Abstract
Piwi-interacting RNAs (piRNAs) and Piwi proteins have the evolutionarily conserved function of silencing of repetitive genetic elements in germ lines. The founder of the Piwi subfamily, Drosophila nuclear Piwi protein, was also shown to be required for the maintenance of germ-line stem cells (GSCs). Hence, null mutant piwi females exhibit two types of abnormalities, overexpression of transposons and severely underdeveloped ovaries. It remained unknown whether the failure of GSC maintenance is related to transposon derepression or if GSC self-renewal and piRNA silencing are two distinct functions of the Piwi protein. We have revealed a mutation, piwiNt, removing the nuclear localization signal of the Piwi protein. piwiNt females retain the ability of GSC self-renewal and a near-normal number of egg chambers in the ovarioles but display a drastic transposable element derepression and nuclear accumulation of their transcripts in the germ line. piwiNt mutants are sterile most likely because of the disturbance of piRNA-mediated transposon silencing. Analysis of chromatin modifications in the piwiNt ovaries indicated that Piwi causes chromatin silencing only of certain types of transposons, whereas others are repressed in the nuclei without their chromatin modification. Thus, Piwi nuclear localization that is required for its silencing function is not essential for the maintenance of GSCs. We suggest that the Piwi function in GSC self-renewal is independent of transposon repression and is normally realized in the cytoplasm of GSC niche cells.
Keywords: oogenesis, short RNA
Drosophila Piwi (P-element induced wimpy testes) protein was first described as a factor required for germ-line stem cell (GSC) maintenance and normal development of the testes and ovaries (1–3). The same function was also demonstrated for Piwi orthologues in the germ lines of other organisms (2, 4). In Drosophila, the ovaries are composed of ovarioles consisting of a germarium and a string of egg chambers at consecutive developmental stages. Oogenesis starts in the anterior germarium containing two or three GSCs in close contact with somatic cap cells, the components of their niche. After stem cell division, the daughter cell adjacent to the cap cell remains within the niche as a stem cell, whereas the other daughter cell initiates differentiation into a cystoblast and eventually an egg (5). The Piwi protein is localized in the nuclei of somatic and germinal ovarian cells, but the production of signals to maintain GSC renewal requires Piwi expression only in the somatic niche (2, 3, 6). As a result of immediate GSC differentiation into cystoblasts, piwi mutant females usually contain germ line-less germaria and no more than two or three egg chambers (1, 2). Although several suppressors of piwi mutations restoring GSC maintenance were identified (7–10), the key niche signal regulated by piwi remains unknown (reviewed in refs. 11, 12). It was also shown that the intrinsic expression of Piwi in GSCs promotes their mitotic divisions (3, 6). Another role of Piwi in germ-line development is related to the formation of maternally inherited pole plasm (13). Finally, piwi mutations lead to transposable element overexpression and cause a transposition burst as a result of the loss of Piwi-interacting RNA (piRNA) silencing (14–18). Piwi is the founding member of the evolutionarily conserved piRNA-binding Piwi protein subfamily, which also includes Aub and Ago3 proteins in Drosophila (18). piRNAs are produced by the primary processing of single-stranded transcripts of heterochromatic master loci or by ping-pong amplification (19–21). Whereas germ cell-specific Aub and Ago-3 proteins are actively involved in the ping-pong cycle, the Piwi protein is mainly loaded with primarily processed piRNAs and represses transposons in germinal and somatic ovarian cells (18, 19, 22). Piwi is a predominantly nuclear protein, whereas most other piRNA machinery proteins are localized in the cytoplasm, particularly in the electron-dense perinuclear nuage organelle of germinal cells (23) and Yb bodies of ovarian somatic cells (24–26).
It has remained unknown whether Piwi functions in GSC self-renewal and piRNA-mediated silencing of transposable elements are interrelated. It has been suggested that a cessation of piRNA function can affect stem cell maintenance (8). Here we show that a mutant cytoplasmic Piwi is capable of supporting GSC self-renewal but loses the ability to repress transposable elements, leading to female sterility. We also show that Piwi-mediated silencing takes place within the nuclei of germinal cells and involves chromatin modification.
Results
Identification of piwiNt Mutation.
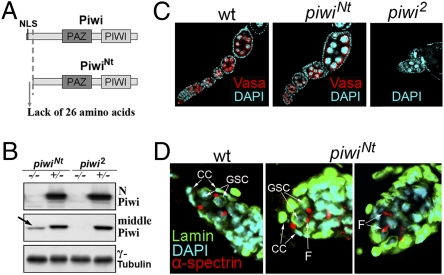
While characterizing a female sterile mutation, hereafter piwiNt (i.e., N-truncated), on chromosome 2, we detected sterile transheterozygotes carrying the piwiNt chromosome and an opposite chromosome with deletions uncovering the region containing piwi and aub genes. Sterility was also observed in flies carrying transheterozygous combinations of piwiNt with piwi2 or piwi3 but not with aub mutations. We revealed a 5′ truncation of the piwi gene as a result of P element vector insertion in the coding region of the first exon (Fig. S1A). RT-PCR analysis demonstrated the presence of the piwi transcript in the mutant ovaries, but 5′-RACE defined its start site at the first intron of piwi (Fig. S1A). This start site position was confirmed by RT-PCR. Analysis of this region using McPromoter software (27) predicted a cryptic promoter at this site. The presence of the ATG codon near the intron 1/exon 2 boundary enables initiation of translation of a shortened Piwi protein lacking 26 N-terminal amino acids including the nuclear localization signal (NLS), but with two additional amino acids (M and Q) encoded by the intron sequence (Fig. S1A). The rest of the piwi gene encoding the PAZ and Piwi domains responsible for short RNA binding and target RNA slicing remained unchanged (Fig. 1A). Antibodies against the extreme N-terminal part of Piwi did not recognize the mutant protein, whereas those against Piwi C-terminal or middle regions showed the presence of the shortened Piwi protein (Fig. 1B and Fig. S1B). Both shortened polypeptide and transcript were not detected in the WT ovaries by Western analysis and 5′-RACE, respectively, and the corresponding sequences were not found in the available databases, indicating that PiwiNt represents a mutant protein but not an isoform encoded by the piwi gene.
Fig. 1.
Flies carrying the piwiNt mutation that leads to the formation of N-truncated Piwi protein have a normal number of egg chambers and germinal stem cells. (A) Truncated PiwiNt protein lacks 26 N-terminal amino acids including the NLS. (B) Antibodies against the middle Piwi region, but not against the N-terminal part (Upper), detect the shortened Piwi (arrow). (C) WT, piwiNt, and piwi2 (null mutant) ovarioles stained for Vasa (red), a marker of germinal cells, and DAPI (blue). (D) Staining of GSCs in the WT (Left) and piwiNt (Middle and Right) germaria by lamin (green) and α-spectrin (red) antibodies marking spectrosomes at the site of their contact with somatic cap cells (CCs); germ cell undergoing division can be observed by an elongated fusome material (“F”).
Loss of Nuclear Piwi Localization Does Not Affect Stem Cell Maintenance.
The null piwi mutants have severely degenerate ovarioles with an extremely small amount of egg chambers because of the complete differentiation of GSCs with no renewal divisions (1, 2). By contrast, the ovaries of homozygous piwiNt, transheterozygous piwiNt/piwi2, or piwiNt/Df(2L)BSC145 females had a near-normal number of egg chambers in the ovarioles (Fig. 1C), thus indicating the piwiNt ability to maintain GSC self-renewal. piwiNt homozygous females aged 1 to 5 d contained an average of 4.3 egg chambers per ovariole (n = 120), and piwiNt/+ heterozygotes had 6.2 chambers (n = 150). The observed slight decrease of egg chamber number is characteristic of piRNA system mutants, which can be explained by a delay in GSC/cystoblast mitotic divisions (28) but not by GSC direct differentiation into cystoblasts. Oogenesis proceeds completely in piwiNt mutants and oocytes are correctly positioned in most piwiNt egg chambers, although some ovarioles (2%) have an abnormal phenotype reflected by characteristics such as fused egg chambers (Fig. S2A). We also observed oocyte axis specification defects typical for piRNA mutants (23), such as abnormal dorsal appendages and mislocalization of the posterior morphogen Oskar (Fig. S2 B and C). Only approximately 30% of piwiNt oocytes (21 of 65) had correctly positioned Piwi and Osk in the oocyte pole plasm.
Whereas the adult ovarioles in the piwi-null mutants contain the germaria depleted of germ-line cells (1, 2), the piwiNt germaria carries developing germ-line cysts and a normal amount of GSCs (two or three per germarium) as visualized by α-spectrin staining of spectrosomes, specific germ cell organelles at the sites of GSC contacts with cap cells (Fig. 1D). Undergoing divisions of germ cells were indicated by the fusome material at the cytoplasmic bridges connecting mother and daughter cells (Fig. 1D, Right).
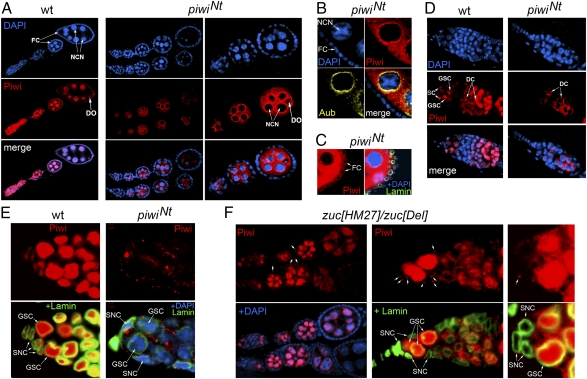
Next we tested the expression pattern and localization of the mutant PiwiNt protein. The requirement of the NLS in the Piwi N-terminal region has been shown previously for Piwi import into the nuclei of cultured ovarian somatic cells (25, 29). In the piwiNt ovaries, the nuclear localization of the mutant Piwi protein was lost completely in all cells in which Piwi is expressed, including germinal nurse cells, developing oocytes, and somatic follicle cells (Fig. 2 A–D and Fig. S3). The PiwiNt protein was detected mainly in the cytoplasm of nurse cells and around their nuclei, similarly to nuage components, e.g., Aub (Fig. 2B). Mutant protein was found in the cytoplasm of follicle somatic cells (Fig. 2C) but at a significantly lower level than in germ cells.
Fig. 2.
Localization of the Piwi protein in the ovaries of piwiNt and zuc mutants. (A) Piwi (red) and DAPI (blue) staining of the WT (Left) and piwiNt (Middle and Right) ovarioles. Piwi protein is known to be localized in the nuclei of nurse cells (NCN), developing oocytes (DO), and somatic follicle cells (FC) enveloping egg chambers. In the piwiNt mutant, Piwi nuclear localization is lost completely. The mutant Piwi protein is mainly localized in germinal cells and is undetectable in somatic cells by the microscope setup. (B) Fragment of piwiNt egg chamber demonstrating partial colocalization of cytoplasmic Piwi (red) and Aub protein (yellow) around the nurse cell nucleus. (C) PiwiNt protein can be observed in the cytoplasm of follicle somatic cells by using the microscope setup for enhanced detection. (D) Piwi staining of the WT (Left) and piwiNt (Right) germaria. SC, somatic cells; DC, differentiating cysts. Piwi mutant protein is detected basically in DC. (E) PiwiNt mutant protein localization in the anterior part of the germarium with the GSC niche. In the somatic niche cells (SNC), the PiwiNt protein accumulates in cytoplasmic granules. (F) Left: Normal number of egg chambers in zuc[HM27]/zuc[Del] ovarioles. Piwi accumulates in follicle cell cytoplasm (arrows). Middle and Right: Piwi is lost from the nuclei of zuc mutant niche cells (SNC) and accumulates in cytoplasmic granules (arrows).
Thus, our data provide an unexpected conclusion that cytoplasmic Piwi is capable of supporting GSC maintenance. This process is known to depend on Piwi expression only in somatic cells forming the GSC niche at the anterior-most end of the germarium (2, 6). The GSC niche consists of the terminal filament, escort cells, and cap cells, which directly associate with GSCs via adherens junctions (11, 30). In the WT germarium, Piwi is known to be strongly stained in the nuclei of niche cells and GSCs, weakly stained in cystoblasts and early mitotic cysts, and strongly stained in late mitotic and differentiating 16-cell cysts (3, 6). We observed PiwiNt in cytoplasmic inclusions in niche cells (Fig. 2E), although its amount in the anterior part of germarium was drastically lowered compared with germ-line cysts (Fig. 2D).
It has been reported that the depletion of the Zuc protein, a piRNA system component, caused Piwi disappearance from follicle cell nuclei and its accumulation in cytoplasmic Yb bodies (24, 25). We analyzed Piwi localization in the germaria of zuc[HM27]/zuc[Del] mutant flies and observed a complete loss of Piwi from the nuclei of GSC niche cells (Fig. 2F, Middle and Right), whereas zuc mutants showed no GSC deficiency phenotype (Fig. 2F, Left) (31). This observation also argues for the ability of cytoplasmic Piwi to provide a signal for GSC maintenance.
Piwi Nuclear Localization Is Indispensable for Transposon Silencing in Germ Cells.
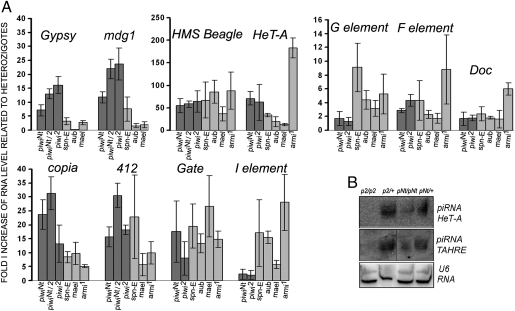
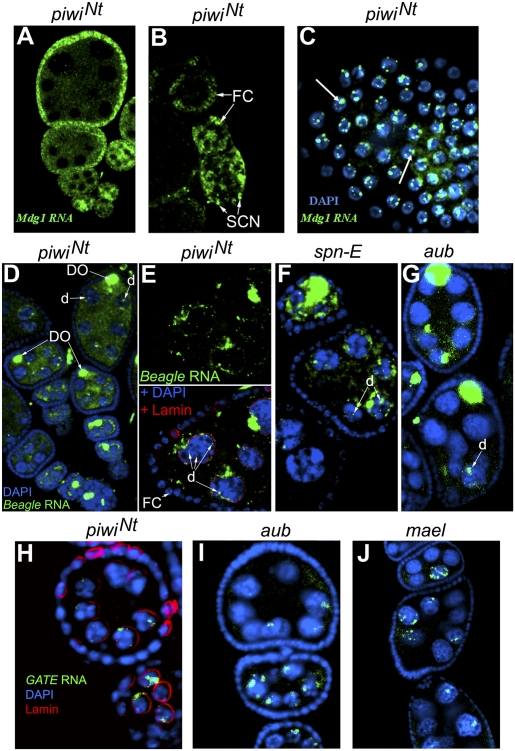
We revealed drastic transposon derepression in the piwiNt ovaries (Fig. 3A). To analyze the specificity of this effect on piRNA silencing, we compared it with that of the piwi2-null mutation and with aub, mael, spn-E, and armi mutations affecting the cytoplasmic components of the piRNA machinery, some of which have been shown to be responsible for the activation of a number of transposons tested in our study (16, 32–38) (Fig. 3A). Transcript level of the Gypsy and mdg1 retrotransposons increased 10- to 20-fold in the ovaries of homozygous piwiNt and piwi2 or transheterozygous piwi2/piwiNt flies relative to the corresponding heterozygotes (Fig. 3A). In line with the current view (19, 24), these elements known to be expressed mainly in the somatic ovarian cells (18, 19, 37, 38) (Fig. S4A) showed no or minor up-regulation caused by aub, mael, and spn-E mutations affecting the proteins of the germ-line piRNA machinery (except the effect of spn-E on mdg1 expression; Fig. 3A). We found mdg1 transcripts localized largely in the nuclei of somatic cells including GSC niche cells in the germaria of the piwiNt ovaries (Fig. 4 A–C). The quantitative effects on the germ line expressed transposons observed in piwiNt homozygotes and piwi2/piwiNt transheterozygotes were more similar to the effects of piwi2 than to those of the aub, mael, spn-E, and armi mutations (Fig. 3A). Hence, piwiNt seems to abolish the silencing function of the Piwi protein completely and to be peculiar to the piwi loss of function mutation. Indeed, I and G elements, whose complementary piRNAs were preferentially found in the Aub and Ago3 complexes than in Piwi ones (18), showed a weak up-regulation both in piwi2 and piwiNt mutants and a stronger up-regulation in aub, mael, spn-E, and armi mutants (Fig. 3A). At the same time, the aub, mael, and spn-E mutations exhibited similar to piwiNt and piwi2 quantitative effects on the derepression of HMS-Beagle and Gate retrotransposons (Fig. 3A) and caused a distribution of their transcripts in germ-line cells similar to that of piwiNt (Fig. 4 D–J). As a result of the derepression, the HMS-Beagle RNA accumulated mainly in the developing oocytes and nurse cells (Fig. 4 D–G and Fig. S4B), where it amassed in the form of cytoplasmic clouds around the nuclei and as separate small dots within the nuclei (Fig. 4 E–G). The Gate transcripts predominantly accumulated in the nuclei of nurse cells (Fig. 4 H–J and Fig. S4C). These data indicate Piwi intranuclear activity contributing to transposon repression and also suggest that cytoplasmic piRNA pathway proteins (Aub, Mael, and Spn-E) may influence the Piwi-mediated silencing process in the germ-line nuclei. What underlies different ability of transposon RNAs for nuclear export remains unclear. Most likely, it is determined by the degeneration of transposon RNA sequences involved in the interaction with the nuclear export machinery.
Fig. 3.
The piwiNt mutation leads to transposon desilencing. (A) Quantitative RT-PCR evaluation of the fold accumulation of transposon transcripts in the homozygous piwiNt and piwi2 or transheterozygous piwiNt/piwi2 ovaries relative to heterozygotes (mean ± SD; n = 3). (B) Northern blot detection of piRNAs complementary to the germ line expressed HeT-A and TAHRE retrotransposons in ovarian RNA of piwi2 (p2) and piwiNt (pNt) mutants and the corresponding heterozygotes, normalized to U6 RNA.
Fig. 4.
Localization of mdg1 (A–C), HMS-Beagle (D–G), and Gate (H–J) transposon transcripts (green) by RNA in situ hybridization in the piwiNt ovaries. In WT and heterozygotes, no signal was observed with the same microscope settings. (A and B) mdg1 is expressed predominantly in follicle cells (FC) and niche somatic cells (SCN). (C) Dot-like mdg1 RNA accumulation in the nuclei of follicle cells (indicated by arrows) stained with DAPI (blue). (D–G) HMS-Beagle retrotransposon transcripts accumulate in the developing oocytes (DO) and in nurse cells in the ovaries of piwiNt (D and E), spn-Е (F), and aub (G) mutants. In nurse cells, the transcripts amassed as cytoplasmic clouds and distinct nuclear dots (d). (E) Fragment of the egg chamber of the piwiNt mutant stained for HMS-Beagle RNA (Upper) and for Lamin and DAPI (Lower). (H–J) Gate retrotransposon transcripts are observed mainly in the nuclei of nurse cells.
By Northern analysis, we detected germ line-specific piRNAs species which were absent in the piwi2 mutant (Fig. 3B), suggesting that piRNA biogenesis and its loading into Piwi complexes occur in the cytoplasm of germ-line cells. piRNA amount decreased compared with the heterozygotes (Fig. 3B), which can be attributed to a lower total amount of PiwiNt protein than in the WT (Fig. 1B and Fig. S1B).
Piwi Is Involved in Chromatin Silencing of Transposons in Ovaries.
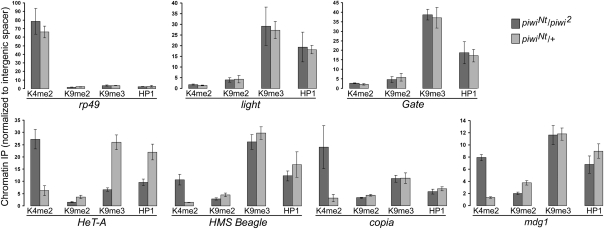
The role of Piwi in heterochromatin formation in somatic cells as well as the direct interaction of Piwi with chromatin and the HP1 protein have been reported (8, 39–44). However, it remained unknown whether Piwi-mediated intranuclear silencing of transposons in the germ line is based on chromatin regulation. Previously, we have shown that the spn-E mutation leads to the opening of retrotransposon chromatin structure in the ovaries (36). As Piwi is a single Drosophila piRNA-interacting protein with nuclear localization, it is the most probable effector of the putative piRNA-mediated chromatin silencing pathway. The close to normal phenotype of the piwiNt/piwi2 ovaries allowed us to carry out an adequate analysis of HP1 and histone mark occupancies in transposon sequences by ChIP in Piwi-depleted nuclei, which would be impossible in severely underdeveloped ovarian tissues of the piwi-null mutants. ChIP analysis of whole ovaries mainly reflects the chromatin state in the nurse cells, whereas follicle cell chromatin can also have its contribution, because a mature egg chamber contains 15 polyploid nurse cells with a DNA content of as much as 2,048C (45) and approximately 1,000 follicle cells, which undergo polyploidization up to 4C to 16C (46, 47). We found that the piwiNt mutation increased the active H3K4me2 mark but decreased repressive H3K9me3/me2 modifications and HP1 enrichment in the chromatin of telomeric HeT-A element (Fig. 5) compared with the heterozygotes. In the chromatin of the HMS-Beagle, mdg1, and copia nontelomeric retroelements, we observed a mutation-induced increase of the H3K4me2 mark, but in contrast to the telomeric elements, no significant changes in the abundance of H3K9me2/me3 or HP1 were detected (Fig. 5). The mutation had no effect on both analyzed histone marks and HP1 content in the chromatin related to two different regions of the Gate retrotransposon (Fig. 5), despite the accumulation of its transcripts mainly in the nucleus (Fig. 4H). Overall, these data suggest that Piwi is involved in transposon repression by altering their chromatin structure, whereas some other Piwi-mediated nuclear mechanisms of silencing may occur related specifically to a transposon type.
Fig. 5.
ChIP analysis of transposon chromatin status. The bars indicate quantitative PCR-measured occupancies of HP1 and the modified histones in piwiNt/piwi2 transheterozygotes (dark) and piwiNt/+ heterozygotes (light) normalized to the corresponding values for the intergenic 60D cluster (mean ± SD; n = 3). The ribosomal rp49 and heterochromatic light genes were used as examples of actively transcribed and repressed chromatin, respectively.
Discussion
Drosophila Piwi protein is known to be implicated in GSC self-renewal (2, 6), as well as in piRNA-mediated repression of transposable elements (19–21, 23). Here we describe the phenotype of a unique mutation in the piwi gene that leads to the formation of cytoplasmic PiwiNt (i.e., N-truncated Piwi protein) lacking the NLS. The properties of this mutant made the direct influence of the piRNA pathway on GSC maintenance unlikely, as the piwiNt mutant displayed normal GSC self-renewal (Fig. 1 C and D) but lost Piwi-mediated transposon repression completely in ovarian cells (Fig. 3A) including niche cells responsible for GSC self-renewal signaling (Fig. 4B). Thus, the Piwi regulatory function in GSC maintenance is distinct from the nuclear piRNA silencing mechanism.
The details of Piwi-mediated transposon silencing process as well as the interactions between Piwi and other piRNA machinery proteins remain poorly understood. We found that the aub, spn-E, and mael mutations, affecting cytoplasmic and nuage components of the piRNA system in germinal ovarian cells (19, 35, 48–50), lead to the nuclear accumulation of transcripts of HMS-Beagle and Gate transposons similarly to the effect of piwi mutations (Fig. 4 D–J and Fig. S4 B and C). Probably, nuage proteins, which are dispensable for Piwi nuclear import (19) (Fig. S5), may nevertheless ensure proper Piwi protein function in nuclear silencing. The reasons why Piwi nuclear localization is required for silencing of a large number of transposons in the germinal cells are not entirely clear. Possibly, particular Piwi-regulated transposons avoid the degradation by Aub/Ago-3 cytoplasmic piRNA machinery as a result of their masking by cytoplasmic proteins. Several reports have implicated Piwi in chromatin status maintenance (8, 39–43). We revealed that the loss of the nuclear Piwi in the piwiNt ovaries enriches the chromatin context of telomeric transposons with H3K4me2 euchromatic marks and decreases the occupancy of heterochromatic marks such as H3K9me2/3 histones and HP1 protein (Fig. 5). These effects resemble those caused by the spn-E mutation (36). The HP1 level in HeT-A repeats was also shown to be decreased in the ovaries of aub and armi piRNA system mutants (51). The piwiNt mutation also leads to enrichment of the HMS-Beagle and mdg1 transposons in the H3K4me2 modification (Fig. 5). These elements are highly up-regulated in the mutant ovaries (Fig. 3A), and a portion of their transcripts amassed as discrete dots within nuclei (Fig. 4 C and E), suggesting their elevated transcription. However, no significant changes in the repressive H3K9me2/me3 marks and HP1 abundance were observed in the HMS-Beagle and mdg1 chromatin (Fig. 5). The absence of a negative correlation between the H3K9me2/me3 and H3K4me2 modifications in the chromatin context of these elements due to the piwiNt mutation might result from a preferential derepression of euchromatic transposon copies lacking the H3K9me2/me3 modifications. In the chromatin context of the Gate element, we detected no changes in active and repressive marks in the piwiNt mutant (Fig. 5), although Gate transcription was also drastically up-regulated according to RT-PCR (Fig. 3A), and was shown to actively accumulate in the nuclei (Fig. 4 H–J and Fig. S4С). These data suggest that Piwi may induce nuclear transposon silencing not only by chromatin-based repression but also by other mechanisms such as nuclear posttranscriptional RNA degradation. This suggestion is in concert with the observation that the nucleoplasm is enriched in the Piwi protein compared with DNA-containing areas of ovarian nuclei (3). Although the presence of RNA degradation complexes including decapping and exoribonuclease activities associated with the piRNA system in the cytoplasm of Drosophila germ-line cells has been established (50), the partners of Piwi in chromatin repression and intranuclear transposon transcript elimination remain to be elucidated.
The principal result of this study is that the cytoplasmic, but not nuclear, Piwi protein is required for GSC maintenance. It is known that GSC self-renewal requires Piwi expression only in somatic niche cells (2, 6), and piwi overexpression in these cells increases the number of GSC-like cells (3). Recent studies have revealed a possibility of Piwi transient localization within cytoplasmic Yb bodies in somatic ovarian cells (24, 25). Presumably, the Yb body mediates piRNA biogenesis and loading into Piwi complexes, which is a prerequisite for Piwi nuclear import (24, 25). Yb bodies are also present in niche cells (52), and their key component, the Yb protein, is indispensable for GSC maintenance (26, 52–54). Defects of GSC self-renewal observed in the Yb mutant ovaries lacking Yb bodies are very similar to those in piwi mutants (26, 52–54). Piwi protein is uniformly distributed throughout the cytoplasm of Yb-deficient somatic cells (24, 26). Thus, it is likely that a signal for GSC maintenance may be produced by the Piwi-Yb complex compartmentalized in Yb bodies of niche cells. This suggestion is in line with the observed granular localization of the PiwiNt protein in the niche cell cytoplasm (Fig. 2E). Although the amount of the PiwiNt protein in niche cells is drastically lower compared with the WT Piwi level (Fig. 2 D and E), it is sufficient to maintain GSCs. Similarly, only a tiny portion of the Piwi protein in WT ovarian cells seems to be located in cytoplasmic Yb bodies, where it was detected only by coimmunoprecipitation experiments (24, 25, 55). An additional argument for the role of cytoplasmic Piwi in GSC maintenance is provided by the phenotype of the zuc mutants, which show no evidence of stem cell renewal disturbance (31) (Fig. 2F), whereas Piwi is absent from the nuclei of niche cells and accumulates in Yb bodies (Fig. 2F). The loading of piRNAs into Piwi complexes was shown to be prevented in somatic cells lacking Zuc (24, 25, 55). In light of our results, this observation argues for the piRNA-independent mechanism of Piwi function in GSC maintenance, although we cannot be certain that short RNAs are completely absent from niche cells of zuc mutants. Genetic screens revealed that a mutation in the corto gene, which encodes a chromodomain protein, restored GSC division in both piwi and Yb mutants (10). It can be suggested that a putative GSC signal produced by niche cells may be regulated both in the nucleus by Corto and in the cytoplasm by Piwi/Yb.
Taking into account the evolutionarily conserved role of Piwi orthologs in GSC maintenance, further studies of the molecular mechanism of Piwi functions in the signaling pathways unrelated to piRNA silencing appear to be very intriguing.
Materials and Methods
Drosophila Strains.
Flies were maintained at 25 °C on standard medium. The ovaries from 1- to 6-d-old flies were dissected. Oregon-R and y1 w67c23 strains were used as a WT control. The piwiNt cn1 bw1 chromosome was kept over the CyO balancer. Homozygous piwiNt flies were rare and transheterozygous piwiNt/piwi2 flies were also used to check piwiNt phenotype. Df(2L)BSC145 and Df(2L)BSC213 deletions in the 32C1;32C5 region containing the piwi gene were obtained from the Drosophila stock center (Bloomington, IN). Other mutant fly stocks used in this work are indicated in SI Materials and Methods.
Molecular Characterization of piwiNt Mutation, RT-PCR Analysis, ChIP, Immunohistochemistry, and Western Blot.
Methods for molecular characterization of piwiNt mutation, quantitative RT–PCR analysis, ChIP, immunohistochemistry, and Western blotting are provided in SI Materials and Methods.
RNA in Situ Hybridization.
RNA in situ hybridization using DIG-labeled strand-specific riboprobes was performed basically as previously described (32). For the synthesis of the mdg1 riboprobe by T7 in vitro transcription, the plasmid containing the cloned PCR fragment of mdg1 retrotransposon (15) was used. PCR products carrying T7 promoters were used for T7 transcription of riboprobes to detect transcripts of HMS-Beagle and Gate transposons as well as piwi. Additional details can be found in SI Materials and Methods.
Short RNA Detection.
Detection of short RNA was performed by Northern blot essentially as described previously (56). Additional details can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Alexey Aravin, Gregory Hannon, Haruhiko Siomi, and Mikiko Siomi for providing antibodies; Alla Kalmykova for the mdg1-containing plasmid; and Trudi Schupbach for zuc flies. This work was supported by the Molecular and Cell Biology Program of the Russian Academy of Sciences, Grants МK-65160.2010.4 from the President of the Russian Federation, and Russian Foundation for Basic Research Grants 10-04-01812-a and 11-04-12027.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106676108/-/DCSupplemental.
References
- 1.Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- 2.Cox DN, et al. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- 4.Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spradling A. In: The Development of Drosophila melanogaster. Bate M, Arias AM, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1993. [Google Scholar]
- 6.Szakmary A, Cox DN, Wang Z, Lin H. Regulatory relationship among piwi, pumilio, and bag-of-marbles in Drosophila germline stem cell self-renewal and differentiation. Curr Biol. 2005;15:171–178. doi: 10.1016/j.cub.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Smulders-Srinivasan TK, Lin H. Screens for piwi suppressors in Drosophila identify dosage-dependent regulators of germline stem cell division. Genetics. 2003;165:1971–1991. doi: 10.1093/genetics/165.4.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- 9.Chen D, McKearin D. Gene circuitry controlling a stem cell niche. Curr Biol. 2005;15:179–184. doi: 10.1016/j.cub.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Smulders-Srinivasan TK, Szakmary A, Lin H. A Drosophila chromatin factor interacts with the Piwi-interacting RNA mechanism in niche cells to regulate germline stem cell self-renewal. Genetics. 2010;186:573–583. doi: 10.1534/genetics.110.119081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dansereau DA, Lasko P. The development of germline stem cells in Drosophila. Methods Mol Biol. 2008;450:3–26. doi: 10.1007/978-1-60327-214-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang N, Xie T. In: Regulatory Networks in Stem Cells. Turksen K, Rajasekhar VK, Vemuri MC, editors. New York: Humana; 2009. [Google Scholar]
- 13.Megosh HB, Cox DN, Campbell C, Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr Biol. 2006;16:1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 14.Sarot E, Payen-Groschêne G, Bucheton A, Pélisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–1321. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalmykova AI, Klenov MS, Gvozdev VA. Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res. 2005;33:2052–2059. doi: 10.1093/nar/gki323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vagin VV, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 17.Saito K, et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 19.Malone CD, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 21.Ghildiyal M, Zamore PD. Small silencing RNAs: An expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khurana JS, Theurkauf W. piRNAs, transposon silencing, and Drosophila germline development. J Cell Biol. 2010;191:905–913. doi: 10.1083/jcb.201006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 2010;29:3301–3317. doi: 10.1038/emboj.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saito K, et al. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 2010;24:2493–2498. doi: 10.1101/gad.1989510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi H, et al. The Yb body, a major site for Piwi-associated RNA biogenesis and a gateway for Piwi expression and transport to the nucleus in somatic cells. J Biol Chem. 2011;286:3789–3797. doi: 10.1074/jbc.M110.193888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohler U. Identification of core promoter modules in Drosophila and their application in accurate transcription start site prediction. Nucleic Acids Res. 2006;34:5943–5950. doi: 10.1093/nar/gkl608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pek JW, Kai T. A role for vasa in regulating mitotic chromosome condensation in Drosophila. Curr Biol. 2011;21:39–44. doi: 10.1016/j.cub.2010.11.051. [DOI] [PubMed] [Google Scholar]
- 29.Saito K, et al. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- 30.Spradling A, Drummond-Barbosa D, Kai T. Stem cells find their niche. Nature. 2001;414:98–104. doi: 10.1038/35102160. [DOI] [PubMed] [Google Scholar]
- 31.Pane A, Wehr K, Schüpbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007;12:851–862. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kogan GL, et al. The GATE retrotransposon in Drosophila melanogaster: Mobility in heterochromatin and aspects of its expression in germline tissues. Mol Genet Genomics. 2003;269:234–242. doi: 10.1007/s00438-003-0827-1. [DOI] [PubMed] [Google Scholar]
- 33.Vagin VV, et al. The RNA interference proteins and vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster. RNA Biol. 2004;1:54–58. [PubMed] [Google Scholar]
- 34.Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006;20:345–354. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci USA. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klenov MS, et al. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35:5430–5438. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pélisson A, Sarot E, Payen-Groschêne G, Bucheton A. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J Virol. 2007;81:1951–1960. doi: 10.1128/JVI.01980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Desset S, Buchon N, Meignin C, Coiffet M, Vaury C. In Drosophila melanogaster the COM locus directs the somatic silencing of two retrotransposons through both Piwi-dependent and -independent pathways. PLoS One. 2008;3:e1526. doi: 10.1371/journal.pone.0001526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lei EP, Corces VG. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat Genet. 2006;38:936–941. doi: 10.1038/ng1850. [DOI] [PubMed] [Google Scholar]
- 40.Pal-Bhadra M, et al. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 41.Grimaud C, et al. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell. 2006;124:957–971. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 42.Brower-Toland B, et al. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kavi HH, Birchler JA. Interaction of RNA polymerase II and the small RNA machinery affects heterochromatic silencing in Drosophila. Epigenetics Chromatin. 2009;2:15. doi: 10.1186/1756-8935-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moshkovich N, Lei EP. HP1 recruitment in the absence of argonaute proteins in Drosophila. PLoS Genet. 2010;6:e1000880. doi: 10.1371/journal.pgen.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dej KJ, Spradling AC. The endocycle controls nurse cell polytene chromosome structure during Drosophila oogenesis. Development. 1999;126:293–303. doi: 10.1242/dev.126.2.293. [DOI] [PubMed] [Google Scholar]
- 46.Lilly MA, Duronio RJ. New insights into cell cycle control from the Drosophila endocycle. Oncogene. 2005;24:2765–2775. doi: 10.1038/sj.onc.1208610. [DOI] [PubMed] [Google Scholar]
- 47.Shcherbata HR, Althauser C, Findley SD, Ruohola-Baker H. The mitotic-to-endocycle switch in Drosophila follicle cells is executed by Notch-dependent regulation of G1/S, G2/M and M/G1 cell-cycle transitions. Development. 2004;131:3169–3181. doi: 10.1242/dev.01172. [DOI] [PubMed] [Google Scholar]
- 48.Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development. 2003;130:859–871. doi: 10.1242/dev.00310. [DOI] [PubMed] [Google Scholar]
- 49.Snee MJ, Macdonald PM. Live imaging of nuage and polar granules: Evidence against a precursor-product relationship and a novel role for Oskar in stabilization of polar granule components. J Cell Sci. 2004;117:2109–2120. doi: 10.1242/jcs.01059. [DOI] [PubMed] [Google Scholar]
- 50.Lim AK, Tao L, Kai T. piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J Cell Biol. 2009;186:333–342. doi: 10.1083/jcb.200904063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khurana JS, Xu J, Weng Z, Theurkauf WE. Distinct functions for the Drosophila piRNA pathway in genome maintenance and telomere protection. PLoS Genet. 2010;6:e1001246. doi: 10.1371/journal.pgen.1001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szakmary A, Reedy M, Qi H, Lin H. The Yb protein defines a novel organelle and regulates male germline stem cell self-renewal in Drosophila melanogaster. J Cell Biol. 2009;185:613–627. doi: 10.1083/jcb.200903034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.King FJ, Lin H. Somatic signaling mediated by fs(1)Yb is essential for germline stem cell maintenance during Drosophila oogenesis. Development. 1999;126:1833–1844. doi: 10.1242/dev.126.9.1833. [DOI] [PubMed] [Google Scholar]
- 54.King FJ, Szakmary A, Cox DN, Lin H. Yb modulates the divisions of both germline and somatic stem cells through piwi- and hh-mediated mechanisms in the Drosophila ovary. Mol Cell. 2001;7:497–508. doi: 10.1016/s1097-2765(01)00197-6. [DOI] [PubMed] [Google Scholar]
- 55.Haase AD, et al. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev. 2010;24:2499–2504. doi: 10.1101/gad.1968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kotelnikov RN, et al. Peculiarities of piRNA-mediated post-transcriptional silencing of Stellate repeats in testes of Drosophila melanogaster. Nucleic Acids Res. 2009;37:3254–3263. doi: 10.1093/nar/gkp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.