Abstract
The predominant mode of HIV-1 infection is heterosexual transmission, where a genetic bottleneck is imposed on the virus quasispecies. To probe whether limited genetic diversity in the genital tract (GT) of the transmitting partner drives this bottleneck, viral envelope sequences from the blood and genital fluids of eight transmission pairs from Rwanda and Zambia were analyzed. The chronically infected transmitting partner's virus population was heterogeneous with distinct genital subpopulations, and the virus populations within the GT of two of four women sampled longitudinally exhibited evidence of stability over time intervals on the order of weeks to months. Surprisingly, the transmitted founder variant was not derived from the predominant GT subpopulations. Rather, in each case, the transmitting variant was phylogenetically distinct from the sampled locally replicating population. Although the exact distribution of the virus population present in the GT at the time of transmission cannot be unambiguously defined in these human studies, it is unlikely, based on these data, that the transmission bottleneck is driven in every case by limited viral diversity in the donor GT or that HIV transmission is solely a stochastic event.
HIV heterosexual transmission is the primary mode of infection worldwide, and according to Joint United Nations Program on HIV/AIDS reports, the overall epidemic is increasingly driven by this type of transmission, particularly in developed countries (1). However, the biological mechanisms underlying heterosexual transmission are poorly understood. Limited availability of samples during early infection, especially from the genital mucosa, and a lack of knowledge about the source of transmitted variants contribute to the challenge of deciphering the events that occur at the time of transmission.
Most transmission studies have relied primarily on men who have sex with men and female sex worker cohorts (2–8), and, generally, the transmitted viruses could not be directly compared with the virus population in the transmitting partner (donor) from which they were derived. In our previous studies, we evaluated blood samples from epidemiologically linked transmission pairs during acute infection and found evidence that a single variant establishes infection (9, 10). Other studies of acutely infected individuals have confirmed this genetic bottleneck in the recipient blood, thus suggesting selection for particular variants during transmission (11–16).
Understanding the origin of the genetic bottleneck that occurs during heterosexual transmission requires characterizing the source of the transmitted virus in the donor genital tract (GT). Recent studies have begun to answer the question of whether viral variants are sequestered in the genital tissue (17–22) and show that distinct genetic lineages can propagate there. Here, we studied the genetic relationship of viruses in the blood and genital fluids from the chronically infected donor partners of eight Rwandan and Zambian transmission pairs and show that although distinct viral subpopulations are present in the GT, the transmitting founder variant did not originate from the predominant sampled population. These results provide evidence that transmission is not solely stochastic and may involve selection of a more transmissible genetic variant.
Results
Donor env V1–V4 Sequences Are Heterogeneous in Blood and the GT.
Genital samples (vaginal swabs or semen) were collected from eight chronically infected donor partners, with concomitant blood samples from both donor and recipient, at the time the recipient partner was identified as p24 ELISA- or HIV antibody-positive (Table 1). Estimated days to infection (EDI) and most recent common ancestor (MRCA) were determined in our previous study (10). EDI for the recipients was estimated to be 17–86 d, and inference of MRCA for each recipient indicated that this time period ranged from 5 to 66 d (11). Collectively, these data indicate that the samples represent very recent transmission events. Transmission pairs RW36 and RW56 from Rwanda and ZM292 from Zambia were infected with subtype A HIV-1, whereas the five remaining transmission pairs were from Zambia and were infected with subtype C HIV-1. Six of the eight pairs analyzed were female-to-male (FTM) transmissions, and two were male-to-female (MTF) transmissions.
Table 1.
Summary of human subject data
| No. sequences analyzed |
|||||||||||||||
| Coded ID | Partner status | PB | PL | SW | CA | CF | Total | Env | p24 | Sero date | Genital fluid sample date | EDI | MRCA | VL (copies/mL) | VL sample date |
| FTM | |||||||||||||||
| RW36F | D | 20 | 20 | 12 | 6 | 58 | A1 | Mar 7, 2005 | 283,391 | Mar 7, 2005 | |||||
| RW36M | R | 20 | 21 | 41 | A1 | ND | Mar 7, 2005 | <86 | 21 | 113,510 | Mar 7, 2005 | ||||
| ZM201F | D | 16 | 21 | 19 | 20 | 17 | 93 | C | Feb 7, 2003 | 471,382 | Feb 7, 2003 | ||||
| ZM201M | R | 17 | 29 | 46 | C | Feb 7, 2003 | Feb 7, 2003 | 31 | 35 | 750,000 | Feb 7, 2003 | ||||
| ZM216F | D | 23 | 17 | 17 | 9 | 9 | 75 | C | Jan 17, 2004 | 371,901 | Jan 17, 2004 | ||||
| ZM216M | R | 19 | 40 | 59 | C | Jan 17, 2004 | Jan 17, 2004 | 31 | 66 | 750,000 | Jan 17, 2004 | ||||
| ZM221F | D | 21 | 16 | 34 | 1 | 0 | 72 | C | Mar 8, 2003 | 62,741 | Mar 8, 2003 | ||||
| ZM221M | R | 26 | 25 | 73 | C | Mar 7, 2003 | Mar 7, 2003 | 31 | 23 | 750,000 | Mar 7, 2003 | ||||
| ZM238F | D | 21 | 20 | 30 | 1 | 3 | 75 | C | Oct 29, 2002 | 297,927 | Oct 29, 2002 | ||||
| ZM238M | R | 21 | 18 | 39 | C | Oct 11, 2002 | Nov 29, 2002 | 40 | 5 | 90,791 | Nov 29, 2002 | ||||
| ZM292F | D | 17 | 20 | 13 | 13 | 8 | 71 | A1 | May 24, 2005 | 14,301 | May 24, 2005 | ||||
| ZM292M | R | 17 | 22 | 39 | A1 | May 18, 2005 | May 24, 2005 | 28 | 44 | 129,545 | May 24, 2005 | ||||
| MTF | |||||||||||||||
| RW56M | D | 20 | 20 | 11 | 7 | 58 | A1 | Aug 24, 2005 | 44,160 | Aug 23, 2005 | |||||
| RW56F | R | 20 | 20 | 40 | A1 | ND* | Jul 21, 2005 | 17 | 18 | 693 | Jul 21, 2005 | ||||
| ZM242M | D | 19 | 20 | 7 | 0 | 46 | C | Jan 25, 2003 | 1,019,048 | Jan 25, 2003 | |||||
| ZM242F | R | 13 | 18 | 31 | C | Jan 25, 2003 | Jan 25, 2003 | 31 | 30 | 2,974 | Jan 25, 2003 | ||||
The numbers of sequences analyzed from FTM and MTF transmission pairs are identified for each chronically infected donor (D) and recipient partner (R). Donor and recipient blood PB and PL were collected at either the first p24 antigen-positive date (p24) or seroconversion date (Sero date). The p24 antigen date was not available in some cases [not determined (ND)]. Donor genital fluids were collected near or on the same dates (Genital fluid sample date), and the numbers of sequences analyzed from vaginal swab (SW), CA, and CF fractions are shown. Envelope subtype (Env), EDI, and MRCA were determined. PL viral loads (VL) were determined on the sample date shown (VL sample date).
*One individual was identified as being viral RNA-positive.
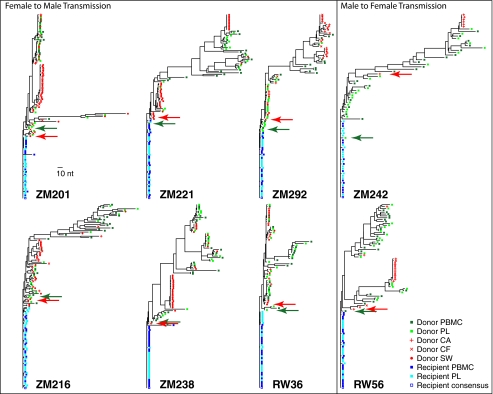
Nucleic acids were extracted from blood and the GT samples of the chronically infected donor partners and from the blood of the recipient partners. To investigate the genetic diversity of viruses in the different compartments, single-genome PCR amplification (SGA) of the envelope-encoding region was performed, followed by direct amplicon sequencing. Donor and recipient blood env sequences included in this study were determined and reported in our previous study (10). Maximum likelihood (ML) trees generated from the V1–V4 sequences for each transmission pair revealed a genetically diverse population derived from the donor blood, with sequences from plasma (PL) and peripheral blood mononuclear cells (PB) intermingled on the tree (Fig. 1). Donor GT sequences, cell-associated (CA), cell-free (CF), and swab-associated (SW), were also genetically diverse but exhibited a different distribution in the phylogenetic trees. Some GT sequences intermingled with blood sequences, but the majority (up to 86%) was limited to distinct branches, some of which contained identical or nearly identical sequences. This is consistent with restricted movement of virus between the blood and GT compartments and with some localized replication, which we formally address below (Fig. 2 and Fig. S1). For example, the majority (55%) of GT sequences from ZM201F (female donor) clustered on a single branch of the donor tree, whereas the remaining GT sequences were intermingled with the blood sequences (Fig. 2A). Although 12 of the 31 sequences on this branch are identical, overall diversity exceeds 1.85%, indicating sustained GT evolution. A similar pattern was also observed for female donor ZM238F, where a single branch containing 21 identical and highly related sequences made up 64% of amplified sequences (Fig. 2B). Donor ZM221F exhibited two distinct GT branches consisting of 16 somewhat diverse and 11 identical GT sequences, respectively (Fig. 2C). Based on the phylogenetic structure of these branches, it seems likely that those containing identical sequences originate from some form of transient clonal amplification (see below), whereas the more developed branches reflect established GT infection, consistent with findings reported for seminal PL samples from subtype C HIV-1–infected males in Malawi (18). Evidence of distinct GT sequences was also observed in a majority of the remaining five transmission pairs (Fig. S1), although because of the difficulty in amplifying from seminal samples of the MTF transmission pairs available (ZM242 and RW56), a more limited sampling of male GT sequences was available.
Fig. 1.
Phylogenetic analysis of env V1–V4 sequences from the blood and GT in eight heterosexual transmission pairs. Env V1–V4 nucleotide sequences were aligned for each linked transmission pair, and ML trees were drawn to reveal sequence heterogeneity and homogeneity. Donor blood sequences are shown in green (dark green-filled squares, PB; light green-filled squares, PL), donor GT sequences are shown in red (red +, CA; red x, CF; red-filled circles, SW), and recipient blood sequences are shown in blue (dark blue-filled squares, PB; light blue-filled squares, PL). Highlighter analysis revealed the donor GT (red arrow) and blood sequence (green arrow) most related to the consensus recipient sequence (blue open square). Horizontal branch lengths are drawn to scale, with the bar representing 10-nt changes.
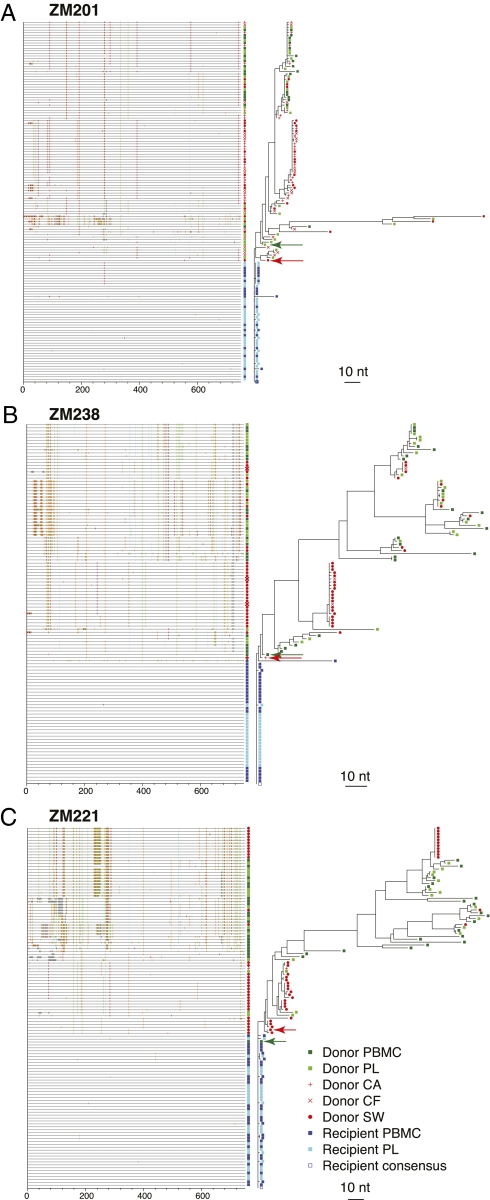
Fig. 2.
Highlighter and phylogenetic analysis reveal distinct GT populations. Aligned Env V1–V4 nucleotide sequences for transmission pairs were analyzed by the Los Alamos Highlighter tool. (A–C) Output files with Highlighter plots aligned to the phylogenetic trees. Tick marks indicate nucleotide differences from the recipient consensus sequence (blue open square). Nucleotide differences are color-coded and marked according to their genetic location along the length of V1–V4. Colors are as follows: green, A; red, T; orange, G; blue, C; gray, gaps. Arrows point to those variants in the blood (green) and GT (red) most closely related to the transmitted founder virus.
Donor GT Sequences Exhibit Evidence of Compartmentalization.
To define further the nature of GT sequence compartmentalization in the donor, we used the Slatkin–Maddison (SM) strategy (23) to test the hypothesis that GT sequences cluster together more with each other than they do with sequences from the blood, and therefore constitute a compartmentalized subpopulation (18, 23, 24). The SM test compares the observed number of migration events with a null distribution from permuted tree labels to evaluate whether evidence for compartmentalization exists in the phylogeny (Table 2).
Table 2.
Compartmentalization between blood and GT
| Full-length (HXB2 6600..7478) |
Partitioned at GARD breakpoints |
|||||||||
| Identical sequences present |
Identical sequences removed |
|||||||||
| Donor ID | s | p(s’≤ s)* | q(s’ ≤ s)† | s | p(s’ ≤ s) | q(s’ ≤ s) | Region | s | p(s’ ≤ s) | q(s’ ≤ s) |
| RW36F | 15 | 0.3902 | 0.2149 | 10 | 0.5976 | 0.2724 | 5′: 6600..7000 | 3 | 0.0225 | 0.0279 |
| 3′: 7001..7478 | 7 | 0.4059 | 0.2205 | |||||||
| RW56M | 5 | <0.0001 | 0.0004 | 4 | <0.0001 | 0.0004 | 5′: 6600..6827 | 4 | 0.0298 | 0.0348 |
| Mid: 6828..7193 | 4 | 0.1932 | 0.1321 | |||||||
| 3′: 7194..7478 | 3 | 0.0023 | 0.0057 | |||||||
| ZM201F | 20 | 0.0005 | 0.0018 | 16 | 0.0032 | 0.0067 | 5′: 6600..6879 | 13 | 0.0592 | 0.0471 |
| 3′: 6880..7478 | 13 | 0.2361 | 0.1419 | |||||||
| ZM216F | 19 | 0.0073 | 0.0107 | 17 | 0.0535 | 0.0442 | 5′: 6600..6888 | 13 | 0.0507 | 0.0428 |
| 3′: 6889..7478 | 15 | 0.0405 | 0.0376 | |||||||
| ZM221F | 7 | <0.0001 | 0.0004 | 7 | <0.0001 | 0.0004 | 5′: 6600..6898 | 5 | <0.0001 | 0.0004 |
| 3′: 6899..7478 | 6 | 0.0017 | 0.0045 | |||||||
| ZM238F | 11 | <0.0001 | 0.0004 | 9 | 0.0036 | 0.0067 | 5′: 6600..6925 | 7 | 0.0274 | 0.0329 |
| 3′: 6926..7478 | 7 | 0.4429 | 0.2374 | |||||||
| ZM242M | 2 | <0.0001 | 0.0004 | 2 | 0.0376 | 0.0366 | 5′: 6600..6791 | 2 | 1.0000 | 0.4089 |
| Mid: 6792..7183 | 2 | 0.0004 | 0.0016 | |||||||
| 3′: 7184..7478 | 2 | 1.0000 | 0.4089 | |||||||
| ZM292F | 13 | <0.0001 | 0.0004 | 9 | <0.0001 | 0.0004 | 5′: 6600..7186 | 10 | 0.0035 | 0.0067 |
| 3′: 7187..7478 | 10 | 0.0713 | 0.0534 | |||||||
The SM test was performed on donor partners (Donor ID) using full-length envelope V1–V4 sequences (6600-7478, HXB2 nucleotide coordinates), full-length with identical sequences removed, and alignment regions (region) partitioned at genetic algorithm for recombination detection (GARD) breakpoints with identical sequences removed. The SM analysis used the observed number of migration events (s) to determine the P value (p).
*SM P value is the proportion of relabeled trees with as many or fewer migration events (s) as observed: p(s’ ≤ s), where s’ denotes results from 10,000 compartment-label permutations on the fixed tree, shown in bold, where P < 0.05.
†Storey and Tibshirani (25) q value from SM P value, shown in bold, where q < 0.05.
Because GT-enriched branches in several individuals contained identical sequences, which are potentially transient, we considered whether the presence of these monotypic sequences influence the outcome of the SM analysis by performing the test two ways: first, on the full-length sequences with monotypic variants included and, second, on full-length sequences with identical sequences removed. For the latter dataset, there was evidence for recombination, so we further analyzed sequence alignments partitioned at recombination breakpoints to determine whether recombination was influencing the outcome of the analysis. Both with and without the exclusion of sequences found in multiple copies, seven of eight subjects show evidence of compartmentalization (Table 2). Further, we have applied multiple test corrections to false-positive rates (P values) that compute false discovery rates (q values) using the method of Storey and Tibshirani (25), and we note that the distribution of significant results is unchanged at threshold significance levels of 0.05 (Table 2). This evidence for compartmentalization of GT variants is also supported by previous studies in both male and female subjects, where non-SGA analyses were performed (2–7, 17, 19, 20, 24, 26–30).
Distribution of env V1-V4 Sequences in the Female Donor GT Exhibits Evidence of Stability over Time.
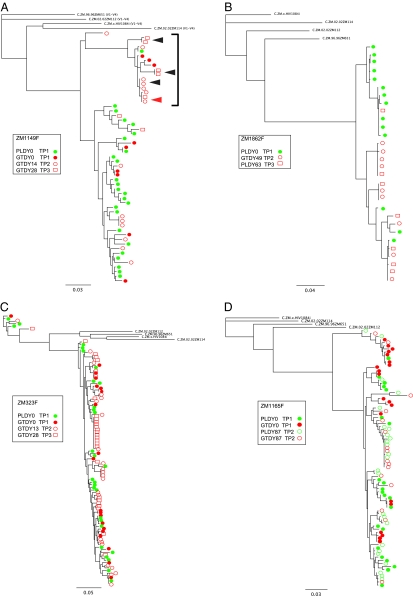
Because it is impossible to sample the diversity and compartmentalization of GT virus at the exact time of transmission, we examined whether the distribution of GT sequences is stable over a length of time similar to the window between transmission and sampling. We performed a longitudinal analysis of GT sequences using samples collected from four chronically infected females. Overall, when all sequences were combined, a distribution similar to that of the eight chronically infected donor partners was observed (Fig. 3). However, when individual time points were analyzed, relatively stable populations were observed in two of the four individuals. For example, in ZM1149F, the predominant GT branch of the phylogenetic tree consisted of sequences from three time points spanning 28 d (Fig. 3A, bracket); for time points 2 and 3, these represent 50% or more of amplified sequences [7 of day 14 (DY14) samples and 5 of 7 DY28 samples], whereas for the initial time point, they represent 33%. Within this cluster (delineated by the bracket in Fig. 3A), we observed identical and near-identical sequences from both the same time points (filled arrowheads) and different time points (open arrowheads), suggesting persistent clonal amplification in this tissue over at least 14 d (Fig. 3A). An SM analysis of these sequences also provided support for compartmentalization for a majority of time points, whether or not identical sequences were included (Table S1).
Fig. 3.
Phylogenetic analysis of longitudinal samples. Longitudinal sequences (V1–V4) from chronically infected females, blood PL and GT, were aligned, and ML phylogenetic trees were drawn for chronically infected female subjects ZM1149F (A), ZM1862F (B), ZM323F (C), and ZM1165F (D). Blood-derived sequences are shown in green, and GT-derived sequences are shown in red. Closed arrowheads in A identify identical or nearly identical sequences from both the same time points [GTDY28 time point 3 (TP3)], and open arrowheads identify identical or nearly identical sequences from different time points [GTDY14 time point 2 (TP2) and GTDY28 time point 3 (TP3)]. Four Zambian subtype C reference sequences were used to root the tree. Horizontal branch lengths are drawn to the scale shown.
In ZM1862F, evidence for a persistent GT population was also observed. In samples from this individual, two distinct populations were observed (Fig. 3B). One was represented primarily by PL virus sequences, and the other was composed mainly of GT variants. Within the latter population, sequences from both time points (time points 2 and 3) were equally represented on the subbranches (Fig. 3B), and although the SM analysis of full-length sequences did not reveal significant compartmentalization at the second time point, a trend to significance was observed when recombination break points were taken into account (Table S1).
In the remaining two females (ZM323F and ZM1165F), there was less evidence for stable GT populations. In ZM323F, sequences in blood and GT at the first two time points were evenly distributed across the tree (equilibrated), with no statistical support for compartmentalization, whereas at time point 3, there was evidence for clonal expansion in the GT with a predominant rake of identity present. This is consistent with such amplified sequences being transient in some cases. Sequences from ZM1165F did show both phylogenetic and statistical evidence of compartmentalization at time point 1, with a major subpopulation (35%) on a discrete branch. At time point 2, however, only 15% of the sequences were associated with this branch of the tree (Fig. 3 and Table S1).
Donor Variants Very Similar to the Recipient Founder Virus Are Present in Both Blood and GT.
In contrast to the heterogeneity of the donor env V1–V4 sequences described above, the recipient sequences from blood were homogeneous in all cases and emanated from one branch of the donor tree, consistent with a single donor genetic variant establishing infection (Fig. 1). To identify the sequence in the donor quasispecies that was most closely related to a consensus of the recipient sequences, which represents the transmitted founder env, the highlighter analysis tool from Los Alamos National Laboratories (http://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter.html) and matrices of intersequence distances were used (10, 11). The sequence information present in insertions was considered as well as base substitutions to identify this relationship. As an example, in ZM201, an FTM transmission, 29 (64%) of 45 recipient blood sequences were identical throughout V1–V4, with an additional 14 (31%) sequences having a single-nucleotide change (Fig. 2A). The highlighter analysis illustrates the heterogeneity in the donor female blood sequences, the similarly heterogeneous interspersed donor GT sequences, and a cluster of highly related GT sequences resulting from localized replication that correspond to the GT-enriched branches shown on the phylogenetic tree. Highlighter plots for all eight transmission pairs revealed similar patterns (Fig. 2 and Fig. S1).
Variants that were highly related to the founder virus in the recipient were found in both blood and GT of the donor partner for each of the eight transmission pairs. Inspection of donor ZM201F sequences showed that the variant that most closely resembled the recipient founder sequence was derived from blood and had five nucleotide mismatches, whereas an SW sequence from the donor GT differed by six nucleotides from the recipient founder sequence (Table S2). In a similar analysis of nucleotide differences for ZM238 and ZM221, we found that the closest blood and GT variants differed by four (PB) and three (CA) nucleotides for the former and zero (PB) and six (SW) nucleotides for the latter.
An analysis of all eight transmission pairs revealed that for five pairs (ZM201, ZM221, ZM292, RW56, and ZM242), a blood variant was most closely related to the recipient founder sequence. In three of these five cases, the source of the most highly related sequence was PB, whereas in two cases, the source was PL virus (Table S2).
Dominant Viral Population in the Donor GT at the Time of Sampling Is Not the Source of the Transmitted Variant.
To define further the genetic bottleneck associated with transmission in these couples, we assessed the closest donor variant's relatedness to the founder on the phylogenetic tree and to other donor blood and GT sequences. This analysis revealed that the donor GT variant closest to the founder virus was distinct from the predominant GT populations and was not associated with the GT enriched populations in any case. As an example, female donor ZM238F had a very obvious branch of the tree that comprised the bulk of the GT sequences [21 (62%) of 34] (Fig. 2B). The GT sequence most closely related to the recipient founder sequences, on the other hand, was clearly distant from this GT subpopulation. This obvious pattern of exclusion was also found in ZM201F, with the bulk of the donor genital sequences clustering together [31(55%) of 56] but not with the transmitted donor variant (Fig. 2A). The phylogenetic tree of ZM221F had two distinct branches that contributed to the bulk of the GT population, but, again, neither included the transmitted donor variant (Fig. 2C). This finding was consistent across all eight transmission pairs in both MTF and FTM transmissions (Fig. 2 and Fig. S1).
To determine whether the founder-like GT variant in the donor is statistically distinct from the defined donor GT virus subpopulations in these eight transmission pairs, we used a test of random transmission (ToRT) for the probability that the transmitted sequence is sampled from outside a dominant GT population. We objectively defined GT clusters by grouping together sequences more related than a threshold distance (D), which we varied systematically from 1 to 10 nucleotides. Among GT sequences from each of the eight donors, the variant most closely related to the recipient consensus was distinct from the clonally amplified GT clusters, defined by no nucleotide differences, (i.e., a “rake of identity” among two or more sequences in the tree) at a probability significantly greater than chance (P = 0.002; Table 3). When clades were defined by including increasingly distant sequences from the MRCA of the cluster, a second peak of significance was observed with a value for D of 8. To reduce the likelihood that we were comparing potentially transient clonally amplified populations with the founder-related sequence, a second analysis was performed in which all identical sequences were excluded from the analysis. In this case, although significance was reduced for all values of D, a significant P value (0.046) was observed when the GT-specific subpopulations were defined as variants differing by less than nine nucleotides. This is consistent with the subpopulations observed visually in the individuals (ZM201, ZM221, and ZM238) described above. These analyses thus provide objective evidence that, when considering the data across all eight transmission pairs, the GT sequence that was most related to the founder variant was preferentially not part of a defined GT cluster.
Table 3.
Test of random transmission
| No duplicates* |
||||
| D† | nobs‡ | p(D)§ | nobs | p(D) |
| 1 | 8 | 0.002 | — | — |
| 2 | 6 | 0.010 | 6 | 0.239 |
| 3 | 5 | 0.030 | 5 | 0.284 |
| 4 | 5 | 0.023 | 5 | 0.228 |
| 5 | 5 | 0.013 | 5 | 0.123 |
| 6 | 5 | 0.012 | 5 | 0.114 |
| 7 | 5 | 0.012 | 5 | 0.114 |
| 8 | 5 | 0.003 | 5 | 0.046 |
| 9 | 4 | 0.014 | 4 | 0.113 |
| 10 | 2 | 0.238 | 2 | 0.546 |
The ToRT identified donor GT sequences more closely related than a given nucleotide difference threshold (D) as clusters and identified the number of donors (n) in which the GT variant most like the founder virus was not a part of the cluster identified for D. p(D) determined the statistical significance that the founder-like GT variant was distinct from the GT virus clusters.
*To evaluate the influence of the presence of identical sequences, the test was repeated with identical sequences removed.
†Threshold distance, in nucleotides, below which sequences are clustered together.
‡Number of transmission events, from a total of eight, in which the transmitted variant is observed to occur outside of a cluster of sequences related by distance D.
§Probability that the observed number of transmission events with sequences transmitted outside a cluster (i.e., not in a rake) is significantly different from what would be expected if transmission were equally likely among all sequences. Shown in bold are P values < 0.05.
Discussion
HIV heterosexual transmission originates from viruses present in the donor GT. Therefore, the characterization of the viruses within this compartment in the context of transmission pairs is critical to our understanding of the genetic bottleneck and for developing strategies aimed at reducing virus spread (9–16). The genetic bottleneck could result from at least four possibilities that are not mutually exclusive: (i) limited diversity of the virus quasispecies in the transmitting partner's GT, (ii) inefficient trafficking of viruses across the genital mucosa of the uninfected partner, (iii) inefficient dissemination of viruses from the initial infection of the genital mucosa to the periphery, and (iv) a phenotypic trait that is essential for establishment and dissemination of infection. In this study, we have used end-point dilution PCR and direct sequencing of viral envelope coding sequences in blood and genital fluids from eight epidemiologically linked FTM and MTF heterosexual transmission pairs to investigate the origin of the transmission bottleneck. This allowed us to define the viral diversity in the GT of the transmitting partner and to identify the virus variant in the donor that was most similar to the virus that established de novo infection.
Our comparative analyses of HIV-1 env V1–V4 sequences in the blood (PB and PL) and GT (semen and vaginal fluids) identified genetically distinct viral populations within the GT. A majority of GT-derived sequences frequently clustered together on distinct branches, and there was statistically significant evidence for compartmentalization between virus in the GT and blood, whether or not identical sequences were removed from the analysis. Evidence for clonal expansion within the GT was also observed, in which rakes of identical or near-identical sequences formed distinct clusters that likely evolved from a single ancestor. The distinct GT clusters consisted of sequences from both CA and CF virus, and it is therefore likely that they were generated through local amplification. This is supported by our longitudinal analysis in which viruses from more than one time point were present in the same rake (Fig. 3 A–C). These results are similar to those described recently by Anderson et al. (18) for seminal PL and consistent with other studies that have investigated the relationship between blood and genital viral populations (5, 17–21).
If transmission were a stochastic sampling of the donor GT virus population, the most common viral variants in the donor GT should have the greatest opportunity to interact with the recipient genital mucosa and be favored for transmission. By contrast, the absence of the most abundant GT variants after transmission in eight transmission pairs argues for selection of a variant that is more fit for transmission. This observation is strengthened by our ToRT analysis, which objectively determined that for a majority of transmission pairs, the founder variant was distinct from GT clusters at a probability significantly greater than by chance, irrespective of whether identical sequences were removed from the analysis. It is difficult, if not impossible, to establish unequivocally the distribution of variants at the actual time of transmission in human studies; however, given that some individuals exhibit evidence of stability in the GT over time, it is remarkable that locally replicating viruses within phylogenetically distinct GT subpopulations at the time of sampling were also genetically distinct from the virus establishing infection in all eight cases. Therefore, despite uncertainties about the exact genetic makeup of the virus population at the time of heterosexual transmission, these findings point to the selection of a minor GT variant with properties favoring transmission.
It will be critical, in future studies, to elucidate the phenotypic properties of the viruses that appear to be uncommon in the GT yet establish infection in the new host and to compare those properties with those of viruses that are associated with stable subpopulations but are apparently not transmitted. We and others have reported that founder viruses generally have fewer glycosylation sites and more compact Envs (9, 31–33), and our earlier studies also demonstrated that newly transmitted viruses were not escape variants from PL-neutralizing antibodies in the donor (9). Recent studies have suggested that subtype A and C founder viruses bind efficiently to the gut homing receptor α4β7 expressed on a subset of highly susceptible CD4 T lymphocytes and that this property can be reduced with evolution of the virus after transmission (34). Collectively, the current study and these previous studies point to selection for a virus variant with a set of essential biological properties during transmission.
Methods
Study Subjects.
Cohabiting HIV-1–serodiscordant heterosexual couples enrolled in the Zambia Emory HIV Research Project and Projet San Francisco in Lusaka, Zambia and Kigali, Rwanda were tested quarterly with HIV rapid antibody assays and an HIV-1 p24 antigen ELISA (Beckman Coulter) (35–38). Individuals identified as being antigen-positive returned within 1 wk and were tested using an HIV rapid antibody to diagnose acute infection (37, 39). Informed consent and human subject protocols were approved by the Emory University Institutional Review Board, the University of Zambia School of Medicine Research Ethics Committee, and the Rwanda Ethics Committee.
The algorithm used to determine EDI was previously described by Haaland et al. (10). The MRCA was determined as previously described (11). HIV-1 viral loads from blood were determined by an HIV-1 RNA RT-PCR assay (Amplicor HIV-1 Monitor Test, v1.5, standard version; Roche Diagnostics) performed by the Emory Center for AIDS Research.
Nucleic Acid Extractions from Blood and Genital Samples, Generation of HIV-1 env Single-Genome Amplicons.
Venipuncture blood samples were centrifuged to separate PL and PB and were stored at −80 °C. Whole semen was immediately aliquoted into 250-μL cryovials and stored at −80 °C. Vaginal fluids were collected using Dacron swabs (Fisher Scientific), immediately placed in 1.5 mL of RNAlater RNA stabilization solution (Ambion), and stored at −80 °C. Blood viral RNA and genomic DNA were extracted from PL and PB per the manufacturer's instructions using the QIAamp RNA (Qiagen) and the QIAamp DNA (Qiagen) kits, respectively. Vaginal swabs (SW) were removed from RNAlater and set aside while the remaining vaginal fluid in RNAlater or 250 μL of whole semen was centrifuged at 1,500 × g for 5 min at room temperature to collect CA virus. The supernatant fraction from above was ultracentrifuged at 114,000 × g for 1 h at 4 °C, and CF virus was collected. Vaginal and seminal nucleic acids were extracted from CA, CF, and SW per the instructions of the manufacturer of the NucliSens Isolation Kit (bioMérieux, Inc.), with the exception of increasing the amount of silica beads to 200 μL (40, 41). Our preliminary studies showed that the SW retained ∼50% of virus collected and could therefore not be identified as either CA or CF.
Viral DNA was used to establish epidemiological linkage and Env subtype as previously described (9, 10, 42). cDNA synthesis and SGA of the env gene from PB, PL, and genital fluids (SW, CA, and CF) was performed using end point-limiting dilution nested PCR to produce full-length gp160 (9–11, 14, 15, 18, 43). To confirm that no proviral DNA was amplified as CF viral RNA, the SGA protocol was performed on RT-minus PL and genital SW, CA, and CF samples.
Approximately 40 viral amplicons from blood and 20–30 from the GT were analyzed (10, 11) (Table 1). Amplification from whole semen was inefficient and resulted in a smaller number of amplicons for analysis compared with female vaginal fluids.
Viral Sequence Analysis.
PCR amplicons were purified and sequenced as previously described (10). Sequences were trimmed from the second cysteine in V1 to the last cysteine in V4, codon-aligned, and then hand-aligned using Geneious bioinformatics software (Biomatters Ltd., Aukland, New Zealand). All V1–V4 env sequences from each newly infected and chronically infected individual were deposited in GenBank under accession numbers FJ185853-FJ187678 and HQ858065-HG858607.
Diversity of donor and recipient viral populations was examined using ML phylogenies (44). Datamonkey (www.datamonkey.org) was used to select the appropriate substitution model (generalized time reversible), and two unrelated outgroup sequences from the same clade were included as tree reference sequences.
Highlighter (hiv.lanl.gov) alignment views allowed us to visualize sequence polymorphisms, with the predominant sequence in the recipient blood, representing the transmitted variant, as the reference sequence. Coupling this with the phylogenies depicts donor and recipient env diversity in blood and genital compartments (Fig. 2 and Fig. S1).
Statistical Data Analysis.
We used the SM test to evaluate whether HIV env sequences replicate in separate subpopulation (blood, GT) compartments or migrate freely between compartments (18, 23, 24). Statistical support was determined per donor by computing the number of migration events in the observed tree and a null distribution of migration events from 10,000 trees with randomly permuted compartment labels. Evidence for compartmentalization was inferred when the observed number of migration events was smaller than 5% of the cumulative null distribution (P < 0.05). Because the SM test can be adversely influenced by recombination, it was performed both on full-length alignments and with alignments partitioned at recombination breakpoints found via GARD analysis (www.datamonkey.org) (45).
We repeated compartmentalization testing with the Hudson (46) nearest-neighbor statistic using the TrN93 distance metric and saw similar patterns of strong support for compartmentalization by that distance-based test. Other established compartmentalization tests [i.e., FST (Wright’s measure of population subdivision), Pearson correlation, the F statistic in analyses of molecular variance (AMOVA)] examined by Zárate et al. (47) work most reliably on Gaussian intersequence distance distributions, which is unlikely to occur in all cases, and thereby yield discordant outcomes.
Transmission Analysis.
Given n = 8 transmission pairs, a ToRT was developed to evaluate whether the donor variant most similar to the predominant recipient variant in each transmission pair was sampled randomly from the donor population (null hypothesis) or was more likely to come from outside a cluster of sequences sharing close similarity. We repeated the analysis for different cluster sizes (details are provided in SI Methods).
Supplementary Material
Acknowledgments
The investigators thank all the volunteers in Rwanda and Zambia who participated in this study and all staff members at Projet San Francisco in Kigali and the Zambia Emory HIV Research Project in Lusaka who made this study possible. This study was funded by National Institutes of Health Grant AI51231 and the Bill and Melinda Gates Foundation Grand Challenges Program (no. 37874/Shaw, to E.H.); National Institutes of Health Grants AI23980 and AI40951, National Institute of Child Health and Human Development Grant R01 40125, National Institute of Mental Health Grant R01 66767, Fogarty International Center Grant D43 TW001042, and the International AIDS Vaccine Initiative (to S.A.); and National Institutes of Health Grant AI067854(CHAVI) (to B.T.K.). This work was supported, in part, by the Virology Core at the Emory Center for AIDS Research (Grant P30 AI050409). We also thank Dr. Hayley Crawford for her critical review of this manuscript.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. FJ185853–FJ187678 and HQ858065–HG858607).
*This Direct Submission article had a prearranged editor.
See Author Summary on page 18589.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103764108/-/DCSupplemental.
References
- 1.UNAIDS Global report 2010, Geneva. 2010. Available at http://www.unaids.org/glovalreport/documents/20101123_GlovalReport_full_en.pdf. Accessed November, 2010.
- 2.Delwart EL, et al. Human immunodeficiency virus type 1 populations in blood and semen. J Virol. 1998;72:617–623. doi: 10.1128/jvi.72.1.617-623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta P, et al. Human immunodeficiency virus type 1 shedding pattern in semen correlates with the compartmentalization of viral quasi species between blood and semen. J Infect Dis. 2000;182:79–87. doi: 10.1086/315644. [DOI] [PubMed] [Google Scholar]
- 4.Paranjpe S, et al. Subcompartmentalization of HIV-1 quasispecies between seminal cells and seminal plasma indicates their origin in distinct genital tissues. AIDS Res Hum Retroviruses. 2002;18:1271–1280. doi: 10.1089/088922202320886316. [DOI] [PubMed] [Google Scholar]
- 5.Zhu T, et al. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: Evidence for viral compartmentalization and selection during sexual transmission. J Virol. 1996;70:3098–3107. doi: 10.1128/jvi.70.5.3098-3107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Overbaugh J, Anderson RJ, Ndinya-Achola JO, Kreiss JK. Distinct but related human immunodeficiency virus type 1 variant populations in genital secretions and blood. AIDS Res Hum Retroviruses. 1996;12:107–115. doi: 10.1089/aid.1996.12.107. [DOI] [PubMed] [Google Scholar]
- 7.Poss M, et al. Diversity in virus populations from genital secretions and peripheral blood from women recently infected with human immunodeficiency virus type 1. J Virol. 1995;69:8118–8122. doi: 10.1128/jvi.69.12.8118-8122.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sagar M, et al. Human immunodeficiency virus type 1 (HIV-1) diversity at time of infection is not restricted to certain risk groups or specific HIV-1 subtypes. J Virol. 2004;78:7279–7283. doi: 10.1128/JVI.78.13.7279-7283.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derdeyn CA, et al. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 10.Haaland RE, et al. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog. 2009;5:e1000274. doi: 10.1371/journal.ppat.1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keele BF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ritola K, et al. Multiple V1/V2 env variants are frequently present during primary infection with human immunodeficiency virus type 1. J Virol. 2004;78:11208–11218. doi: 10.1128/JVI.78.20.11208-11218.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frost SD, et al. Characterization of human immunodeficiency virus type 1 (HIV-1) envelope variation and neutralizing antibody responses during transmission of HIV-1 subtype B. J Virol. 2005;79:6523–6527. doi: 10.1128/JVI.79.10.6523-6527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salazar-Gonzalez JF, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abrahams MR, et al. CAPRISA Acute Infection Study Team Center for HIV-AIDS Vaccine Immunology Consortium Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J Virol. 2009;83:3556–3567. doi: 10.1128/JVI.02132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer W, et al. Transmission of single HIV-1 genomes and dynamics of early immune escape revealed by ultra-deep sequencing. PLoS ONE. 2010;5:e12303. doi: 10.1371/journal.pone.0012303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellerbrock TV, et al. Cellular replication of human immunodeficiency virus type 1 occurs in vaginal secretions. J Infect Dis. 2001;184:28–36. doi: 10.1086/321000. [DOI] [PubMed] [Google Scholar]
- 18.Anderson JA, et al. Center for HIV/AIDS Vaccine Immunology, translator. HIV-1 populations in semen arise through multiple mechanisms. PLoS Pathog. 2010;6:e1001053. doi: 10.1371/journal.ppat.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bull M, et al. Compartmentalization of HIV-1 within the female genital tract is due to monotypic and low-diversity variants not distinct viral populations. PLoS ONE. 2009;4:e7122. doi: 10.1371/journal.pone.0007122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diem K, et al. Male genital tract compartmentalization of human immunodeficiency virus type 1 (HIV) AIDS Res Hum Retroviruses. 2008;24:561–571. doi: 10.1089/aid.2007.0115. [DOI] [PubMed] [Google Scholar]
- 21.Nickle DC, et al. Evolutionary indicators of human immunodeficiency virus type 1 reservoirs and compartments. J Virol. 2003;77:5540–5546. doi: 10.1128/JVI.77.9.5540-5546.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitney JB, et al. Genital tract sequestration of SIV during acute infection. PLoS Pathog. 2011;7:e1001293. doi: 10.1371/journal.ppat.1001293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slatkin M, Maddison WP. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics. 1989;123:603–613. doi: 10.1093/genetics/123.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poss M, et al. Evolution of envelope sequences from the genital tract and peripheral blood of women infected with clade A human immunodeficiency virus type 1. J Virol. 1998;72:8240–8251. doi: 10.1128/jvi.72.10.8240-8251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Byrn RA, Kiessling AA. Analysis of human immunodeficiency virus in semen: Indications of a genetically distinct virus reservoir. J Reprod Immunol. 1998;41:161–176. doi: 10.1016/s0165-0378(98)00056-4. [DOI] [PubMed] [Google Scholar]
- 27.Byrn RA, Zhang D, Eyre R, McGowan K, Kiessling AA. HIV-1 in semen: An isolated virus reservoir. Lancet. 1997;350:1141. doi: 10.1016/S0140-6736(97)24042-0. [DOI] [PubMed] [Google Scholar]
- 28.Coombs RW, et al. Association between culturable human immunodeficiency virus type 1 (HIV-1) in semen and HIV-1 RNA levels in semen and blood: Evidence for compartmentalization of HIV-1 between semen and blood. J Infect Dis. 1998;177:320–330. doi: 10.1086/514213. [DOI] [PubMed] [Google Scholar]
- 29.Pillai SK, et al. Semen-specific genetic characteristics of human immunodeficiency virus type 1 env. J Virol. 2005;79:1734–1742. doi: 10.1128/JVI.79.3.1734-1742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ping LH, et al. Effects of genital tract inflammation on human immunodeficiency virus type 1 V3 populations in blood and semen. J Virol. 2000;74:8946–8952. doi: 10.1128/jvi.74.19.8946-8952.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chohan B, et al. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1-V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J Virol. 2005;79:6528–6531. doi: 10.1128/JVI.79.10.6528-6531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Go EP, et al. Characterization of glycosylation profiles of HIV-1 transmitted/founder envelopes by mass spectrometry. J Virol. 2011;85:8270–8284. doi: 10.1128/JVI.05053-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, et al. Env length and N-linked glycosylation following transmission of human immunodeficiency virus type 1 subtype B viruses. Virology. 2008;374:229–233. doi: 10.1016/j.virol.2008.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nawaz F, et al. The genotype of early-transmitting HIV gp120s promotes α (4) β(7)-reactivity, revealing α (4) β(7) +/CD4+ T cells as key targets in mucosal transmission. PLoS Pathog. 2011;7:e1001301. doi: 10.1371/journal.ppat.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chomba E, et al. Rwanda Zambia HIV Research Group Evolution of couples’ voluntary counseling and testing for HIV in Lusaka, Zambia. J Acquir Immune Defic Syndr. 2008;47:108–115. doi: 10.1097/QAI.0b013e31815b2d67. [DOI] [PubMed] [Google Scholar]
- 36.McKenna SL, et al. Rapid HIV testing and counseling for voluntary testing centers in Africa. AIDS. 1997;11(Suppl 1):S103–S110. [PubMed] [Google Scholar]
- 37.Fideli US, et al. Virologic and immunologic determinants of heterosexual transmission of human immunodeficiency virus type 1 in Africa. AIDS Res Hum Retroviruses. 2001;17:901–910. doi: 10.1089/088922201750290023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allen S, et al. Effect of serotesting with counselling on condom use and seroconversion among HIV discordant couples in Africa. BMJ. 1992;304:1605–1609. doi: 10.1136/bmj.304.6842.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dunkle KL, et al. New heterosexually transmitted HIV infections in married or cohabiting couples in urban Zambia and Rwanda: An analysis of survey and clinical data. Lancet. 2008;371:2183–2191. doi: 10.1016/S0140-6736(08)60953-8. [DOI] [PubMed] [Google Scholar]
- 40.Delany S, et al. Comparison of cervicovaginal lavage, cervicovaginal lavage enriched with cervical swab, and vaginal tampon for the detection of HIV-1 RNA and HSV-2 DNA in genital secretions. J Acquir Immune Defic Syndr. 2008;49:406–409. doi: 10.1097/qai.0b013e31818c7f75. [DOI] [PubMed] [Google Scholar]
- 41.Lorello G, et al. Discordance in HIV-1 viral loads and antiretroviral drug concentrations comparing semen and blood plasma. HIV Med. 2009;10:548–554. doi: 10.1111/j.1468-1293.2009.00725.x. [DOI] [PubMed] [Google Scholar]
- 42.Trask SA, et al. Molecular epidemiology of human immunodeficiency virus type 1 transmission in a heterosexual cohort of discordant couples in Zambia. J Virol. 2002;76:397–405. doi: 10.1128/JVI.76.1.397-405.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simmonds P, Balfe P, Ludlam CA, Bishop JO, Brown AJ. Analysis of sequence diversity in hypervariable regions of the external glycoprotein of human immunodeficiency virus type 1. J Virol. 1990;64:5840–5850. doi: 10.1128/jvi.64.12.5840-5850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 45.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol. 2006;23:1891–1901. doi: 10.1093/molbev/msl051. [DOI] [PubMed] [Google Scholar]
- 46.Hudson RR. A new statistic for detecting genetic differentiation. Genetics. 2000;155:2011–2014. doi: 10.1093/genetics/155.4.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zárate S, Pond SL, Shapshak P, Frost SD. Comparative study of methods for detecting sequence compartmentalization in human immunodeficiency virus type 1. J Virol. 2007;81:6643–6651. doi: 10.1128/JVI.02268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]