Abstract
The product of the CLU gene promotes or inhibits tumourigenesis in a context-dependent manner. It has been hypothesised that different CLU isoforms have different and even opposing biological functions, but this theory has not been experimentally validated. Here we show that molecules involved in survival pathways are differentially modulated by the intracellular or secreted forms of CLU. Secreted CLU, which is selectively increased after transformation, activates the survival factor AKT, whereas intracellular CLU inhibits the activity of the oncogenic transcription factor nuclear factor kappa B. Furthermore, intracellular CLU is inactivated by the pro-proliferative and pro-survival activity of the chaperone protein HSP60 in neuroblastoma cells by forming a physical complex. Thus, localisation is key for CLU physiology, explaining the wide range of effects in cell survival and transformation.
Keywords: Apolipoprotein J, HSP60, apoptosis, neuroblastoma, AKT, NF-κB
CLU (also known as Clusterin, Apolipoprotein J, TRPM-2 and SGP-2) is a ubiquitously expressed protein involved in the regulation of cell survival and modulation of tumourigenesis.1 The 449-amino-acid primary polypeptide chain of human CLU is proteolytically cleaved into the alpha and beta chains forming an heterodimer held together by disulphide bridges. CLU is glycosylated before secretion and is highly conserved in different species, showing a 70–80% protein homology in mammals.2 CLU expression is low in normal conditions but is induced by stress stimuli, suggesting that its function could be directly or indirectly related to the stress response. Mouse development is not affected by genetic inactivation of CLU; however, CLU-null mice show increased sensitivity to autoimmune myocarditis, suggesting a role for CLU in protecting the heart tissue from postinflammatory destruction.3 In contrast, neuronal cells are sensitised by CLU to hypoxic injury.4 In many instances, CLU has been shown to be antiapoptotic, protecting cells against a variety of death signals. Exogenous CLU protects tumour cells from cytokine- or drug-induced apoptosis, and inhibition of CLU results in the increased sensitivity of cancer cells to chemotherapeutic drugs.5, 6, 7, 8, 9, 10, 11
How CLU exerts its function has been matter of speculation. One hypothesis is that CLU is involved in the clearance of toxic substances from extracellular spaces through its ability to bind to unfolded proteins and cell debris or immune complexes.12, 13 CLU binds to the endocytic receptor megalin (LRP-2), and the clearance effect could be achieved by internalisation of the receptor–ligand complex and lysosomal degradation of the toxic substances.1, 2 Apart from its role as a ‘extracellular chaperone', CLU may act as a signalling molecule: CLU has been shown to activate or suppress phosphoinositide-3-kinase-AKT or MAP/ERK signalling pathways in different studies.11, 14, 15, 16, 17
The ability of CLU to modulate proliferative and survival pathways probably explains the plethora of biological functions attributed to this molecule. For example, CLU can relieve disease-induced inflammation by interfering with nuclear factor kappa B (NF-κB) activity via direct physical contact with inhibitors of Kappa B.18 In cancer, we have shown that CLU exerts a tumour-suppressive role by suppressing NF-κB.19, 20, 21 NF-κB is a transcription factor central to cellular processes such as cell survival and proliferation, immunity and tumourigenesis.22 Mouse embryo fibroblasts with a disrupted CLU gene show IKB destabilisation, which results in increased NF-κB activity and modulation of NF-κB-target genes.21 CLU KO mice show increased susceptibility to cancer,19, 20, 23 but the tumour-suppressive function of CLU is sometimes ad odds with reports in which CLU has been shown to be increased in aggressive forms of human cancer. How can CLU function as a tumour suppressor and oncogene at the same time?
One hypothesis investigated in this study is that CLU exerts distinct functions in different locations. For example, nuclear CLU has been shown to promote apoptosis, and to inhibit prostate cancer cell proliferation.24, 25, 26 In neuroblastoma, the expression of nuclear CLU is marginal and we have only observed this variant in terminally necrotic or apoptotic cells, suggesting that its expression is a consequence rather a cause of cell death in physiological conditions.20 Instead, CLU is detectable in the cytoplasm, extracellular spaces or the Golgi apparatus and it is not known whether its localisation also affects its biological function.20 We hypothesised that the controversial role that CLU has in cancer could be explained if the expression of intracellular and extracellular CLU has different biological outcomes.
The purpose of this study was to understand the function of intracellular and secreted CLU in the context of neuroblastoma. We show here that secreted and intracellular CLU has distinct properties, which could explain, at least in part, the enigmatic role of CLU in cancer.
Results
Intracellular and secreted CLU isoforms modulate distinct signalling molecules and their relative expression changes during transformation
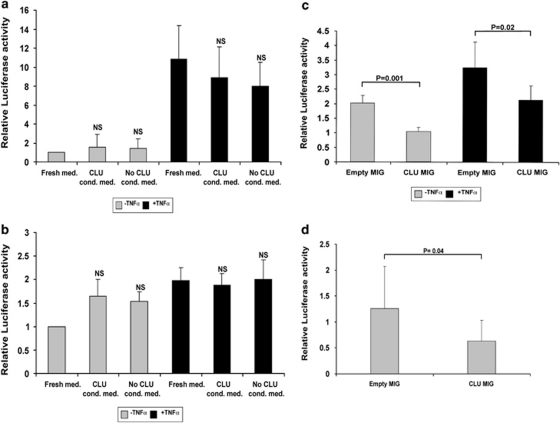
To investigate whether CLU signalling is determined by its intracellular or extracellular localisation, we prepared supernatants from 293 cells transfected with a CLU expression vector that we used as a source of extracellular CLU (Supplementary Figure 1), and used these supernatants in luciferase assays in which we assessed the activity of a NF-κB reporter vector. Tumour necrosis factor (TNF) was used to activate NF-κB activity and served as a positive control. Secreted CLU was unable to inhibit basal or TNF-induced NF-κB activity in two neuroblastoma cell lines, although LA-N-1 cells were unresponsive to TNF (Figures 1a and b). This experiment was further validated by culturing cells in the presence of purified CLU obtained from a commercial source with the identical results (not shown).
Figure 1.
Secreted CLU is unable to modulate NF-κB activity. (a) SHSY5Y or (b) LA-N-1 neuroblastoma cells were transfected with a NF-κB luciferase reporter vector, incubated with control or CLU-conditioned medium, as indicated in the bottom of the graphs, and subjected to luciferase assays. Activation of NF-κB by TNF served as a positive control for the assay. Relative luciferase activity is indicated on the y axis. (c) SHSY5Y or (d) LA-N-1 cells were co-transfected with the NF-κB reporter vector and a control (empty MIG), or CLU, expressing plasmid with or without TNF. Error bars indicate standard deviations, and significance is indicated by the P. NS indicates that the difference is not significant
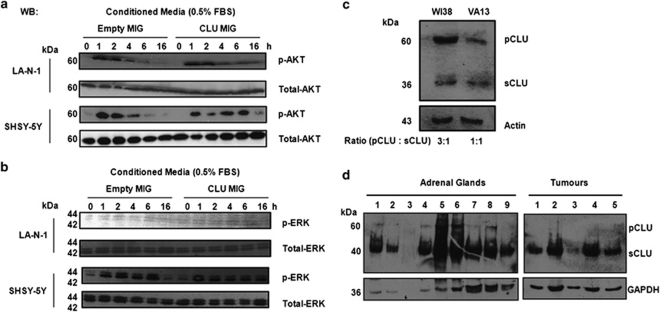
In contrast, NF-κB activity was inhibited after transiently transfecting the CLU expression plasmid into the same neuroblastoma cell lines, suggesting that only intracellular CLU is able to inhibit NF-κB (Figures 1c and d). Next, we investigated whether lack of signalling activity of secreted CLU was intrinsic or related to the extracellular localisation. We stimulated two neuroblastoma cell lines with control or CLU-containing medium and monitored the activation of the signalling molecules AKT and ERK. Extracellular CLU caused sustained activation of AKT, but not of ERK, suggesting that extracellular CLU, as previously reported,6, 14 is a specific activator of the pro-survival Pi3K–AKT axis (Figures 2a and b). Again, the result was validated with purified CLU (data not shown). To investigate whether the expression of intracellular and secreted CLU changes during transformation, we monitored the ratio of the 60 kDa intracellular precursor and 35 kDa secreted CLU isoforms in isogenic variants of primary human fibroblasts before and after SV40 transformation. Interestingly, expression of the intracellular precursor is dramatically decreased in transformed VA13 compared with non-transformed WI38 cells (Figure 2c). Furthermore, expression of precursor CLU is undetectable in primary neuroblastoma tumours from MYCN transgenic mice, whereas it is clearly visible in adrenal glands, the corresponding normal tissue (Figure 2d).
Figure 2.
Secreted CLU modulates AKT and is increased relative to intracellular CLU during transformation. (a) Western blot analysis of phosphorylated and total AKT in the indicated neuroblastoma cell lines. Hours of stimulation with control or CLU-conditioned media (CLU-MIG) are indicated on the top of the blots. (b) Western blot analysis of phosphorylated and total ERK in the indicated neuroblastoma cell lines. Hours of stimulation with the control or CLU-conditioned media are indicated on the top of the blots. (c) Western blot analysis of CLU expression in the isogenic cell lines WI38 (primary human fibroblasts) and VA13 (SV40-transformed WI38). (d) Western blot analysis of the expression of CLU in the adrenal glands and tumours resected from MYCN transgenic mice. pCLU, precursor CLU; sCLU, secreted CLU
Identification of proteins interacting with CLU in neuroblastoma cells
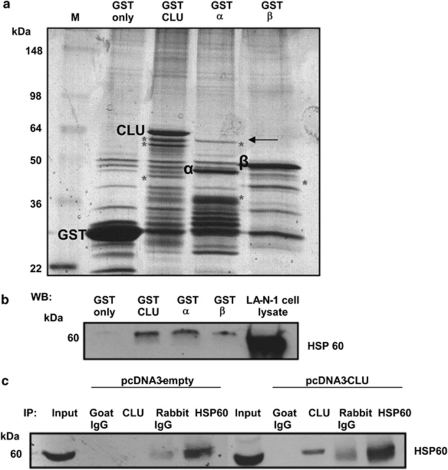
To gain more insights on the role of intracellular CLU in molecular signalling, we carried out pull-down experiments to determine which cellular proteins bind to CLU in neuroblastoma cells. Full length or the alpha and beta chain CLU were fused to glutathione S transferase (GST), and cellular proteins were pulled down and visualised by silver staining. Several cellular proteins (indicated by an asterisk) bound the fusion proteins, but not GST alone. We decided to further investigate a protein of about 60 kDa that was binding to the full-length CLU polypeptide and its alpha chain but not the beta chain (Figure 3a). Protein bands obtained from the pull down with full length and alpha chain were excised and subjected to mass spectrometry analysis.
Figure 3.
Identification of HSP60 as a CLU-interacting protein. (a) proteins pulled down by the indicated GST fusion proteins were visualised by colloidal Coomassie staining. A protein of ∼60 kDa interacting with full length and alpha chain CLU (indicated by the arrow) was excised from the gel and subjected to mass spectrometry analysis. (b) Proteins pulled down by the indicated fusion proteins were subjected to western blot analysis with a HSP60 antibody. (c) Co-immunoprecipitation assay. 293 cells were transfected with control or CLU plasmids, lysed and immunoprecipitated with CLU, HSP60 or control antibodies. After transfer to nitrocellulose, immune complexes were subjected to western blot analysis with a HSP60 antibody. Specific binding proteins are indicated by an asterisk
As expected, more than one polypeptide was present in the excised bands and the presence of keratin, a classical contaminant in pull-down assays, was not further investigated. A fragment of LRP2, also known as megalin and a known CLU receptor, was identified in the band excised from full-length CLU. The presence of megalin was an indication of the success of the assay. We next focused our interest on HSP60, which was found in the protein bands pulled down by both full-length- and alpha chain CLU–GST fusion proteins (Supplementary Figure 2). HSP60 is a chaperone molecule with a role in tumourigenesis and has been recently shown to be an activator of NF-κB.27, 28 Thus, its presence in the CLU-binding assay suggested that the interaction could be physiologically relevant.
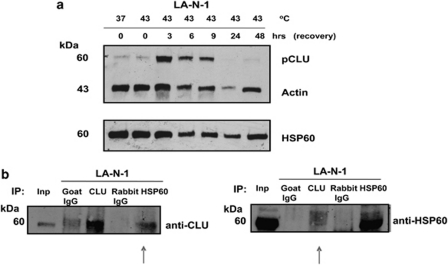
Firstly, we mapped the domain of CLU responsible for the interaction with HSP60 by pull-down and western blot assay with a HSP60 antibody. We observed that CLU full length and alpha chain were more efficient than the beta chain in binding to HSP60, confirming that the alpha chain of CLU is the major interacting domain. As expected GST alone did not bind to HSP60 further demonstrating that the interaction is specific to CLU (Figure 3b). We next assessed whether the interaction was detectable by co-immunoprecipitation assays in living cells. We transfected CLU into 293 cells and observed that exogenously expressed CLU co-immunoprecipitated with endogenous HSP60 in vivo (Figure 3c). Detection of a native CLU/HSP60 complex in neuroblastoma cells is complicated by the very low expression levels of CLU in this tumour.20 To overcome this problem, we subjected LA-N-1 neuroblastoma cells to heat shock, which greatly enhances the expression of endogenous CLU (Figure 4a). In these conditions, a native complex containing HSP60 and CLU is detectable in neuroblastoma cells using reciprocal antibodies demonstrating that their interaction is genuine (Figure 4b).
Figure 4.
HSP60 and CLU form a complex in neuroblastoma cells. (a) Western blot analysis of LA-N-1 cell lysate showing expression of CLU at different time points after a sub-lethal heat shock. Actin and HSP60 expression were used as a loading control. (b) Reciprocal co-immunoprecipitation of heath shocked LA-N-1 cells with CLU, HSP60 and control antibodies. The native CLU/HSP60 complex is indicated by the arrows. Inp, input
The CLU-interacting protein HSP60 promotes cell survival and is a marker of poor prognosis in neuroblastoma
We firstly investigated whether HSP60 could have a role in neuroblastoma by assessing the effect of its knockdown using RNA interference. To this end, we generated SHSY5Y-independent clones with reduced expression of HSP60. In all, three out of three of the interfered cells lines proliferated less than a scrambled control clone (Figures 5a–c). It is likely that reduced proliferation is caused by increased apoptosis, as the HSP60-interfered clones showed enhanced annexin staining as compared with control cells (Supplementary Figure 3). We extended the significance of these results by inducing HSP60 downregulation in primary human neuroblastoma cells isolated from a metastasis of a stage 4 patient. We confirmed that HSP60 was expressed in these primary cells, and the infection with a lentiviral shRNA vector caused a drastic downregulation of HSP60 expression (Figure 5d). Ablation of HSP60 caused reduced proliferation, increased fragmentation of DNA, diagnostic of apoptosis, but no changes in the cell cycle profiles, further suggesting that a primary role of HSP60 is to promote cell survival (Figures 5e and f).
Figure 5.
HSP60 positively regulates proliferation and survival of established and primary neuroblastoma cell lines. (a) Western blot analysis to monitor the expression of HSP60 in three SHSY-5Y cell clones obtained after infection with shRNA control (+) or HSP60 (−) targeting viruses. Quantification of HSP60 expression relative to actin is shown in the bottom of the gel. (b) Fluorescence microscopy analysis to visualise GFP expression of the SHSY-5Y clones shown in (a). Pictures were taken the day after infections (t 0) or after 6 days in culture (t 6). (c) Quantification of the experiment shown in (b). (d) Western blot analysis of primary human neuroblastoma cells infected with a control (+) or HSP60 (−) shRNA lentivirus. Relative expression of HSP60 is indicated at the bottom of the gel. (e) Proliferation assay of primary human neuroblastoma cells after ablation of HSP60 expression. (f) PI staining and FACS analysis to assess cell cycle profile and DNA fragmentation in primary human neuroblastoma cells after downregulation of HSP60. Percentages of cells in the different phases of the cell cycle or subG1 DNA (fragmented DNA) are indicated
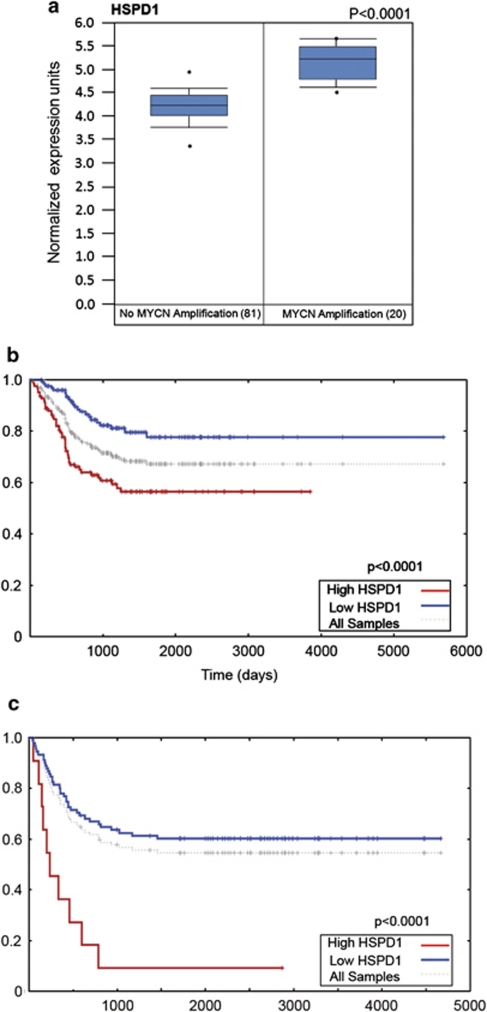
To investigate whether HSP60 expression is clinically relevant, we verified that expression of HSP60 is higher in patients with amplification of the adverse prognostic marker MYCN (Figure 6a). Furthermore, HSP60 expression is associated with poor survival in neuroblastoma patients (Figures 6b and c).
Figure 6.
HSP60 is a negative prognostic marker in human neuroblastoma. (a) Box plot showing the significant association between MYCN amplification and HSP60 expression in 101 neuroblastoma patients. Data were mined through the Oncomine website (http://www.Oncomine.org). Kaplan–Meier survival curves were generated from the Oncogenomics website (http://pob.abcc.ncifcrf.gov/cgi-bin/JK) using the data extracted from the Oberthuer. Black dots indicate outliers. (b) and Seeger (c) affymetrix datasets
HSP60 promotes neuroblastoma cell survival by antagonising CLU
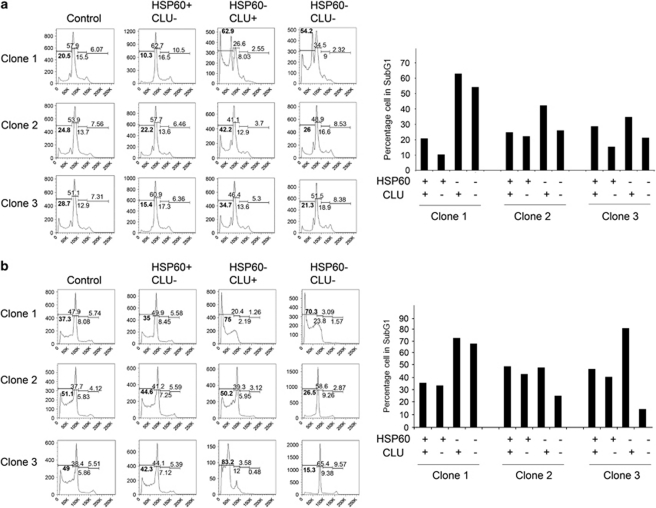
As expression of CLU is inversely correlated with that of MYCN and it marks patients with better probability of survival,20 we hypothesised that the physical interaction between CLU and HSP60 could be antagonistic. To directly address this point, we generated neuroblastoma cell lines with downregulation of HSP60, CLU or both HSP60 and CLU (Supplementary Figure 4). If CLU is a downstream target of HSP60, its knockout should rescue the apoptotic phenotype that follows HSP60 ablation. Indeed, we observed that DNA fragmentation caused by reduced levels of HSP60 was generally rescued by ablation of CLU in SHSY5Y clones in two independent experiments, further corroborating the hypothesis that CLU is a critical downstream target of HSP60 antiapoptotic activity (Figures 7a and b).
Figure 7.
Downregulation of CLU rescues apoptosis induced by ablation of HSP60. Cell cycle profiles and DNA fragmentation (indicated by percentages of subG1 DNA, in bold) were assessed by flow cytometry after knockdown of CLU and HSP60 in SHSY-5Ycell clones. The experiment was repeated twice. (a) experiment 1; (b) experiment 2. Quantification of the percentages of cells showing fragmented DNA in the different conditions is shown in the right of each panel
HSP60 is an activator of NF-κB in mammalian cells and, in keeping with these results, we observed that reducing HSP60 levels causes decreased NF-κB activity in both SHSY-5Y and primary neuroblastoma cells (Figures 8a and b). Interestingly, analysis of affymetrix databases available through the Oncomine website shows that expression of HSP60 is associated with that of NF-κB target genes in neuroblastoma patients, suggesting that one of the mechanisms by which HSP60 behave as a neuroblastoma oncogene could be related to its ability to activate NF-κB signalling (Figure 8c). In agreement with this hypothesis, knockdown of HSP60 causes upregulation of CLU expression indicating that, in addition to other mechanisms, HSP60 could activate NF-κB by antagonising CLU (Figure 8d). A model of the HSP60–CLU axis operating in neuroblastoma is shown in Figure 8e.
Figure 8.
HSP60 signalling is upstream of CLU and NF-κB in neuroblastoma. (a) SHSY5Y or (b) primary human neuroblastoma cells (HNB) were stably infected with scrambled or HSP60 shRNA lentiviruses and subsequently transfected with a NF-κB reporter vector. Luciferase assays show reduced NF-κB activity in the HSP60-interfered cells as compared with the scrambled infected, control cells. Western blot analyses to verify HSP60 downregulation are shown in the right. (c) Analysis of co-expression of HSP60 and NF-κB target genes (boxed) in neuroblastoma patients. The analysis was carried out using tools in the Oncomine website and is based upon the affymetrix datasets indicated in the legend. (d) Western blot analysis showing increased CLU expression in the primary neuroblastoma cells (HNB) after shRNA-induced downregulation of HSP60. (e) Model of the HSP60–CLU axis operating in neuroblastoma cells and the outcome of its manipulation
Discussion
A plethora of conflicting observations about the physiological role of CLU has complicated the understanding of its function in tumourigenesis. Although some of the discrepancies could be caused by the use of different experimental settings and reagents, we speculated that CLU exerts pleiotropic effects in cancer that could be strictly context dependent.29 A further level of complexity results from the intricate regulation of CLU isoforms, which can be found in multiple cellular compartments (i.e., nucleous, cytoplasm, mitochondria, Golgi, endoplasmic reticulum) or in the extracellular spaces and the body fluids.
In this study we tried to dissect the physiological role of intracellular and extracellular CLU. The hypothesis that the biological function of secreted CLU could be different from that of intracellular CLU is based on several points of evidence. For example, in prostate and bladder cancer, secreted CLU is associated with tumour progression and used as a biomarker of the disease.5, 30 Secreted CLU could be involved in survival signalling by modulating the Pi3K pathways or by behaving like an extracellular chaperone, as described in this study and previous investigations.2, 14 However, genetic ablation of the CLU gene, which causes the loss of both intracellular and extracellular CLU isoforms, causes increased tumourigenesis in mice, indicating that the intracellular form could have a dominant tumour-suppressive role.19, 20 In neuroblastoma, secreted and intracellular CLU are likely to have a different role. We have previously observed that in the presence of a CLU-blocking antibody, neuroblastoma cells are sensitised to doxorubicin killing, suggesting that secreted CLU is a survival factor and potential oncogene.31 In spite of this, disruption of the CLU gene causes increased tumourigenesis in neuroblastoma prone mice.20
The logical conclusion of these observations is that intracellular and extracellular CLU must have an opposing role in tumourigenesis and cell signalling, and the purpose of this study was to validate this hypothesis in a coherent system.
The observations in the literature and those presented in this work support the hypothesis that secreted and intracellular CLU have different biological functions: the former activates a survival programme, whereas the latter interferes with the activity of protooncogenes such as NF-κB and HSP60. By mass spectrometry and co-immunoprecipitation experiments, we have identified HSP60 as a new CLU-interacting partner and a potential neuroblastoma oncogene. HSP60 is a chaperonin protein generally increased in cancer and its cytosolic form inhibits apoptosis by activating NF-κB.27, 28, 32 We show that ablation of HSP60 by RNA interference causes increased apoptosis upstream of CLU, demonstrating that in neuroblastoma there is an HSP60–CLU axis with an antagonistic role in NF-κB regulation. This axis is likely to have clinical relevance, as shown by the significant association of elevated HSP60 and NF-κB target genes with disease outcome.
Another interesting observation presented in this study is that CLU isoforms are modulated during tumourigenesis. After transformation, human and mouse cells show increased expression of secreted CLU at the expense of the intracellular precursor, corroborating the hypothesis that secreted CLU is oncogenic, whereas intracellular CLU is tumour suppressive.
We believe that the observations presented in this study are a further step towards a better understanding of the role of CLU in cancer, reconciling some of the controversies surrounding this interesting molecule.
Materials and Methods
Cell culture
WI-38, VA-13, SHSY-5Y, LA-N-1 and 293FT were all obtained from the American Type Culture Collection, ATCC (Middlesex, UK) and maintained in culture with Dulbecco's Modified Eagle Medium supplemented with 10% heat-inactivated foetal bovine serum (FBS), 2 mM -glutamine, penicillin (100 mg/ml), streptomycin (100 mg/ml) and 1% (10 mM) MEM non-essential amino acid (NEAA). All cell lines were purchased from the Primary human neuroblastoma (IP-hNB1, hereafter referred to as HNB) cells were isolated from a tumour metastasised in the neck of a 3-year-old male patient. HNB cells were cultured in RPMI-1640 medium supplemented with 20% heat-inactivated foetal calf serum, 2 mM -glutamine, 10 μM 2-mercaptoethanol, 1 mM sodium pyruvate, penicillin (100 mg/ml), streptomycin (100 mg/ml) and 1% (10 mM) NEAA. All media and sera were purchased from Invitrogen (Paisley, UK).
Transfection and generation of conditioned media
293 FT cells (1 × 106 cells per 100 mm dish) were plated and transiently transfected with 15 μg empty MIG or CLU-MIG vectors using lipofectamine 2000, as described in the manufacturer's protocol. After 16 h, medium (supplemented with 0.5% FBS) was replaced and cells were kept in a humidified incubator at 37°C and 5% CO2 for 24 h before conditioned media were collected and used in the luciferase assay experiments.
Purified human recombinant CLU was purchased from Alexis (Exeter, UK).
Lentiviral constructs and infections
For lentiviral shRNA expression, viral particles were produced in 293FT cells transfected with appropriate packaging plasmids. pGIPZ lentiviral vector was obtained from Open Biosystems, Surrey, UK. The non-silencing shRNAmir construct (scrambled shRNA) served as the negative control. After 24 h of transfection, supernatants were collected, supplemented with 8 mg/ml of polybrene (Sigma, Dorset, UK) and filtered through a 0.45-mm filter unit to remove cell debris. Target cells were infected for 24 h at 37°C.
Immunoblotting
Proteins were quantified by the dye-fixation Bradford method (Sigma-Aldrich, Poole, UK), and the protein bands were resolved by 10% SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (GE Healthcare, Buckinghamshire, UK). After blocking with 5% dry milk in PBS containing 0.1% NP40, membranes were incubated with the following antibodies diluted in blocking buffer: clusterin (M-18, Santa Cruz Biotechnology, Santa Cruz, CA, USA) (1 : 500 dilution), actin (I-19, Santa Cruz Biotechnology) (1 : 500 dilution), HSP60 (H-300, Santa Cruz Biotechnology) (1 : 500 dilution), phospho-AKT (SER473, Cell Signaling Technology, Danvers, MA, USA) (1 : 1000 dilution), AKT (Cell Signaling Technology) (1 : 1000 dilution), GAPDH (Cell Signaling Technology) (1 : 1000 dilution), phospho-ERK (Thr202/Tyr204, Cell Signaling Technology) (1 : 2000 dilution) and ERK (Cell Signaling Technology) (1 : 1000 dilution). The membranes were then incubated with a secondary antibody: HRP-conjugated anti-mouse IgG (1 : 10 000 dilution), HRP-conjugated anti-rabbit IgG (1 : 10 000 dilution) or HRP-conjugated anti-goat IgG (1 : 10 000 dilution), all by Amersham Bioscience (Buckinghamshire, UK). Antibody binding was detected by enhanced chemiluminescence (GE Healthcare).
Preparation of tissues from MYCN transgenic mice
Mice were killed by cervical dislocation or CO2 asphyxiation when a palpable tumour was detected or at the first signs of discomfort or stress, which is indicative of disease. Four abdominal tumours originating in the adrenal glands of two females and two males mice were excised and snap-frozen in liquid nitrogen for subsequent protein analysis. Adrenal glands were isolated from four males and four females non-transgenic CBA mice <3 months old. All experimental procedures involving mice were approved by the University College London and were conducted under the Animal (Scientific Procedures) Act, 1986 (United Kingdom).
Propidium iodide staining and FACS analysis
Cells were fixed in cold 70% ethanol for at least 30 min, centrifuged at 2000 r.p.m. for 5 min at room temperature and washed twice in 1 ml phosphate–citrate buffer (0.2 M Na2HPO4 and 0.1 M citric acid) at room temperature. Cells were resuspended in PBS containing ribonuclease A and propidium iodide (PI). Cells were incubated for 30 min on ice. Samples were analysed using a cytofluorimeter BD LSR II (Becton Dickinson, Oxford, UK).
GST fusion protein and pull-down assay
To generate GST fusion constructs, PCR products containing the full-length human CLU, or the alpha and beta chains were digested and ligated into pGEX4T-1 (GST) empty vectors to create GST fusion protein expression vectors (pGEX4T-1–CLU, pGEX4T-1–alpha, pGEX4T-1–beta). PCR was performed with the following primers: CLU forward 5′-ACGCGTCGACACATGTCCAATCAGGGAAGTAAG-3′ CLU reverse 5′-TTCGCCGGCGTCTCACTCCTCCCGGTGCTTTTT-3′ CLU (beta chain) forward 5′-ACGCGTCGACACATGAGCTTGATGCCCTTCTCTCCG-3′ CLU (alpha chain) reverse 5′-TTCGGCGGCGTCTCAGCGGACGATGCGGGACTTGGG-3′. The PCR products contained the restriction site SalI and NotI (underlined).
GST, GST–CLU, GST–alpha (α) and GST–beta (β) were transformed into the Escherichia coli strain BL21. Bacterial lysates were clarified by centrifugation, and GST fusion proteins were purified on glutathione-Sepharose beads following manufacturer's instructions (GE Healthcare).
For pull-down assays, LA-N-1 (8 × 106) cells were harvested and resuspended on ice in 200 μl of bead-binding buffer A (50 mM KH2PO4 (pH 7.5), 150 mM KCl, 1 mM MgCl2) containing protease inhibitor (Roche, West Sussex, UK) and sonicated for 10 s. After sonication, 200 μl of bead-binding buffer B (50 mM KH2PO4 (pH 7.5), 150 mM KCl, 1 mM MgCl2, 20% glycerol and 2% triton- × 100) containing protease inhibitor (Roche) was added to the cells. Approximately 30 μg of total cellular proteins were first precleared with glutathione-Sepharose beads and then incubated for 1 h at 4°C with immobilized GST fusion proteins. Unbound proteins were removed by washing four times with 1 ml of bead-binding buffer C (50 mM KH2PO4 (pH 7.5), 150 mM KCl, 1 mM MgCl2, 10% glycerol and 1% triton- × 100) containing protease inhibitors (Roche). After the final wash, GST fusion protein beads were boiled in 1 × SDS sample-loading buffer and resolved on a 10% polyacrylamide gel. Brilliant blue G-colloidal concentrate was purchased from Sigma for the staining of proteins in the polyacrylamide gels. The staining was performed according to the manufacturer's protocol. Subsequently, the gel was destained in 10% acetic acid in 25% (v/v) methanol for 60 s with shaking. The gel was rinsed twice with 25% methanol and scanned at 600 nm by Bio-Rad (Hertfordshire, UK) quantity one programme.
Mass-spectrometry
Bands of interest were excised and an in-gel trypsin digestion was carried out. Each band was destained using 200 mM ammonium bicarbonate with 20% acetonitrile, followed by reduction with 10 mM dithiothreitol (Melford Laboratories Ltd, Suffolk, UK), alkylation with 100 mM iodoacetamide (Sigma) and enzymatic digestion with sequencing grade modified porcine trypsin (Promega, Southampton, UK) using an automated digest robot (Multiprobe II Plus EX, Perkin Elmer, Cambridge, UK).
Liquid Chromatography with tandem mass spectrometry (LC-MS/MS) was carried out upon each sample using a 4000 Q-Trap mass spectrometer (Applied Biosystems, Warrington, UK). Peptides resulting from in-gel digestion were loaded at high-flow rate onto a reverse-phase trapping column (0.3 mm i.d. × 1 mm), containing 5 μm C18 300 ˚ Acclaim PepMap media (Dionex, Surrey, UK) and eluted through a reverse-phase capillary column (75 μm i.d. × 150 mm) containing Waters Symmetry C18 100 ˚ media (Waters, Hertfordshire, UK) that was self-packed using a high pressure packing device (Proxeon Biosystems, Odense, Denmark). The output from the column was sprayed directly into the nanospray ion source of the 4000 Q-Trap mass spectrometer.
Fragment ion spectra generated by LC-MS/MS were searched using the MASCOT search tool against the UniProtKB/Swiss-Prot protein database using appropriate parameters. The criteria for protein identification were based on the manufacturer's definitions (Matrix Science Ltd, London, UK). Basically candidate peptides with probability-based Mowse scores exceeding threshold (P<0.05), and thus indicating a significant or extensive homology were referred to as ‘hits'. Protein identifications were only considered if they contained three or more peptides with scores >P<0.05 threshold.
Co-immunoprecipitation
293FT cells (1 × 106 cells in 100 mm dish) were transiently transfected with pcDNA3-empty or pcDNA3-CLU (full length) using LipofectAMINE 2000 (Invitrogen). The culture media were replaced on the next day with additional of 20 μM MG132 (C2211, Sigma) and 48 h after transfection, cells were rinsed in ice-cold PBS twice before 1 ml of ice-cold non-denaturing lysis buffer (50 mM Tris-Cl, (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% (v/v) Triton-X100 containing protease and phosphatase inhibitors (Roche) was added to the plates. The cell lysate was precleared with Protein G sepharose 4B (GE Healthcare) for 30 min on a rotator at 4°C to minimise non-specific binding. After preclearing, immunoprecipitation was performed by incubating equal amount of whole-cell protein extracts (15 μg) with 2 μg of anti-CLU (M-18), anti-HSP60 (H-300) antibodies or normal goat and rabbit IgGs (Santa Cruz Biotechnology) for 1 h on a rotator at 4°C. The immune complexes were collected by incubating protein G-sepharose 4B for 16 h on a rotator at 4°C and then washing it five times in PBS. The bound proteins were resuspended in 1 × SDS sample buffer and applied to western blot with anti-HSP60 antibody. All steps were carried out on ice.
Luciferase assays
For dual luciferase assay, cells (5 × 104/well in a 24-well plate) were plated 24 h before being transiently cotransfected with 200 ng of the NF-κB LUC reporter plasmid and 5 ng of renilla luciferase vector (Promega) with or without 400 μg of empty MIG or CLU-MIG plasmids21 using LipofectAMINE 2000 (Invitrogen). After 16 h, the transfection mixture media was replaced by fresh media containing TNF-α (10 mg/ml) (Invitrogen). Cells were harvested 8 h later and luciferase assays were carried out with a kit (Promega) following manufacturer's instructions. Light emission was measured with a Lumat LB9507 (Berthold Technologies, Hertfordshire, UK).
Annexin-V staining
Annexin V conjugate (Annexin V AlexaFluor 647, Invitrogen) was diluted with Annexin V-binding buffer (BD Biosciences Pharmingen, San Diego, CA, USA) at the ratio of 1 : 200. The cell pellets were then resuspended in 300 μl of the Annexin V conjugate/binding buffer mixture. Cells were mixed gently to ensure equal staining and the samples were kept on ice in the dark for 20 min. Before FACS analysis, 30 μl of 50 μg PI solution was added to each sample. The samples were analysed by a BD LSR II (Becton Dickenson, Oxford, UK).
Statistical analysis
All experiments were repeated in triplicate (n=3), unless otherwise stated. Statistical analysis was carried out using a Student-t-test (unpaired, two-tailed). P-values of ≤0.05 were considered significant.
Acknowledgments
This study was supported by a grant from the Olivia Hodson Cancer Fund and the Wellcome Trust to AS.
Glossary
- NF-κB
nuclear factor kappa B
- GST
glutathione S transferase
- PI3K
phosphoinositide-3-kinase
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by V De Laurenzi
Supplementary Material
References
- Shannan B, Seifert M, Leskov K, Willis J, Boothman D, Tilgen W, et al. Challenge and promise: roles for clusterin in pathogenesis, progression and therapy of cancer. Cell Death Differ. 2006;13:12–19. doi: 10.1038/sj.cdd.4401779. [DOI] [PubMed] [Google Scholar]
- Jones SE, Jomary C. Clusterin. Int J Biochem Cell Biol. 2002;34:427–431. doi: 10.1016/s1357-2725(01)00155-8. [DOI] [PubMed] [Google Scholar]
- McLaughlin L, Zhu G, Mistry M, Ley-Ebert C, Stuart WD, Florio CJ, et al. Apolipoprotein J/clusterin limits the severity of murine autoimmune myocarditis. J Clin Invest. 2000;106:1105–1113. doi: 10.1172/JCI9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BH, DeMattos RB, Dugan LL, Kim-Han JS, Brendza RP, Fryer JD, et al. Clusterin contributes to caspase-3-independent brain injury following neonatal hypoxia-ischemia. Nat Med. 2001;7:338–343. doi: 10.1038/85487. [DOI] [PubMed] [Google Scholar]
- Gleave M, Chi KN. Knock-down of the cytoprotective gene, clusterin, to enhance hormone and chemosensitivity in prostate and other cancers. Ann NY Acad Sci. 2005;1058:1–15. doi: 10.1196/annals.1359.001. [DOI] [PubMed] [Google Scholar]
- Jun HO, Kim DH, Lee SW, Lee HS, Seo JH, Kim JH, et al. Clusterin protects H9c2 cardiomyocytes from oxidative stress-induced apoptosis via Akt/GSK-3beta signaling pathway. Exp Mol Med. 2011;43:53–61. doi: 10.3858/emm.2011.43.1.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch-Brandt C, Morgans C. Clusterin: a role in cell survival in the face of apoptosis. Prog Mol Subcell Biol. 1996;16:130–149. doi: 10.1007/978-3-642-79850-4_8. [DOI] [PubMed] [Google Scholar]
- Miyake H, Hara I, Gleave ME. Antisense oligodeoxynucleotide therapy targeting clusterin gene for prostate cancer: Vancouver experience from discovery to clinic. Int J Urol. 2005;12:785–794. doi: 10.1111/j.1442-2042.2005.01173.x. [DOI] [PubMed] [Google Scholar]
- Miyake H, Hara I, Kamidono S, Gleave ME, Eto H. Resistance to cytotoxic chemotherapy-induced apoptosis in human prostate cancer cells is associated with intracellular clusterin expression. Oncol Rep. 2003;10:469–473. [PubMed] [Google Scholar]
- Sallman DA, Chen X, Zhong B, Gilvary DL, Zhou J, Wei S, et al. Clusterin mediates TRAIL resistance in prostate tumor cells. Mol Cancer Ther. 2007;6:2938–2947. doi: 10.1158/1535-7163.MCT-07-0345. [DOI] [PubMed] [Google Scholar]
- Shim YJ, Shin YJ, Jeong SY, Kang SW, Kim BM, Park IS, et al. Epidermal growth factor receptor is involved in clusterin-induced astrocyte proliferation. Neuroreport. 2009;20:435–439. doi: 10.1097/WNR.0b013e3283262df8. [DOI] [PubMed] [Google Scholar]
- Wilson MR, Easterbrook-Smith SB. Clusterin is a secreted mammalian chaperone. Trends Biochem Sci. 2000;25:95–98. doi: 10.1016/s0968-0004(99)01534-0. [DOI] [PubMed] [Google Scholar]
- Wyatt AR, Yerbury JJ, Berghofer P, Greguric I, Katsifis A, Dobson CM, et al. Clusterin facilitates in vivo clearance of extracellular misfolded proteins Cell Mol Life Sci 2011. e-pub ahead of print 20 April 2011. [DOI] [PMC free article] [PubMed]
- Ammar H, Closset JL. Clusterin activates survival through the phosphatidylinositol 3-kinase/Akt pathway. J Biol Chem. 2008;283:12851–12861. doi: 10.1074/jbc.M800403200. [DOI] [PubMed] [Google Scholar]
- Chou TY, Chen WC, Lee AC, Hung SM, Shih NY, Chen MY. Clusterin silencing in human lung adenocarcinoma cells induces a mesenchymal-to-epithelial transition through modulating the ERK/Slug pathway. Cell Signal. 2009;21:704–711. doi: 10.1016/j.cellsig.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Criswell T, Beman M, Araki S, Leskov K, Cataldo E, Mayo LD, et al. Delayed activation of insulin-like growth factor-1 receptor/Src/MAPK/Egr-1 signaling regulates clusterin expression, a pro-survival factor. J Biol Chem. 2005;280:14212–14221. doi: 10.1074/jbc.M412569200. [DOI] [PubMed] [Google Scholar]
- Jo H, Jia Y, Subramanian KK, Hattori H, Luo HR. Cancer cell-derived clusterin modulates the phosphatidylinositol 3′-kinase-Akt pathway through attenuation of insulin-like growth factor 1 during serum deprivation. Mol Cell Biol. 2008;28:4285–4299. doi: 10.1128/MCB.01240-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devauchelle V, Essabbani A, De Pinieux G, Germain S, Tourneur L, Mistou S, et al. Characterization and functional consequences of underexpression of clusterin in rheumatoid arthritis. J Immunol. 2006;177:6471–6479. doi: 10.4049/jimmunol.177.9.6471. [DOI] [PubMed] [Google Scholar]
- Bettuzzi S, Davalli P, Davoli S, Chayka O, Rizzi F, Belloni L, et al. Genetic inactivation of ApoJ/clusterin: effects on prostate tumourigenesis and metastatic spread. Oncogene. 2009;28:4344–4352. doi: 10.1038/onc.2009.286. [DOI] [PubMed] [Google Scholar]
- Chayka O, Corvetta D, Dews M, Caccamo AE, Piotrowska I, Santilli G, et al. Clusterin, a haploinsufficient tumor suppressor gene in neuroblastomas. J Natl Cancer Inst. 2009;101:663–677. doi: 10.1093/jnci/djp063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santilli G, Aronow BJ, Sala A. Essential requirement of apolipoprotein J (clusterin) signaling for IkappaB expression and regulation of NF-kappaB activity. J Biol Chem. 2003;278:38214–38219. doi: 10.1074/jbc.C300252200. [DOI] [PubMed] [Google Scholar]
- Gilmore TD. Introduction to NF-kappaB: players, pathways, perspectives. Oncogene. 2006;25:6680–6684. doi: 10.1038/sj.onc.1209954. [DOI] [PubMed] [Google Scholar]
- Thomas-Tikhonenko A, Viard-Leveugle I, Dews M, Wehrli P, Sevignani C, Yu D, et al. Myc-transformed epithelial cells down-regulate clusterin, which inhibits their growth in vitro and carcinogenesis in vivo. Cancer Res. 2004;64:3126–3136. doi: 10.1158/0008-5472.can-03-1953. [DOI] [PubMed] [Google Scholar]
- Caccamo AE, Scaltriti M, Caporali A, D'Arca D, Corti A, Corvetta D, et al. Ca2+ depletion induces nuclear clusterin, a novel effector of apoptosis in immortalized human prostate cells. Cell Death Differ. 2005;12:101–104. doi: 10.1038/sj.cdd.4401491. [DOI] [PubMed] [Google Scholar]
- Caccamo AE, Scaltriti M, Caporali A, D'Arca D, Scorcioni F, Candiano G, et al. Nuclear translocation of a clusterin isoform is associated with induction of anoikis in SV40-immortalized human prostate epithelial cells. Ann NY Acad Sci. 2003;1010:514–519. doi: 10.1196/annals.1299.095. [DOI] [PubMed] [Google Scholar]
- Yang CR, Leskov K, Hosley-Eberlein K, Criswell T, Pink JJ, Kinsella TJ, et al. Nuclear clusterin/XIP8, an X-ray-induced Ku70-binding protein that signals cell death. Proc Natl Acad Sci USA. 2000;97:5907–5912. doi: 10.1073/pnas.97.11.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappello F, Conway de Macario E, Marasa L, Zummo G, Macario AJ. Hsp60 expression, new locations, functions and perspectives for cancer diagnosis and therapy. Cancer Biol Ther. 2008;7:801–809. doi: 10.4161/cbt.7.6.6281. [DOI] [PubMed] [Google Scholar]
- Chun JN, Choi B, Lee KW, Lee DJ, Kang DH, Lee JY, et al. Cytosolic Hsp60 is involved in the NF-kappaB-dependent survival of cancer cells via IKK regulation. PLoS One. 2010;5:e9422. doi: 10.1371/journal.pone.0009422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala A, Bettuzzi S, Pucci S, Chayka O, Dews M, Thomas-Tikhonenko A. Regulation of CLU gene expression by oncogenes and epigenetic factors implications for tumorigenesis. Adv Cancer Res. 2009;105:115–132. doi: 10.1016/S0065-230X(09)05007-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazzaa SM, Elashry OM, Afifi IK. Clusterin as a diagnostic and prognostic marker for transitional cell carcinoma of the bladder. Pathol Oncol Res. 2010;16:101–109. doi: 10.1007/s12253-009-9196-3. [DOI] [PubMed] [Google Scholar]
- Cervellera M, Raschella G, Santilli G, Tanno B, Ventura A, Mancini C, et al. Direct transactivation of the anti-apoptotic gene apolipoprotein J (clusterin) by B-MYB. J Biol Chem. 2000;275:21055–21060. doi: 10.1074/jbc.M002055200. [DOI] [PubMed] [Google Scholar]
- Cappello F, David S, Peri G, Farina F, Conway de Macario E, Macario AJ, et al. Hsp60: molecular anatomy and role in colorectal cancer diagnosis and treatment. Front Biosci (Schol Ed) 2011;3:341–351. doi: 10.2741/s155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.