Abstract
Mice deficient for interferon regulatory factor (Irf)2 (Irf2−/− mice) exhibit immunological abnormalities and cannot survive lymphocytic choriomeningitis virus infection. The pancreas of these animals is highly inflamed, a phenotype replicated by treatment with poly(I:C), a synthetic double-stranded RNA. Trypsinogen5 mRNA was constitutively up-regulated about 1,000-fold in Irf2−/− mice compared with controls as assessed by quantitative RT-PCR. Further knockout of IFNα/β receptor 1(Ifnar1) abolished poly(I:C)-induced pancreatitis but had no effect on the constitutive up-regulation of trypsinogen5 gene, indicating crucial type I IFN signaling to elicit the inflammation. Analysis of Ifnar1−/− mice confirmed type I IFN-dependent transcriptional activation of dsRNA-sensing pattern recognition receptor genes MDA5, RIG-I, and TLR3, which induced poly(I:C)-dependent cell death in acinar cells in the absence of IRF2. We speculate that Trypsin5, the trypsinogen5 gene product, leaking from dead acinar cells triggers a chain reaction leading to lethal pancreatitis in Irf2−/− mice because it is resistant to a major endogenous trypsin inhibitor, Spink3.
Keywords: TRIF, IPS-1, Ca2+-binding proteins, cathepsin B
Interferons (IFNs) are cytokines whose actions contribute to the first line of defense against infection. IFNs both render cells resistant to viral attack and regulate cell growth and differentiation (1). IFNs elicit their pleiotropic effects by regulating the expression of many IFN-stimulated genes (ISGs). IFNs themselves are controlled by IFN regulatory factors (IRFs) that also regulate the expression of ISGs. By binding to IFN-stimulated response elements (ISREs) in gene promoters, the nine known IRF family members (IRF1–9) govern the production of cytokines related to inflammation and immune responses.
When pattern recognition receptors (PRRs) such as Toll-like receptors (TLRs) and retinoic acid-inducible gene-I (RIG)-like receptors detect pathogen ligands, these receptors are activated (2) and transduce downstream signaling, activating IRFs and IFNs. Analyses using knockout (KO) mice deficient for various IRFs have revealed their physiological roles. For example, IRF2 functions mainly as a transcriptional repressor by competing for binding to ISREs with other IRFs, especially IRF9 and IRF1 (1).
Irf2-deficient (Irf2−/−) mice spontaneously develop inflammatory skin disease as they age, and die within weeks from lymphocytic choriomeningitis virus (LCMV) infection (3). Ablation of IFNα/β receptor 1 (Ifnar1) or Irf9 ameliorates the skin inflammation of Irf2−/− mice, suggesting that IRF2 negatively regulates gene expression by antagonizing IRF9, which is activated by type I IFN (I-IFN) (4). However, the precise mechanisms underlying the phenotypes of Irf2−/− are not known. In this study, we found that poly(I:C) (pIC) mimicked LCMV-induced pancreatitis, and we have used double KO mice to explore the cause of death in pIC-treated Irf2−/− mice. Our results show that significant trypsinogen5 up-regulation in Irf2−/− mice together with I-IFN-dependent transcriptional activation of dsRNA-sensing PRRs were critical for the pIC-induced death.
Results and Discussion
Irf2−/− Mice Show IFN-Dependent Poly(I:C)-Induced Pancreatitis and IFN-Independent Secretory Dysfunction in Pancreatic Acinar Cells.
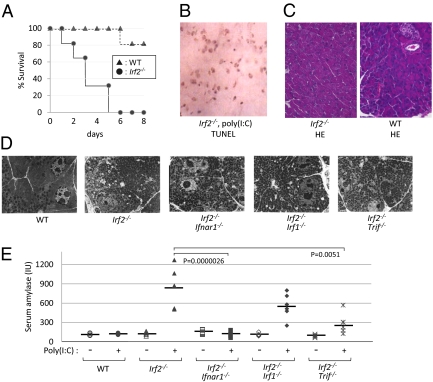
LCMV-infected Irf2−/− mice die within 4 wk postinfection (3), but all Irf2−/− mice challenged intraperitoneally with poly(I:C) (pIC-Irf2−/− mice) died within 1 wk (Fig. 1A). Severe acute pancreatitis was apparent in pIC-Irf2−/− mice, as shown by abundant TUNEL+ apoptotic cells (Fig. 1B). Even in the absence of pIC, however, some abnormalities were detected in Irf2−/− pancreas, as indicated by hematoxylin and eosin staining (Fig. 1C) and electron microscopy (Fig. 1D). A mild infiltration of inflammatory cells (particularly lymphocytes) was noted around Irf2−/− ductal cells, but this pancreatitis was not typical. The pancreatic acinar cells in untreated Irf2−/− mice were filled with eosinophilic secretory granules of heterogeneous size, whereas fewer eosinophilic granules of more uniform size were observed mainly in the apical region of WT acinar cells. Interestingly, treatment of Irf2−/− mice with the stable cholecystokinin (CCK) analog cerulein (5) did not cause acute pancreatitis, as assessed by electron microscopy and serum amylase levels (Fig. S1 A and B). Because mRNA expression of CCK receptors in Irf2−/− mice was normal (Fig. S1C), these results suggest that the secretory and/or vesicle transport systems in Irf2−/− mice are dysfunctional.
Fig. 1.
Irf2 deficiency induces sensitivity to poly(I:C) and pancreatitis. (A) Survival curve after pIC challenge. WT and Irf2-deficient (Irf2−/−) mice were induced by i.p. pIC challenge (250 μg). All of the Irf2−/− mice were deceased within a week, compared with WT mice. (B) Following pIC stimulation, many cells were TUNEL-positive, indicating apoptosis and severe acute pancreatitis in Irf2−/− mice. (C and D) Hematoxylin and eosin (HE) staining (C) and electron microscopic observation (D) were done to examine the pancreas histologically in WT, Irf2−/−, and double KO mice (Irf2−/−Ifnar1−/−, Irf2−/−Irf1−/−, and Irf2−/−Trif−/−). (E) To assess pancreatitis, we monitored serum amylase levels with (+) and without (−) pIC challenge.
The mRNAs encoding the Ca2+-binding proteins Anxa10, Ahsg, and S100-G involved in Ca2+-dependent vesicle transport, sorting, and fusion processes were significantly up-regulated in Irf2−/− pancreas (Table S1). The secretory dysfunction observed in cerulein-treated Irf2−/− mice (6), which is due to an abnormal distribution pattern of normal levels of soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) (6), may be due to the abnormal expression of these Ca2+-binding proteins in the absence of IRF2, because annexin family proteins are known to bind and regulate SNAREs (7).
Skin inflammation in Irf2−/− mice was rescued by abolishing IFN signaling (4). We asked whether the atypical pancreatitis in Irf2−/− mice could be similarly rescued by crossing the Irf2−/− mutants to Ifnar1-, Irf1-, or Trif-deficient mice (3, 8, 9) to generate double knockout mice. Abnormal acinar granule distribution was again observed in Irf2−/−Ifnar1−/−, Irf2−/−Irf1−/−, and Irf2−/−Trif−/− mice (Fig. 1D). Thus, the abnormal acinar structure caused by Irf2 disruption is not mediated by IFN signaling.
To assess pancreatitis in double knockout mice, we measured serum amylase levels before and after pIC challenge (Fig. 1E). Serum amylase was elevated in pIC-Irf2−/− and pIC-Irf2−/−Irf1−/− mice. However, this increase did not occur at all in pIC-Irf2−/−Ifnar1−/− mice, and only to a limited extent in pIC-IRF2−/−TRIF−/− mice. These data indicate that type I IFN signaling via IFNAR1, as well as TLR signaling via the adaptor protein TRIF, are important for the development of pIC-induced pancreatitis in Irf2−/− mice. Moreover, our results show that IRF2 regulates IFN-independent pathways affecting acinar cell secretion as well as IFN-dependent pathways inducing pIC-mediated pancreatitis.
Up-Regulated Trypsinogen5 mRNA in the Pancreas of Irf2−/− Mice.
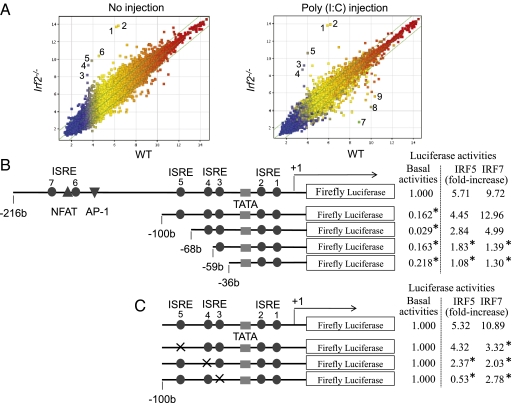
We used an Affymetrix DNA microarray system to compare mRNA expression in the pancreas before and after pIC injection of Irf2−/− and WT mice (Fig. 2A). In Irf2−/− mice, 14 annotated genes were up-regulated and 8 genes were down-regulated more than 10-fold (Table S1) compared with WT mice. The transcriptional profiles of genes important for the etiology of pancreatitis (10, 11) are listed in Table 1. Strikingly, trypsinogen5 mRNA was up-regulated >100-fold in pIC-Irf2−/− pancreas, a noteworthy observation because trypsinogens activate many other pancreatic enzymes, and premature intracellular activation of trypsinogens in pancreatic acinar cells triggers acute pancreatitis (10, 11). There are 20 trypsinogen genes (T1–T20) in the murine T-cell receptor β gene locus (12), 12 of which express trypsinogen proteins (Fig. S2, Right), whereas humans have only 3 trypsinogen genes encoding three proteins: PRSS1, PRSS2, and PRSS3 (Fig. S2, Left) (13). The gene expression profile of pIC-Irf2−/− pancreas is inflammation-prone: Mouse trypsinogen mRNAs of T11 (Prss3) and T4 (Trypsinogen5) were up-regulated (Table 1); the mRNA encoding cysteine protease cathepsin B (Ctsb), an enzyme that can initiate pancreatitis by activating trypsinogens (14–16), was also up-regulated (Table 1). The mRNA encoding chymotrypsin C (Ctrc) was down-regulated and another anti-inflammatory factor inter-α-trypsin inhibitor was also down-regulated, although the mRNA encoding Kazal type 3 (Spink3), a serine protease inhibitor that blocks trypsin activity (17), was slightly up-regulated.
Fig. 2.
Trypsinogen5 is highly expressed in Irf2-deficent mice. (A) Irf2−/− or wild-type mice with or without peritoneal injection of pIC were killed, and the amounts of mRNA from the pancreas were systematically compared using Affymetrix 28,815 gene probes. The points farthest from the diagonal indicate transcripts showing the greatest difference between WT and Irf2−/−. Points 1 and 2, trypsinogen5 with different probes; 3, α-2-HS-glycoprotein (Ahsg); 4, annexin A10 (Anxa10); 5, fetuin-β (Fetub); 6, 3-hydroxy-3-methylglutaryl-CoenzymeA synthase2 (Hmgcs2, HMG-CoA synthase); 7, Ig κ chain variable8 (Igk-V8); 8, unknown; 9, carbonic anhydrase 3 (Car3). (B) A series of deletion mutants of trypsinogen5 proximal promoter region (−216 to +15) was placed upstream of a luciferase reporter gene (1 μg) and analyzed for transcriptional activity in mouse pancreatic acinar cells using a dual luciferase assay at 24 h posttransfection in combination with expression vectors (100 ng) expressing IRF5 or IRF7 or a control vector. The basal luciferase activity of each deletion, measured relative to the −216 to +15 region, and the responses to IRF5 and IRF7 expression vectors are shown as fold increase compared with the control vector. The TATA box, ISRE core, and NFAT- and AP-1 binding sites are indicated. *P < 0.05 versus the −216 to +15 region. (C) Point mutations were introduced into each ISRE site (indicated by x) of the trypsinogen5 promoters as described in Materials and Methods. The promoter activity of each mutant trypsinogen5 was determined with a dual luciferase assay system. *P < 0.05 versus wild type.
Table 1.
Expressions of relevant genes to pancreatitis
| Gene transcripts | WT (−) | WT (pIC) | Irf2−/−(−) | Irf2−/− (pIC) |
| Prss1 (T16, Trypsin1) | 11,161 | 13,863 | 10,388 | 13,788 |
| Prss2 (T20, Trypsin 2) | 16,041 | 15,661 | 15,857 | 15,494 |
| Prss3 (T11, Trypsin 3) | 1,155 | 1,131 | 3,059 ↑ | 2,395 ↑ |
| Trypsinogen5 (T4, 1810009J06Rik) | 70 | 57 | 13,514 ↑ | 14,287 ↑ |
| Chymotrypsin C (Ctrc) | 545 | 368 | 87 ↓ | 119 ↓ |
| Chymotrypsinogen B 1 (Ctrb1) | 19,417 | 18,772 | 20,457 | 19,919 |
| Amylase2-2, pancreatic (Amy2b) | 19,101 | 18,488 | 17,092 | 18,261 |
| Calcium-sensing receptor (Casr) | 37 | 37 | 30 | 26 |
| Cystic fibrosis membrane conductance regulator (Cftr) | 7 | 6 | 11 | 8 |
| Cathepsin B (Ctsb) | 349 | 443 | 848 ↑ | 794 ↑ |
| Serine protease inhibitor, Kazal-type 3 (Spink3) | 4,716 | 3,957 | 7,497 | 7,774 |
| Inter-α-trypsin inhibitor, heavy chain 4 (Itih4) | 375 | 212 | 78 ↓ | 71 ↓ |
| Galanin (Gal) | 879 | 1,057 | 213 ↓ | 71 ↓ |
The levels of gene expression in the pancreas are shown in Affymetrix units. The trypsinogen5 data are Point 1 in Fig. 2.
We examined the tissue specificity and dependency on IRF2 and IFNAR1 of trypsinogen5 expression by quantitative RT-PCR. In untreated WT mice, trypsinogen5 is expressed most highly in pancreas and skin and modestly in spleen (Fig. S3A). In untreated Irf2−/− mice, trypsinogen5 expression in the pancreas was up-regulated ∼1,000-fold compared with controls, and was not affected by IFNAR1 ablation. Trypsinogen5 mRNA was up-regulated in Irf2−/− spleen to a much lower extent than in Irf2−/− pancreas, and was not detectable in liver or lung of WT or Irf2−/− mice.
We examined the effects of various IRFs on the activity of the murine trypsinogen5 promoter, which contains seven ISREs. We cloned a 1.1-kb fragment of the trypsinogen5 promoter region (−1063 to +15) to create a series of promoter deletion construct mutants driving the firefly luciferase reporter gene (Fig. 2B, Left). These were transfected into HEK293T cells along with plasmids overexpressing murine IRF1, human IRF5, IRF7, or MyD88. MyD88 was required for IRF-mediated activation of trypsinogen5 ISREs, and significant promoter activity was observed when IRF1, IRF5, or IRF7 was overexpressed (Fig. S3B). Furthermore, the −216 to +15 promoter region of trypsinogen5 was sufficient for responses to IRF1 or IRF7 stimulation (Fig. S3C). Overexpression of IRF2 inhibited IRF1- or IRF7-stimulated promoter activity in a dose-dependent manner (Fig. S3D). These data suggest that IRF2 binds to the proximal promoter of trypsinogen5 and inhibits the access of IRF1, IRF5, and IRF7 to ISRE sites in this region.
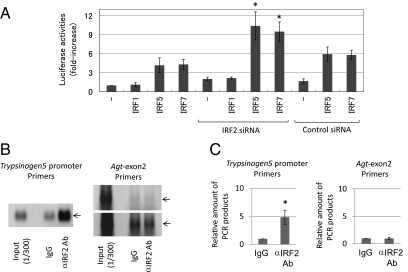
To confirm this hypothesis, we transfected TGP49 cells, a mouse acinar cell line, with trypsinogen5 promoter deletion series reporters as well as with plasmids expressing IRF1, -5, or -7, and assessed the promoter activities (Fig. 2B, Right). The basal promoter activity was drastically decreased by deleting the −216 to −100 region containing two ISREs, a nuclear factor-activated T cell (NFAT), and an activator protein 1 (AP-1) binding site. In contrast to 293T cells, the trypsinogen5 promoter in TGP49 cells could be activated by exogenously expressed IRF5 or IRF7 without MyD88 (Fig. 3A). The promoter could not be activated by IRF1 even in the presence of MyD88 expression. The regions responsive to IRF5 and IRF7 were confirmed to be ISRE4 (−62 to −59) and ISRE3 (−55 to −49) by site-specific mutation analysis (Fig. 2C). The IRF5- and IRF7-dependent promoter activities were significantly (P < 0.05) enhanced by knocking down Irf2 with specific siRNA compared with control (scrambled) siRNA (Fig. 3A).
Fig. 3.
IRF2 binds to the promoter region of trypsinogen5 gene. (A) The effects of siRNAs (3 μg) specific to IRF2 or a control scrambled sequence on transcriptional activity of the −216 to +15 luciferase reporter in TGP49 acinar cells were measured. *P < 0.05 versus control siRNA. (B) A chromatin immunoprecipitation assay was done using TGP49 acinar cells with the IRF2-specific antibody (5 μg) or the same amount of control nonspecific IgG. The precipitated chromatin fragments were detected by PCR with a trypsinogen5 promoter-specific primer set at 35 cycles or a negative control primer set for angiotensinogen (Agt) exon2 at 30 (Upper) and 35 (Lower) cycles. The input before precipitation indicates the predicted size (Trp5, 229 bp; Agt, 221 bp) of the PCR product. (C) The ChIP assay done in B was quantitatively measured using a real-time PCR method with the same primers. The relative amounts of β-actin were calculated, and the amounts of chromatin fragments precipitated with the anti-IRF2 antibody were shown relative to those with the nonspecific control antibody (IgG). *P < 0.01 versus control IgG.
To confirm IRF2 binding to the proximal promoter of trypsinogen5 in pancreatic acinar cells in vivo, we performed chromatin immunoprecipitation (ChIP) assays in TGP49 cells using specific PCR probes spanning all seven ISREs (−173 to +56) in the trypsinogen5 promoter. Anti-IRF2 antibody specifically precipitated the trypsinogen5 promoter, as determined by semiquantitative PCR (Fig. 3B) and real-time PCR (Fig. 3C). These results suggest that in WT mice, trypsinogen5 expression in pancreatic acinar cells is repressed by the binding of IRF2 to ISREs in the proximal promoter region. However, in Irf2−/− mice, the trypsinogen5 gene is activated because IRF5 and IRF7 can access the ISREs in the absence of IRF2.
IRF5 and IRF7 are critical inducers of the expression of proinflammatory cytokines and type I IFNs, respectively (18, 19), and these activities require MyD88. In WT cells, IRF4 inhibits IRF5 function by sequestering MyD88 (18). IRF2 did not associate with MyD88 (18) but, in our study, it did bind to the ISRE-containing region in the trypsinogen5 promoter (Fig. 3 B and C). Therefore, we postulate that IRF2 inhibits IRF5 and IRF7 activity by competing with them for binding to ISREs, rather than by sequestering MyD88.
Trypsinogen5 Is Resistant to the Trypsin Inhibitor Spink3.
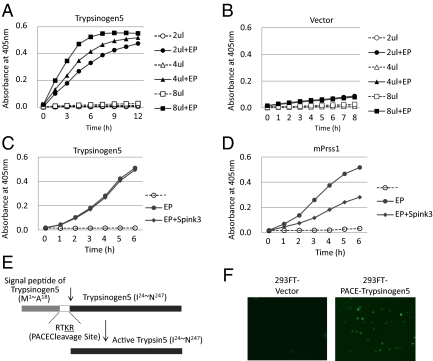
Comparison of mouse trypsinogen5 to other mouse and human trypsinogens (Fig. S4) showed that, although the N-terminal activation peptide sequence (NSDDK-I) in trypsinogen5 differs from that in other trypsinogens (DDDDK-I), other important regions, including the triad amino acid sequence H-D-S, required for enzymatic activity are conserved (10, 11). In addition, tryptic activity in cell lysates of 293FT cells overexpressing trypsinogen5 was dramatically enhanced by treatment with enteropeptidase (Fig. 4 A and B). The trypsinogen5 inhibitor binding site (DSCDGDS), which prevents premature activation, differed from that found in most trypsinogens (DSCQGDS) (10, 11), resembling the inhibitor binding site (DSCQRDS) of the human trypsin inhibitor-resistant PRSS3 enzyme. In addition, the trypsin autolytic cleavage site (Q-V) in trypsinogen5 differed from that in other trypsinogens (R-V), suggesting that trypsinogen5 is resistant to both trypsin inhibitors and self-inactivation. Indeed, trypsinogen5 was resistant to inhibition by Spink3, a major endogenous trypsin inhibitor in mice (Fig. 4 C and D), as well as by soy bean trypsin inhibitor (Fig. S5 A and B). Analysis of the evolutionary pedigree in Fig. S6 showed that mouse trypsinogen5 is most distant from mPrss1 and mPrss2, just as human PRSS3 is most distant from PRSS1 and PRSS2. Therefore, we believe that mouse trypsinogen5 is a homolog of human PRSS3. Moreover, our data suggest that, in the absence of IRF2, trypsinogen5 is highly expressed and exacerbates pIC-induced pancreatitis due to its inhibitor-resistant nature.
Fig. 4.
Trypsinogen activity is activated by proteolytic cleavage. (A and B) A full-length mouse trypsinogen5 cDNA from the mouse pancreas was cloned into pcDNA3 (Invitrogen) and expressed in 293T cells. The indicated amounts of cell lysates (2–8 μL of 5 μg/μL lysates) were mixed with a trypsin-specific substrate (BioVision) in the presence or absence of added enteropeptidase. Tryptic activity was monitored by the amount of released pNA, measuring spectrophotometric units (A405). (C and D) The effects of Spink3 were examined by adding cell lysates expressing Spink3, a major intrinsic trypsin inhibitor in mouse pancreas, to lysates expressing trypsinogen5 (C) or mouse Prss1 (D). (E) The DNA sequence encoding the activation peptide in the trypsinogen5 expression vector was replaced with sequences encoding a PACE cleavage site (-RTKR-) so that tryptic activity is activated by ubiquitously expressed PACE protease. (F) 293FT cells transfected with PACE-trypsinogen5 or control vector were stained with FITC-labeled annexin V to detect apoptosis.
Poly(I:C)-Induced Cell Death Can Be Triggered by a TLR3/TRIF-Dependent Pathway or a RIG-I/MDA5/IPS-1-Dependent Pathway.
Although trypsinogen5 was up-regulated in untreated IRF2−/− mice, only mild inflammation around acinar cells was observed and pancreatitis did not occur. We hypothesize that trypsinogen5 as well as mPrss1, -2, and -3 leaking from dying acinar cells are activated by proteases such as cathepsin B or enteropeptidase, also released from these cells. These activated trypsins trigger signals to induce the death of many acinar cells, a process of cell death amplification we refer to as the “enhancing loop” of acinar cell death. In this way, the initial death of a few cells induced by pIC can precipitate severe pancreatitis. This idea is supported by a report that the extracellular or intracellular treatment of pancreatic acinar cells with active trypsins causes acinar cell death (20). In this study, the enteropeptidase cleavage site (-DDDDK-) of rat trypsinogen was replaced with a cleavage site (-RTKR-) recognized by paired basic amino acid-cleaving enzyme (PACE). This allowed the rat trypsinogen to be activated intracellularly with the ubiquitously expressed PACE enzyme rather than with enteropeptidase, which is expressed mainly in the duodenum. We created a PACE-trypsinogen5 enzyme that successfully induced the apoptosis of 293FT cells when overexpressed (Fig. 4 E and F). These results indicate that proteolytic activation of trypsinogen5 is sufficient to induce cell death.
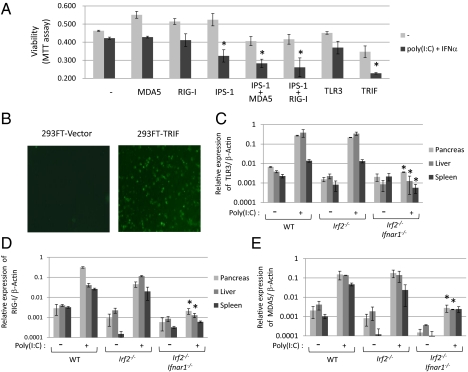
Because pIC-dependent pancreatitis in Irf2−/− mice can be prevented by inactivating IFNAR1 signaling (Fig. 1E), we focused on IFN signaling pathways to identify candidates that might trigger initial cell death following pIC treatment. Indeed, IRF1, IRF7, MyD88, MDA5, RIG-I, and TLR3 gene expression were all up-regulated in the pancreas of pIC-Irf2−/− mice (Table S2). Because these proteins are associated with cell death pathways dependent on TRIF or IPS-1, we examined the effect of IRF2 loss on these well-characterized systems (21, 22). TRIF binds to receptor-interacting proteins and thereby activates caspase8 via FADD to induce cell death (21), whereas the IPS-1-dependent cell death pathway, which is triggered by MDA5 or RIG-I, is reported to activate caspase9 via the mitochondrial pathway dependent on Apaf-1 and cytochrome c (22). We confirmed that 293FT cells transfected with TRIF-expressing plasmid underwent apoptosis, as shown by staining with FITC-labeled annexin V (Fig. 5B). Next, we used the MTT viability assay to quantify the extent of cell death induced by IFN-related molecules in the presence or absence of pIC and IFNα. Exogenous overexpression of IPS-1 or TRIF significantly enhanced the death of pIC- and IFN-treated 293FT cells, and the death-inducing effects of MDA5 and RIG-I were enhanced by cotransfection with IPS-1 (Fig. 5A). These results suggest the existence of at least two pIC-dependent cell death pathways: one TLR3/TRIF-dependent and one RIG-I/MDA5/IPS-1-dependent.
Fig. 5.
Poly(I:C) and IFNα treatment induces cell death through different pathways. (A) Viabilities of 293FT cells transfected with the indicated expression plasmids in the presence or absence of pIC (5 μg/mL) and IFNα (50 ng/mL) for 44 h were quantified with the MTT assay. The values represent the average of at least three separate experiments, with SDs shown by error bars. TRIF and IPS-1 with MDA5 or RIG-I induced significant (*P < 0.02) cell death in response to pIC and IFNα. (B) 293FT cells transfected with TRIF expression vector or vector alone were stained with FITC-labeled annexin V to detect apoptosis. mRNA expression levels of TLR3 (C), RIG-I (D), and MDA5 (E) were measured using real-time PCR with (+) or without (−) i.p. pIC injection (250 μg). mRNAs prepared from pancreas, liver, and spleen of WT, Irf2−/−, and Irf2−/−Ifnar1−/− mice were converted into cDNA, and the amount of cDNA was determined by real-time PCR with the specific primers listed in SI Materials and Methods. The values represent the average of at least two mice, with SDs shown by error bars. *P < 0.05 versus Irf2−/− mice.
We used real-time PCR to examine the induction of TLR3, RIG-I, and MDA5 mRNAs in pIC-treated WT, Irf2−/−, and Irf2−/−Ifnar1−/− mice. The levels of all three mRNAs were induced by nearly 100-fold in both pIC-WT and pIC-Irf2−/− mice, and these increases were abolished by deletion of IFNAR1 (Fig. 5 C–E). The IFN signal activation triggered by pIC is essential to initiate TLR3/TRIF- and RIG-I/MDA5/IPS-1-dependent acinar cell death, but is not sufficient to cause pancreatitis (Table S3). The elevation of trypsinogen5 expression mediated by abolishing IRF2 is also necessary for enhancing the cell death leading to lethal pancreatitis.
Activation Mechanisms of Mouse Trypsinogen5 and Human PRSS3.
Trypsinogens (including trypsinogen5) can be activated in pancreatic acinar cells, or in other cells or tissues by enteropeptidase expressed in nonduodenal cells (23) such as in keratinocytes and oral carcinoma cells (24, 25). It is possible that keratinocyte-expressed enteropeptidase activates the trypsinogen5 expressed in skin (Fig. S3A), promoting age-dependent skin inflammation in Irf2−/− mice (4). Another possibility could be that proteases in addition to enteropeptidase can cleave pancreatic trypsinogen5. We have confirmed that cathepsin B, whose expression was elevated in Irf2−/− mice, can activate trypsinogen5 in vitro (Fig. S5C). The last possibility is that autocatalytic cleavage of trypsinogen, usually restricted under steady-state conditions, is accelerated in response to chemical stress or viral infection. Indeed, the autoactivation of trypsinogen is reportedly accelerated in low pH or by Ca2+ in vitro (26).
In conclusion, this study has identified important genes associated with IRF2 functions in mice. Our results suggest that IRF2 influences the expression of mouse trypsinogen5, whose human counterpart is PRSS3. Our data should therefore help to elucidate new IRF functions in humans.
Materials and Methods
Mice.
Irf1−/− and Irf2−/− mice have been described (3). IFNα/β receptor 1 (Ifnar1)−/− mice were purchased from B&K Universal (8). TRIF−/− mice have been described (9). Irf2−/−Ifnar1−/−, Irf2−/−Irf1−/−, and Irf2−/−Trif−/− double mutant mice were generated by crossing Irf2+/− with Ifnar1−/−, Irf1−/−, and Trif−/− mice, respectively. All mice were maintained under specific pathogen-free conditions and used at 6–12 wk of age. All experiments were performed according to institutional guidelines.
Cells.
Human embryonic kidney (HEK)293T and 293FT (Invitrogen) cells and HeLa cells were cultured in DMEM supplemented with 10% FBS. Mouse pancreatic acinar TGP49 cells were cultured in a 1:1 mixture of DMEM and Ham's F-12 medium supplemented with 10% FBS.
Histological Analysis.
Pancreas tissues were fixed overnight in 10% formalin, embedded in paraffin, sectioned, and stained with hematoxylin (0.4%) and eosin (0.5%) for light microscopic analysis. For electron microscopic analysis, the tissues were fixed in 2.5% glutaraldehyde solution buffered to pH 7.4 with 0.1 M phosphate buffer for 4 h at 4 °C. Postfixation was performed with 2% osmium tetroxide solution buffered to pH 7.4 with the same buffer for 2 h at 4 °C, and they were embedded, sectioned, and doubly stained with uranyl acetate and lead nitrate.
Microarrays.
Total RNAs from the pancreas of wild-type and Irf2−/− mice aged 6 wk, harvested 3 h after no injection or a peritoneal injection with 250 μg poly(I:C), were used in the array studies. The quality of the RNA was assessed with an Agilent 2100 Bioanalyzer, and samples of 100 ng total RNA were reverse-transcribed and then amplified by in vitro transcription according to Affymetrix standard protocols. The mouse Affymetrix GeneChip Mouse Gene 1.0 ST Array was used in all hybridizations. These arrays contain probes representing transcripts for 28,815 mouse gene entities. Microarray data were analyzed using Affymetrix Expression Console software and Gene Spring GX, whereas differentially expressed genes were identified with annotation.
Real-Time RT-PCR.
Total RNA was prepared from tissues using the acid phenol-guanidinium thiocyanate method after immersing the tissues for more than overnight in RNAlater Solution (Ambion). Reverse transcription was conducted for 60 min at 46 °C from 200 ng of purified total RNA using SuperScript III (Invitrogen), followed by 45 cycles of PCR (15-s denaturation at 95 °C, 25-s annealing at 55 °C, and 15-s extension at 72 °C). An SYBR Green PCR Kit (Qiagen) was used to monitor the PCR products on a LightCycler 1.5 and real-time PCR detection system (Roche). Primers designed for the respective genes are listed in SI Materials and Methods.
Plasmid Constructs.
cDNAs encoding human IRF5, IRF7, and IPS-1 were generated from total RNA prepared from 293T cells by RT-PCR using KOD-FX DNA polymerase (Toyobo). Human MDA5, RIG-I, and TLR3 cDNAs were generated from total RNA prepared from THP-1 (a human leukemia cell line) or HeLa cells by RT-PCR. Mouse Trypsinogen5, Prss1, and Spink3 cDNAs were made from total RNA prepared from WT mouse pancreas by PCR. All constructs generated by PCR were confirmed by DNA sequencing. The pTrypsinogen5-Luc reporter plasmid was constructed by inserting the promoter region (−1063 to +15) of the mouse trypsinogen5 gene by PCR into the pGL2-Basic vector. A series of deletion mutants was prepared using proper restriction enzymes (NcoI at −833; SpeI at −579; ScaI at −386; PvuII at −216) and a specific primer for the −100 site. The promoter region (−216 to +15) of the mouse trypsinogen5 gene was used to introduce point mutations into the ISREs. The point mutations of ISRE3 (−55 to −49, ATTGAAA→GTTTGCG), ISRE4 (−62 to −59, TTTC→CGCA), and ISRE5 (−84 to −78, AATGAAA→GATTGCG) were introduced by overlap PCR mutagenesis. All constructs generated by PCR were confirmed by DNA sequencing.
PACE-Trypsinogen5 was constructed by replacing the activation peptide (-NSDDK-) of mouse trypsinogen5 cDNA with the PACE recognition peptide (-RTKR-) by overlap PCR mutagenesis.
Luciferase Reporter Assay.
293T cells (1 × 105 per well) were plated in 24-well plates and transfected 24 h later with 200 ng of the firefly luciferase reporter plasmid pTrypsinogen5-Luc, using FuGENE6 (Roche), along with each expression vector (20 ng unless otherwise stated) as indicated. In all cases, cells were transfected with 20 ng pRL-TK (thymidine kinase promoter-driven Renilla luciferase reporter gene; Promega) to normalize the transfection efficiency. TGP49 cells (1 × 105 per well) were plated in 12-well plates and transfected 24 h later with 1 μg of the firefly luciferase reporter plasmid pTrypsinogen5-Luc using Lipofectamine 2000 (Invitrogen), along with each expression vector (100 ng unless otherwise stated) as indicated. In all cases, cells were transfected with 20 ng pRL-RSV (RSV promoter-driven Renilla luciferase reporter gene). At 26 h posttransfection, luciferase activity was determined with a dual luciferase assay system (Promega). Mouse IRF2-specific and control siRNAs were purchased from Santa Cruz Biotechnology.
Chromatin Immunoprecipitation.
Nuclear extracts from TGP49 cells were subjected to DNA–protein cross-linking with 1% formaldehyde for 5 min. After extensive washing, the samples were suspended in 500 μL of 150 mM NaCl, 25 mM Tris (pH 7.5), 5 mM EDTA, 1% Triton X-100, 0.1% SDS, and 0.5% deoxycholate and sonicated. After centrifugation at 14,000 rpm for 10 min at 4 °C, the supernatants were immunoprecipitated with 0.5 μg anti-IRF2 antibody, or the corresponding IgG (Sigma) (as a control), and Protein A Sepharose4B Fast Flow beads. The amounts of precipitated DNA were quantified by PCR using a pair of mouse Trypsinogen5 promoter-specific primers and Angiotensinogen exon2-specific primers (SI Materials and Methods).
Trypsin Activity Assay.
Trypsin activity was monitored by the amount of released p-nitroanilide (pNA) from a specific substrate, measuring spectrophotometric units at 405 nm (A405) (Trypsin Activity Assay Kit; BioVision). Cell lysates prepared at 48 h posttransfection of the indicated expression plasmids were used with or without enteropeptidase (light chain, porcine; GenScript).
Cell Death Assay.
Pancreatic tissues were used in a TUNEL assay. Briefly, tissue sections were incubated with 20 μg/mL proteinase K for 20 min, followed by inhibition of endogenous peroxidase by incubation with 2% H2O2 for 7 min. TdT (GIBCO-BRL) and biotinylated dUTP (Roche) in TdT buffer [0.1 M potassium cacodylate (pH 7.2), 2 mM CoCl2, 0.2 mM DTT] were added to the sections and incubated in a humid atmosphere at 37 °C for 90 min after immersion in TdT buffer. The reaction was terminated by transferring the slides to TB buffer (300 mM NaCl, 30 mM Na citrate) for 30 min. The sections were covered with 10% rabbit serum for 10 min and then with the avidin–biotin peroxidase complex for 30 min. Finally, 3,3'-diaminobenzidine (DAB) was used as the chromogen. To detect apoptotic cells, FITC-conjugated annexin V (BioVision) was used according to the manufacturer's instruction. An MTT (ICN) assay to assess living cells was performed according to the manufacturer's instruction.
Supplementary Material
Acknowledgments
This work was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (22659092) and by the Global Center of Excellence Program at Nagasaki University.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116273108/-/DCSupplemental.
References
- 1.Savitsky D, Tamura T, Yanai H, Taniguchi T. Regulation of immunity and oncogenesis by the IRF transcription factor family. Cancer Immunol Immunother. 2010;59:489–510. doi: 10.1007/s00262-009-0804-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 3.Matsuyama T, et al. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell. 1993;75:83–97. [PubMed] [Google Scholar]
- 4.Hida S, et al. CD8(+) T cell-mediated skin disease in mice lacking IRF-2, the transcriptional attenuator of interferon-α/β signaling. Immunity. 2000;13:643–655. doi: 10.1016/s1074-7613(00)00064-9. [DOI] [PubMed] [Google Scholar]
- 5.Lampel M, Kern H-F. Acute interstitial pancreatitis in the rat induced by excessive doses of a pancreatic secretagogue. Virchows Arch A Pathol Anat Histol. 1977;373:97–117. doi: 10.1007/BF00432156. [DOI] [PubMed] [Google Scholar]
- 6.Mashima H, et al. Interferon regulatory factor-2 regulates exocytosis mechanisms mediated by SNAREs in pancreatic acinar cells. Gastroenterology. 2011;141:1102–1113. doi: 10.1053/j.gastro.2011.05.051. [DOI] [PubMed] [Google Scholar]
- 7.Gerke V, Creutz C-E, Moss S-E. Annexins: Linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol. 2005;6:449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- 8.Hwang SY, et al. A null mutation in the gene encoding a type I interferon receptor component eliminates antiproliferative and antiviral responses to interferons α and β and alters macrophage responses. Proc Natl Acad Sci USA. 1995;92:11284–11288. doi: 10.1073/pnas.92.24.11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamamoto M, et al. Role of adaptor TRIF in the MyD88-independent Toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 10.Chen J-M, Férec C. Chronic pancreatitis: Genetics and pathogenesis. Annu Rev Genomics Hum Genet. 2009;10:63–87. doi: 10.1146/annurev-genom-082908-150009. [DOI] [PubMed] [Google Scholar]
- 11.Whitcomb D-C. Genetic aspects of pancreatitis. Annu Rev Med. 2010;61:413–424. doi: 10.1146/annurev.med.041608.121416. [DOI] [PubMed] [Google Scholar]
- 12.Spicuglia S, Pekowska A, Zacarias-Cabeza J, Ferrier P. Epigenetic control of Tcrb gene rearrangement. Semin Immunol. 2010;22:330–336. doi: 10.1016/j.smim.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 13.Rowen L, et al. Interchromosomal segmental duplications explain the unusual structure of PRSS3, the gene for an inhibitor-resistant trypsinogen. Mol Biol Evol. 2005;22:1712–1720. doi: 10.1093/molbev/msi166. [DOI] [PubMed] [Google Scholar]
- 14.Figarella C, Miszczuk-Jamska B, Barrett A-J. Possible lysosomal activation of pancreatic zymogens. Activation of both human trypsinogens by cathepsin B and spontaneous acid. Activation of human trypsinogen 1. Biol Chem Hoppe Seyler. 1988;369(Suppl):293–298. [PubMed] [Google Scholar]
- 15.Halangk W, et al. Role of cathepsin B in intracellular trypsinogen activation and the onset of acute pancreatitis. J Clin Invest. 2000;106:773–781. doi: 10.1172/JCI9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meister T, et al. Missorting of cathepsin B into the secretory compartment of CI-MPR/IGFII-deficient mice does not induce spontaneous trypsinogen activation but leads to enhanced trypsin activity during experimental pancreatitis—without affecting disease severity. J Physiol Pharmacol. 2010;61:565–575. [PubMed] [Google Scholar]
- 17.Hashimoto D, et al. Involvement of autophagy in trypsinogen activation within the pancreatic acinar cells. J Cell Biol. 2008;181:1065–1072. doi: 10.1083/jcb.200712156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Negishi H, et al. Negative regulation of Toll-like-receptor signaling by IRF-4. Proc Natl Acad Sci USA. 2005;102:15989–15994. doi: 10.1073/pnas.0508327102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honda K, et al. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature. 2005;434:1035–1040. doi: 10.1038/nature03547. [DOI] [PubMed] [Google Scholar]
- 20.Ji B, Gaiser S, Chen X, Ernst S-A, Logsdon C-D. Intracellular trypsin induces pancreatic acinar cell death but not NF-&kappaB activation. J Biol Chem. 2009;284:17488–17498. doi: 10.1074/jbc.M109.005520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaiser W-J, Offermann M-K. Apoptosis induced by the Toll-like receptor adaptor TRIF is dependent on its receptor interacting protein homotypic interaction motif. J Immunol. 2005;174:4942–4952. doi: 10.4049/jimmunol.174.8.4942. [DOI] [PubMed] [Google Scholar]
- 22.Lei Y, et al. MAVS-mediated apoptosis and its inhibition by viral proteins. PLoS One. 2009;4:e5466. doi: 10.1371/journal.pone.0005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yahagi N, et al. Complementary DNA cloning and sequencing of rat enteropeptidase and tissue distribution of its mRNA. Biochem Biophys Res Commun. 1996;219:806–812. doi: 10.1006/bbrc.1996.0315. [DOI] [PubMed] [Google Scholar]
- 24.Nakanishi J, Yamamoto M, Koyama J, Sato J, Hibino T. Keratinocytes synthesize enteropeptidase and multiple forms of trypsinogen during terminal differentiation. J Invest Dermatol. 2010;130:944–952. doi: 10.1038/jid.2009.364. [DOI] [PubMed] [Google Scholar]
- 25.Vilen S-T, et al. Intracellular co-localization of trypsin-2 and matrix metalloprotease-9: Possible proteolytic cascade of trypsin-2, MMP-9 and enterokinase in carcinoma. Exp Cell Res. 2008;314:914–926. doi: 10.1016/j.yexcr.2007.10.025. [DOI] [PubMed] [Google Scholar]
- 26.Chen J-M, et al. Evolution of trypsinogen activation peptides. Mol Biol Evol. 2003;20:1767–1777. doi: 10.1093/molbev/msg183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.