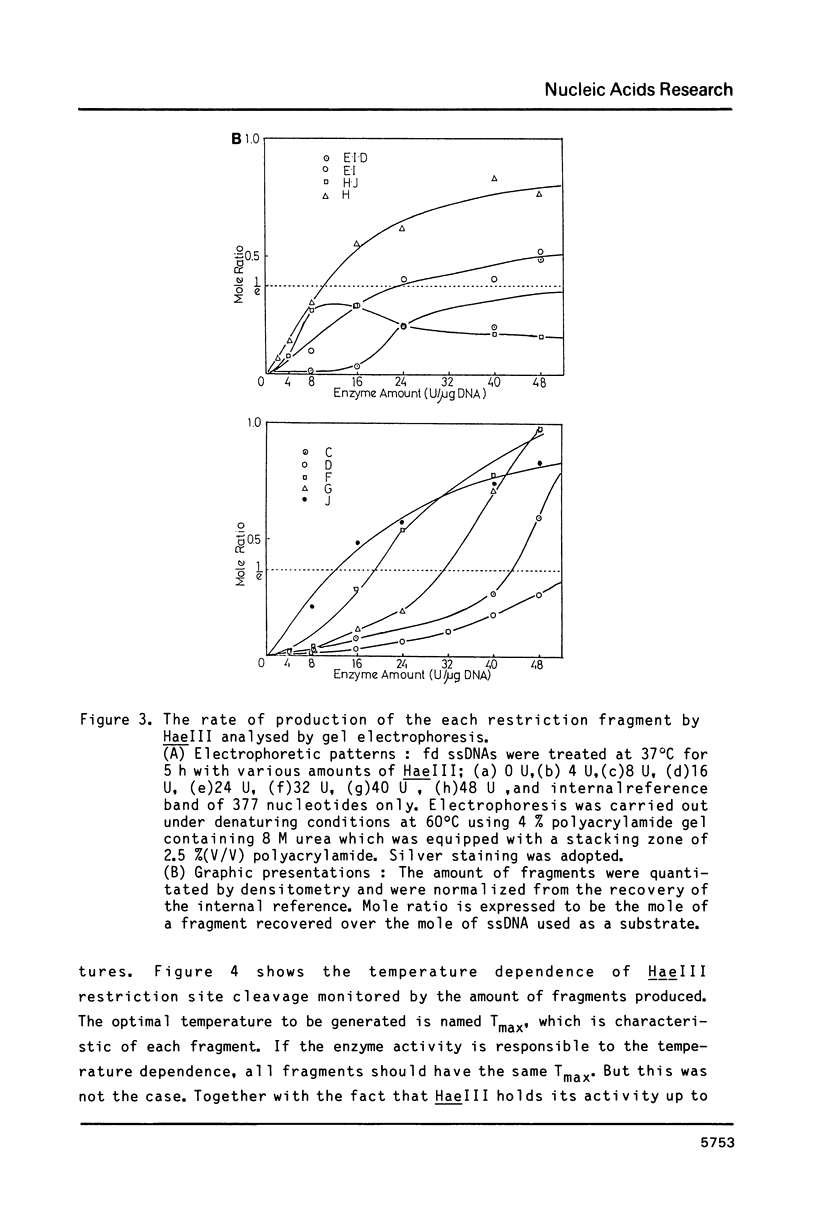
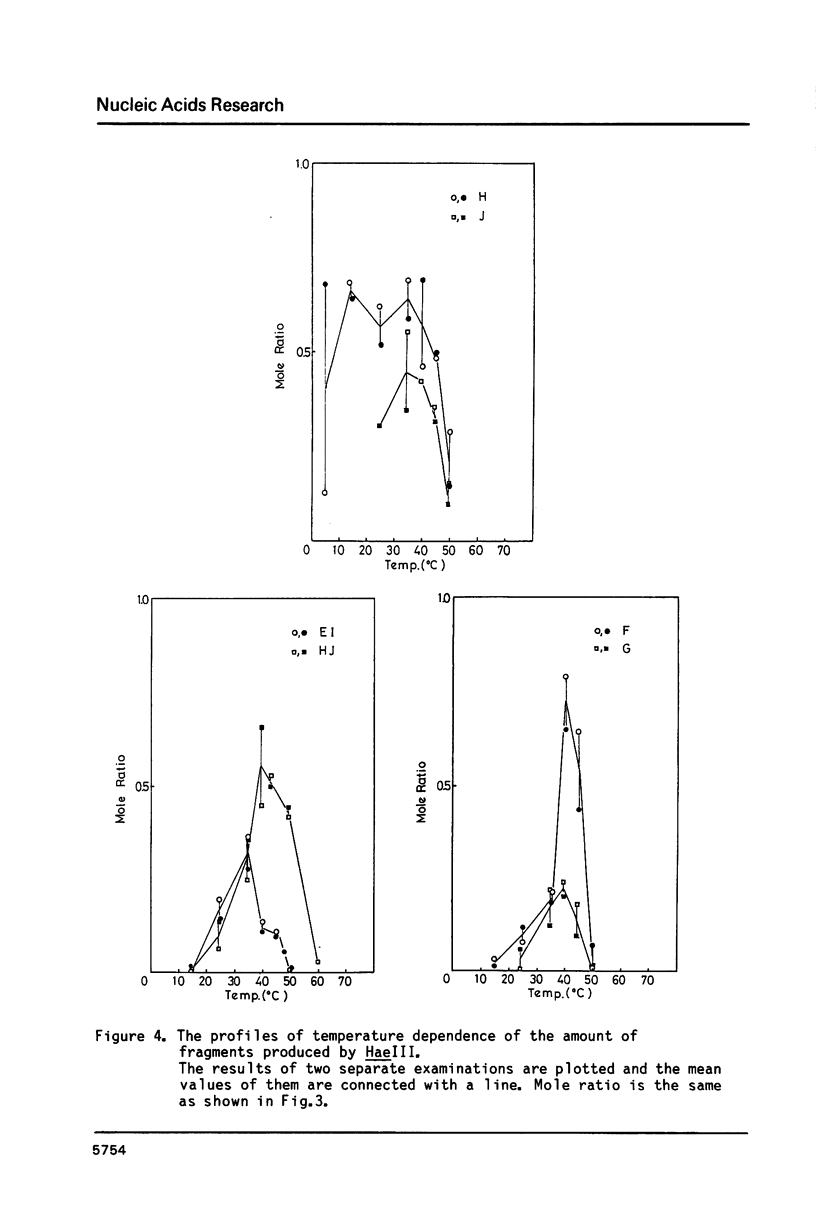
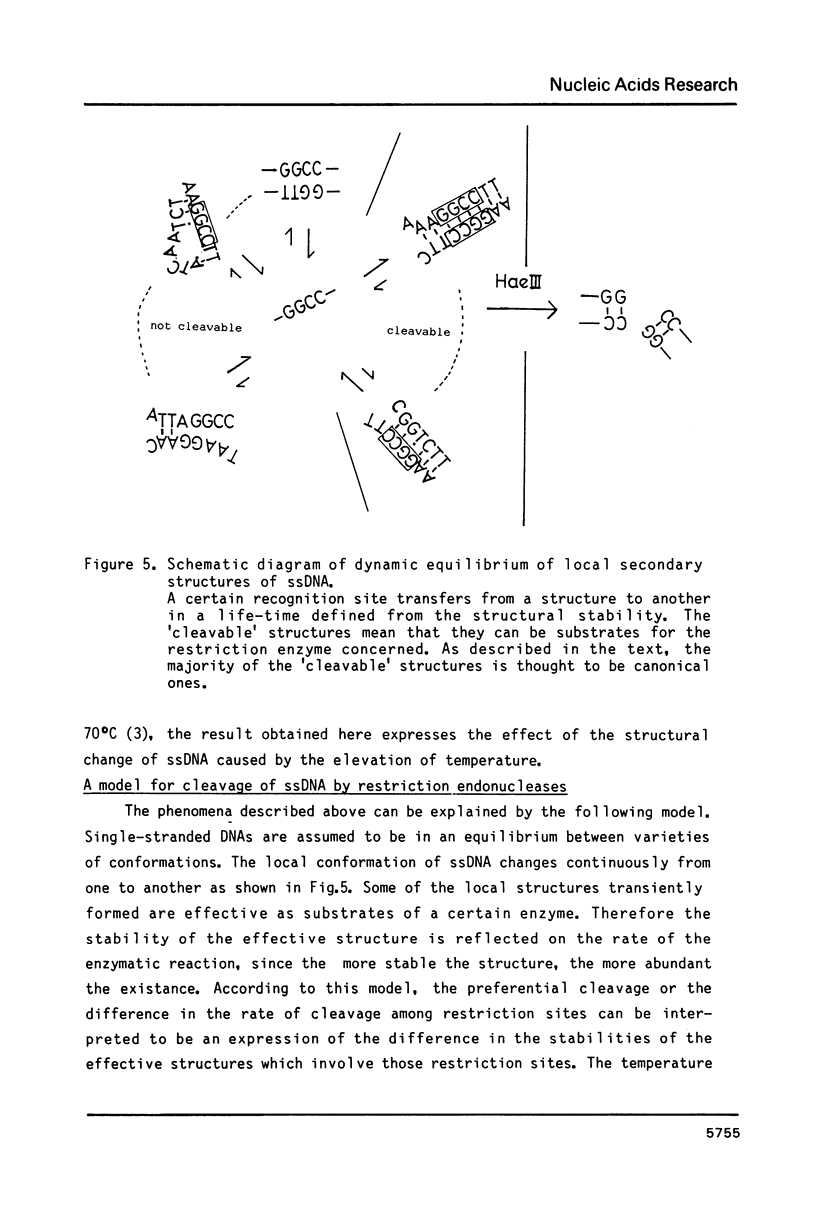
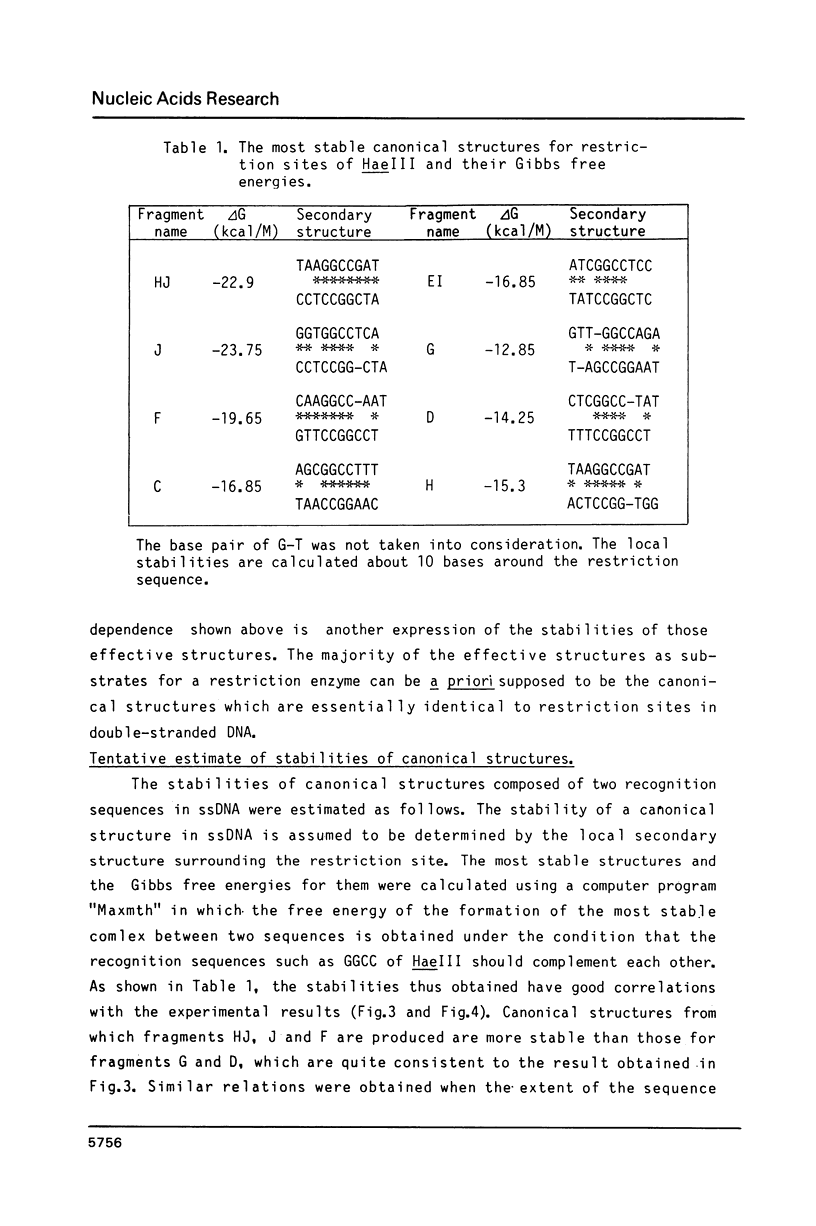



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
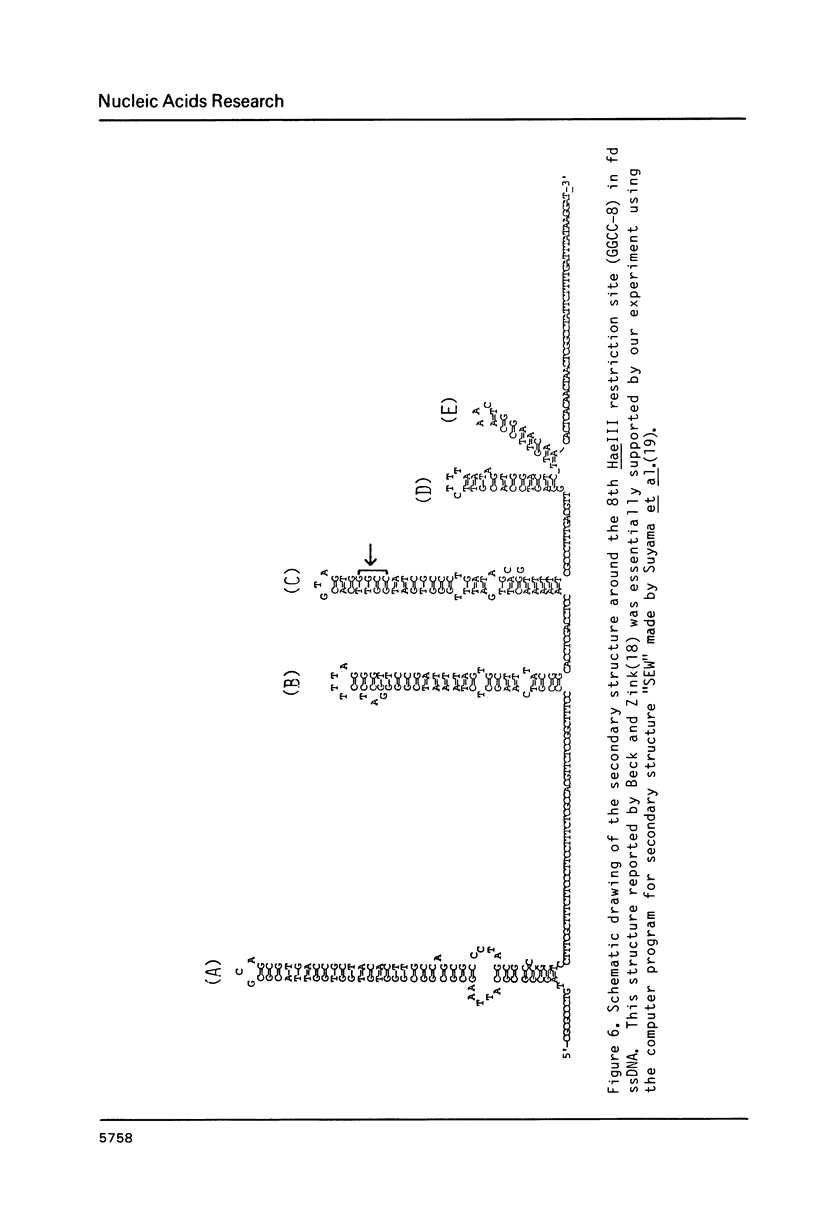
- Beck E., Zink B. Nucleotide sequence and genome organisation of filamentous bacteriophages fl and fd. Gene. 1981 Dec;16(1-3):35–58. doi: 10.1016/0378-1119(81)90059-7. [DOI] [PubMed] [Google Scholar]
- Beidler J. L., Hilliard P. R., Rill R. L. Ultrasensitive staining of nucleic acids with silver. Anal Biochem. 1982 Nov 1;126(2):374–380. doi: 10.1016/0003-2697(82)90530-9. [DOI] [PubMed] [Google Scholar]
- Blakesley R. W., Dodgson J. B., Nes I. F., Wells R. D. Duplex regions in "single-stranded" phiX174 DNA are cleaved by a restriction endonuclease from Haemophilus aegyptius. J Biol Chem. 1977 Oct 25;252(20):7300–7306. [PubMed] [Google Scholar]
- Blakesley R. W., Wells R. D. 'Single-stranded' DNA from phiX174 and M13 is cleaved by certain restriction endonucleases. Nature. 1975 Oct 2;257(5525):421–422. doi: 10.1038/257421a0. [DOI] [PubMed] [Google Scholar]
- Boulikas T., Hancock R. A highly sensitive technique for staining DNA and RNA in polyacrylamide gels using silver. J Biochem Biophys Methods. 1981 Oct;5(4):219–228. doi: 10.1016/0165-022x(81)90046-4. [DOI] [PubMed] [Google Scholar]
- Fuchs R., Blakesley R. Guide to the use of type II restriction endonucleases. Methods Enzymol. 1983;100:3–38. doi: 10.1016/0076-6879(83)00043-9. [DOI] [PubMed] [Google Scholar]
- Godson G. N., Roberts R. J. dna, single stranded/*metab. Virology. 1976 Sep;73(2):561–567. doi: 10.1016/0042-6822(76)90421-9. [DOI] [PubMed] [Google Scholar]
- Hofer B., Ruhe G., Koch A., Köster H. Primary and secondary structure specificity of the cleavage of 'single-stranded' DNA by endonuclease Hinf I. Nucleic Acids Res. 1982 May 11;10(9):2763–2773. doi: 10.1093/nar/10.9.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi K., Zinder N. D. Site-specific cleavage of single-stranded DNA by a Hemophilus restriction endonuclease. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2555–2558. doi: 10.1073/pnas.72.7.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen P. G. Separation and isolation of DNA fragments using linear polyacrylamide gradient gel electrophoresis. Methods Enzymol. 1980;65(1):305–319. doi: 10.1016/s0076-6879(80)65041-1. [DOI] [PubMed] [Google Scholar]
- Modrich P. Studies on sequence recognition by type II restriction and modification enzymes. CRC Crit Rev Biochem. 1982;13(3):287–323. doi: 10.3109/10409238209114231. [DOI] [PubMed] [Google Scholar]
- Moses P. B., Boeke J. D., Horiuchi K., Zinder N. D. Restructuring the bacteriophage f1 genome: expression of gene VIII in the intergenic space. Virology. 1980 Jul 30;104(2):267–278. doi: 10.1016/0042-6822(80)90332-3. [DOI] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Schaller H., Voss H., Gucker S. Structure of the DNA of bacteriophage fd. II. Isolation and characterization of a DNA fraction with double strand-like properties. J Mol Biol. 1969 Sep 28;44(3):445–458. doi: 10.1016/0022-2836(69)90372-6. [DOI] [PubMed] [Google Scholar]
- Shishido K., Ikeda Y. Isolation of double-helical regions rich in guanine-cytosine base pairing from bacteriophage fl DNA. Biochem Biophys Res Commun. 1971 Feb 5;42(3):482–489. doi: 10.1016/0006-291x(71)90396-2. [DOI] [PubMed] [Google Scholar]
- Suyama A., Eguchi Y., Wada A. An algorithm for the bonding-probability map of nucleic acid secondary structure. Nucleic Acids Symp Ser. 1983;(12):217–220. [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]
- Yamazaki K., Imamoto F. [A highly sensitive silver stain for DNA and RNA in agarose gels]. Tanpakushitsu Kakusan Koso. 1982 Aug;27(10):1277–1279. [PubMed] [Google Scholar]
- Yoo O. J., Agarwal K. L. Cleavage of single strand oligonucleotides and bacteriophage phi X174 DNA by Msp I endonuclease. J Biol Chem. 1980 Nov 25;255(22):10559–10562. [PubMed] [Google Scholar]