Abstract
Although in vitro models have been a cornerstone of anti-cancer drug development, their direct applicability to clinical cancer research has been uncertain. Using a state-of-the-art Taqman-based quantitative RT-PCR assay, we investigated the multidrug resistance (MDR) transcriptome of six cancer types, in established cancer cell lines (grown in monolayer, 3D scaffold, or in xenograft) and clinical samples, either containing >75% tumor cells or microdissected. The MDR transcriptome was determined a priori based on an extensive curation of the literature published during the last three decades, which led to the enumeration of 380 genes. No correlation was found between clinical samples and established cancer cell lines. As expected, we found up-regulation of genes that would facilitate survival across all cultured cancer cell lines evaluated. More troubling, however, were data showing that all of the cell lines, grown either in vitro or in vivo, bear more resemblance to each other, regardless of the tissue of origin, than to the clinical samples they are supposed to model. Although cultured cells can be used to study many aspects of cancer biology and response of cells to drugs, this study emphasizes the necessity for new in vitro cancer models and the use of primary tumor models in which gene expression can be manipulated and small molecules tested in a setting that more closely mimics the in vivo cancer microenvironment so as to avoid radical changes in gene expression profiles brought on by extended periods of cell culture.
Keywords: gene signature, NCI-60 panel, translational medicine, gene expression profiling assay, cell culture model
The study of human cancer-derived cell lines has made important contributions to cancer biology and has formed the basis for our current understanding of drug sensitivity and resistance in cancer. Advances in genomics during the last decade have opened new avenues for translational research (1) and allowed the direct evaluation of clinical samples. Based on both in vitro models and clinical studies, the literature is replete with hundreds of prognostic and predictive markers, yet clinical progress in improving cancer treatment has been incremental at best (2). Besides issues associated with the limitations of technology and the selection of patients (3), the clinical relevance and the usefulness of in vitro models for assessing new therapies is controversial (4–8).
High-throughput gene expression profiling has highlighted the genetic and epigenetic heterogeneity among tumors (9, 10) and, therefore, the necessity to study a panel of cell lines for each cancer type to capture this heterogeneity and variability in drug response. For this reason, the National Cancer Institute's Developmental Therapeutics Program (DTP) assembled a panel of 60 cancer cell lines derived from nine different tumor types, termed the NCI-60 panel (11). Because these cells have been extensively characterized, we chose to use them, and additional cancer cell lines, to assess the relevance of cultured cell lines in the study of clinical multidrug resistance (MDR) mechanisms (12).
Over the past 30 y, in vitro studies have led to the enumeration of close to 400 genes whose expression affects response to chemotherapy (13). Among those genes, ATP-binding cassette (ABC) transporters, a superfamily of 48 highly homologous members classified in seven subfamilies, have an important role in the pleiotropic mechanisms mediating MDR by exporting chemotherapeutic agents from the cell (14, 15). Although the roles of 13 ABC transporters in MDR have been fully characterized, recent studies suggest the involvement of up to 30 members of the 48 encoded in the human genome (16, 17). Moreover, besides classical drug efflux, it has also been demonstrated that some of these transporters may mediate the intracellular sequestration of chemotherapeutic drugs (18–20). This intracellular sequestration is the case for ABCA3, which was recently found to be overexpressed in clinical samples of childhood AML and correlated with poor response to treatment (21).
The establishment of a specific and sensitive standard assay, capable of discriminating highly homologous genes, is critical to a better understanding of MDR mechanisms. We and others have shown that Taqman Low Density Arrays (TLDAs) provide the most sensitivity and specificity in measuring the expression patterns of ABC transporter genes (22, 23). Therefore, we chose to configure such a platform to study multidrug resistance mechanisms in clinical cancer specimens.
Results and Discussion
Taqman-Based Quantitative RT-PCR (qRT-PCR) as the State-of-the-Art Assay to Discriminate Between Members of Highly Homologous Gene Families.
We compared the expression profiles of MDR-linked genes obtained by using two main platforms, the oligonucleotide microarray and TaqMan-based qRT-PCR, to determine the best assay for accurately discriminating among the nine cancer types represented in the NCI-60 panel. This analysis (Table 1), which shows the probability of successfully characterizing a cell line as belonging to a specific cancer type, demonstrates that the 380 MDR-linked gene expression profiles obtained from TLDAs, TaqMan-based qRT-PCR, yield a more accurate classification of the NCI-60 panel into the nine cancer types they represent than the profiles obtained by microarrays. This finding highlights a major drawback linked to the technological limitations of the platforms commonly used to determine gene expression profiles. These limitations include inadequate sensitivity of some assays, poor dynamic range, and lack of specificity of the probes, especially in studies investigating MDR mechanisms, because many of the mediators belong to highly homologous gene superfamilies (3, 24).
Table 1.
Accuracy of classification of the nine NCI-60 panel cancer types using various profiling technologies*
| Adjusted P value threshold for gene selection |
||||||||
| Genes analyzed | Gene expression profiling assays | 0.25 | 0.1 | 0.05 | 0.02 | 0.01 | 0.005 | 0.001 |
| 380 MDR-linked genes | TLDA | 0.73 | 0.69 | 0.71 | 0.68 | 0.66 | 0.68 | 0.69 |
| HG-U133A array | 0.71 | 0.64 | 0.66 | 0.66 | 0.61 | 0.59 | 0.61 | |
| ABC transporter genes | TLDA | 0.59 | 0.61 | 0.53 | 0.42 | 0.46 | 0.44 | 0.29 |
| HG-U133A array | 0.37 | 0.39 | 0.36 | 0.37 | 0.31 | 0.18† | 0‡ | |
| SybrGreen-based qRT-PCR | 0.43 | 0.45 | 0.40 | 0.40 | 0.32 | 0.23 | 0.25 | |
| Biomark 48.48 | 0.53 | 0.53 | 0.42 | 0.42 | 0.47 | 0.46 | 0.44 | |
| SLC genes | HG-U133A array | 0.61 | 0.63 | 0.64 | 0.63 | 0.63 | 0.54 | 0.58 |
| 14,500 genes | HG-U133A array | 0.20 | 0.20 | 0.22 | 0.25 | 0.31 | 0.32 | 0.47 |
*Seventy-one percent of the cell lines were correctly classified at P = 0.05 and 69% at P = 0.001 with the TLDA 380 gene MDR set, whereas the expression profiles of the same genes obtained from HG-U133A oligonucleotide microarray analysis classified the 60 cancer cell lines with only 66% accuracy at P = 0.05 and 61% at P = 0.001. Confining the analysis to only ATP-Binding Cassette (ABC) transporter genes, some of the major mediators of multidrug resistance in cultured cells, generates less accurate classification. Only 53% of cell lines were correctly classified at P = 0.05 and 29% at P = 0.001, whereas microarray analysis of the same genes provides the worst results, with 36% accuracy at P = 0.05, with no classification achievable at P = 0.001. ABC transporter gene expression profiling using Sybr Green-based qRT-PCR provides intermediate results with 40% of cell lines properly classified at P = 0.05 and 25% at P = 0.001. Using Biomark 48.48, a high-throughput nanofluidic TaqMan-based qRT-PCR platform, the classification accuracy reaches 44% at P = 0.001. Solute carriers belong to a large family of uptake transporters that are also important MDR mediators. Their expression profiles measured by HG-U133A provide more accurate classification than the ABC transporter genes, with 64% at P = 0.05 and 58% at P = 0.001. Interestingly, the expression profiles of the 14,500 genes on the HG-U133A array do not improve the classification accuracy of the 9 cancer types, as only 22% of the cancer cell lines are correctly classified at P = 0.05, whereas an accuracy of 47% is achieved at P = 0.001. The reason for this apparent paradox is that at lower statistical significance (P < 0.05), more genes are being analyzed and the background noise is greater than at P < 0.001, which reduces the accuracy.
†Three samples unclassified.
‡Fifty-four samples unclassified.
Ovarian Cell Culture Models Failed to Reflect Clinical MDR Gene Expression Patterns.
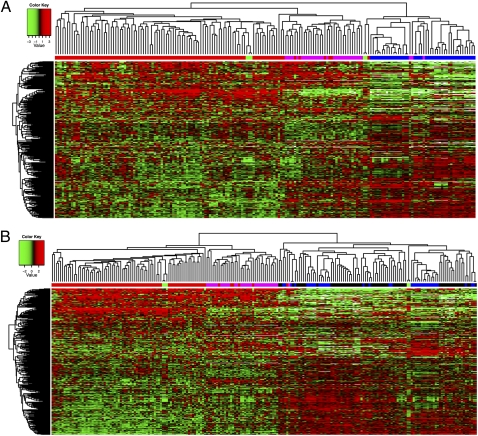
To address the clinical relevance of the NCI-60 panel and other cancer cell lines, we performed comparisons by using the most common ovarian cancer models and clinical samples. We studied a cohort comprised of 80 patients with ovarian primary serous carcinoma. This ovarian cancer type is by far the most common of all ovarian malignancies. The clinical samples from which mRNA was obtained consisted of a minimum of 75% cancer cells, as determined by pathological examination of tissue sections. Our data indicate that 15 ovarian cancer cell lines including 5 from the NCI-60 panel and 10 cisplatin-resistant cell lines, the multidrug-resistant ovarian cancer cell line NCI-ADR-Res (OVCAR8-ADR) and its drug-sensitive counterpart, and 3 established cisplatin-resistant cell lines (25, 26) have a gene expression profile strikingly different from the specimens of untreated ovarian primary serous carcinoma taken from 80 patients (Fig. 1A). Fifty-eight percent of these patients had platinum-resistant cancers defined as the following: progression-free survival <6 mo and no complete response. Although the differences between the platinum-sensitive clinical samples and the deliberately selected cisplatin-resistant cell lines are expected, the ovarian cancer cell lines derived from primary, untreated cancers, to be useful for analyses of mechanisms of drug-resistance, should have MDR gene expression profiles similar to those of the platinum-sensitive clinical specimens, and they do not. We then investigated the effect of 3D culture on the MDR gene profile of the OVCAR-5 cell line, chosen as being representative of the ovarian cancer cell lines. Gene expression profiling of cells grown in this way (including the hanging drop, algimatrix, geltrex, and low attachment plate) did not reveal any major differences in their MDR gene profile compared with 2D cultures. The same observation was made when we injected OVCAR-5 cells grown in both 2D and 3D settings (algimatrix, geltrex, and low attachment plate) into beige-nude-scid mice. Thirty-two effusion samples were also added to our analysis (27). Although all of the clinical samples clustered together, hierarchical clustering illustrates the gene expression pattern differences among those tissues (Fig. 1A). When the eight additional cancer types of the NCI-60 panel were added to the heatmap of Fig. 1A, we made the striking observation that all of the cell lines either grown in vitro or in vivo bear more resemblance to each other, regardless of the tissue of origin, than to the clinical samples that they are supposed to model (Fig. 1B).
Fig. 1.
Hierarchical clustering using the average linkage algorithm and 1-Pearson correlation as the distance measure of the ovarian cancer samples analyzed. (A) The 380 MDR-linked gene expression profile (measured by using TLDA) of ovarian cancer models (in vitro and in vivo) is strikingly different from that of specimens of untreated ovarian primary serous carcinoma taken from 80 patients and 32 effusion samples originating from primary ovarian serous carcinoma. The x axis shows clusters of samples. Red, primary ovarian serous carcinoma; magenta, effusion samples originating from primary ovarian serous carcinoma; green, normal ovarian tissue; blue, in vitro models of ovarian cancer, including xenograft models of ovarian cancer, ovarian cancer cell lines of the NCI-60 panel, and cisplatin-resistant cell lines. The y axis shows gene clustering. (B) When adding the eight additional cancer types of the NCI-60 panel to the heatmap presented in A, the striking observation is made that all of the cell lines either grown in vitro or in vivo bear more resemblance to each other, regardless of the tissue of origin, than to the clinical samples that they are supposed to model. Along the x axis: red, primary ovarian serous carcinoma; magenta, effusion samples originating from primary ovarian serous carcinoma; green, normal ovarian tissue; blue, in vitro models of ovarian cancer; black, cancer cell lines of the eight additional cancer types of the NCI-60 panel. The y axis shows gene clustering.
Established Cancer Cell Lines Are Highly Selected for Expression of Genes Associated with MDR.
Our study identified an important subset of genes up-regulated across the entire set of in vitro models as environmental stress response genes. Subsequent analysis comparing the clinical samples and the ovarian cancer models, including cancer cell lines and xenografts, supports the conclusion that the samples from the three groups, primary ovarian serous carcinoma, effusion samples, and ovarian cancer models, have very different signatures. Two hundred twenty-five of 380 total genes were differentially expressed in the three groups with P < 0.001 (Table S1). All of these genes fulfill the false discovery rate (FDR) criterion of P < 0.001, meaning that the probability of finding 225 genes by chance at the threshold of P < 0.001 is <0.1%. This up-regulation in the expression of specific genes in the cultured cell lines or tumors derived from them across all cancer types is consistent with the idea that their expression is more likely the result of selection pressure and culture conditions, thereby allowing the cells to thrive in their in vitro environment. In other words, the cancer cell lines are highly selected during their establishment for expression of genes associated with MDR. The overexpression of MDR genes associated with response to environmental adversity also argues against the differences in gene expression between clinical cancer and cancer cell lines being due to contamination by noncancer cells of the clinical specimens. Although we did not study mouse models that attempt to recapitulate ovarian carcinoma (28, 29), our data clearly illustrate a need for caution against direct extrapolation of the results to the clinic when incorporating the use of cultured lines in the study of drug resistance in ovarian cancer. These findings may explain, in part, the discrepancies between numerous reported studies highlighting predictive and prognostic markers (30–32).
Cell Culture Models for Five Additional Cancer Types also Failed to Reflect Clinical MDR Gene Expression Patterns.
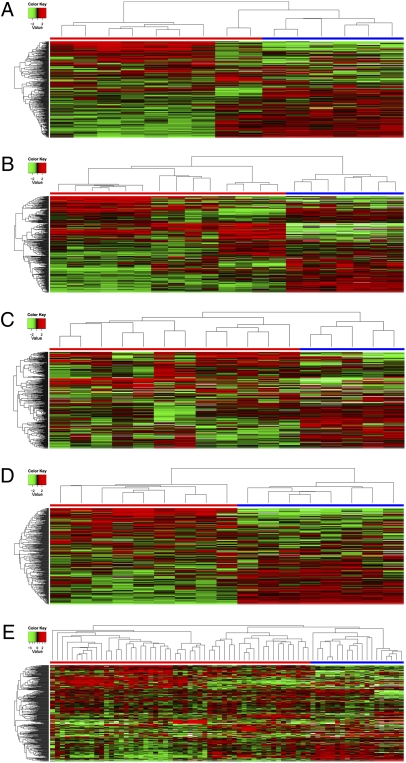
Similar observations were made for five other types of cancer, glioblastoma, colorectal cancer, breast cancer, metastatic melanoma (microdissected from tissue sections), and leukemia, however using fewer samples. The six cell lines representing the CNS in the NCI-60 panel were compared with nine glioblastoma multiforme (GBM) clinical samples, including four primary GBM, three GBM at relapse, and two metastatic GBM. Hierarchical clustering reveals two distinct clusters that discriminate between the in vitro models and the clinical samples (Fig. 2A). Four cell lines, SNB19, U251, SF295, and SF268 were established from glioblastomas, whereas SF539 and SNB75 are gliosarcoma and astrocytoma cell lines, respectively.
Fig. 2.
Hierarchical clustering (using the average linkage algorithm and 1-Pearson correlation as the distance measure) reveals two distinct clusters that discriminate between the in vitro models (cancer cell lines of the NCI-60 panel) and the clinical samples. (A) Heatmap of nine clinical samples of glioblastoma cancer, including four primary tumors, three recurrent tumors, and two metastases. (B) Heatmap of seven colon cancer samples paired with normal colon tissue taken during tumor resection. (C) Heatmap of seven clinical samples of breast cancer, four normal breast tissues, and five cancer cell lines. (D) Heatmap of nine metastatic melanoma samples. (E) Heatmap generated from 23 T-acute lymphoblastic leukemia (12 were untreated, whereas 11 received conventional chemotherapy) and 11 paired samples of acute myeloid leukemia taken at diagnosis and after relapse. x axis: blue, cell lines; red, tumors. The y axis shows gene clustering. Labels for x-axis samples analyzed can be seen in Fig. S7 A–E.
Comparison of clinical samples of colon and breast cancers and metastatic melanoma reveals similar findings with a clear distinction between clinical samples and in vitro models (Fig. 2 B–D). Clinical samples of colon cancer were also paired with normal colon tissue taken during tumor resection.
T-ALL and AML Analyses Revealed that Differences Between Cell Lines and Cancer Cells Are Not Confined to Solid Tumors.
Our analysis of two types of leukemia, T acute lymphoblastic leukemia (T-ALL) and acute myelogenous leukemia (AML), revealed that differences between cell lines and cancer cells are not confined to solid tumors (Fig. 2E). One cluster consisted of AML samples with one T-ALL sample, all untreated. We observed a second major cluster grouping all of the cell lines and the bulk of the T-ALL samples, untreated and treated. Within this cluster, four subgroups were found. Four cell lines grouped together (CML-K562, RPMI8226 myeloma, SR T-ALL, and HL60 AML), whereas two cell lines, Molt4 and CCRF-CEM, clustered with six of the untreated T-ALL clinical samples. Two other subgroups comprise both untreated and treated T-ALL clinical samples.
Heterogeneity Among the Cell Lines Within Tumor Types of the NCI-60 Panel.
We next determined, given the similarity of the MDR signatures among the cultured cells, whether the 380 MDR-linked genes would provide enough information to decipher MDR signatures for each of the nine cancer types represented in the NCI-60 panel. Genes were excluded from the analysis if <20% of the expression data had at least a 1.5-fold change, in either direction, compared with the median gene expression value, bringing the total number of genes to 354. After performing the multivariate permutation test, 185 genes were identified to be differentially expressed within these nine cancer types with P < 0.05 and a FDP < 10% (Table S2). Hierarchical clustering analysis was performed to identify relationships among the cancer cell lines, and a heatmap was produced to illustrate the gene expression of the various cancer types (Fig. S1). There are two main clusters distinguishing the melanoma and leukemia cell lines from the seven other cancer types. The melanoma cell lines clustered together. Similarly, the leukemia cell lines were also in one cluster, with the exception of one colon cancer cell line, SW-620, that clustered with them. The CNS and colon cancer cell lines were also clustered according to cancer type, with few exceptions. The ovarian, breast, renal, lung and prostate cancer cell lines, however, were found to be scattered in the second main cluster (Fig. S1). This gene pattern indicates that four cancer types, melanoma, leukemia, CNS, and colon, are individually composed of a relatively homogenous panel of cell lines. However, there is some heterogeneity among the different cell lines within each of the five other tumor types of the NCI-60 panel. The extent of this heterogeneity could reflect differences in cell type of origin and/or environmental influences on the tumor (e.g., location, primary or metastatic, blood supply of the tumor) that could act as selective factors in vivo. After the identification of the 185 MDR genes that allow the best clustering by type of cancer cell lines, we determined the specific cancer type in which each gene is differentially expressed (Table S3 and Table S4). All of the 185 genes, except ABCA1, APEX1, AQP7, BAX, CYP3A4, GGT1, and NRAS, showed considerable differences in expression among cancer types. The number of genes that are differentially expressed is greatest for the leukemia and melanoma cell lines. Also noteworthy is the observation that many of these genes are differentially expressed in more than one type of cancer tissue type. Both of these findings corroborate what we observed in the previous hierarchical clustering (Fig. S1).
p53 Pathway Activation as the Highest Ranked Network.
Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems) was used to identify connections within the gene signatures revealed for each of the tumor types to see whether there was an obvious explanation for the similarity of all of these gene signatures in the cultured cells. Pathway analysis for the ovarian cell lines generated a pathway centralized on p53 (Fig. S2). The second-ranked pathway for the melanoma cell lines also focused on p53 pathway activation (Fig. S3). Both pathways were further enhanced by using SIMUSITE software (BioPhase Systems), which integrates data from high-throughput expression profiling assays to build system-specific protein interactions. This approach was applied to two cell lines to illustrate the potential of such a method (Fig. S4 A and B). Notably, seven of the nine tumor types show p53 pathway activation as the highest ranked network, whereas the other two types show p53 pathway activation as the second highest ranked network. This result may be attributed to the heterogeneous response of p53 to stress, which depends on cell type, tissue type, and the nature of the stressor (33). Moreover, p53 can regulate the expression of hundreds of genes in response to stress (34). It is clearly possible that p53 pathway activation results from selective pressure in the tissue culture environment, in turn causing the up-regulation of stress response genes across all cell lines and contributing to the uniformity of the MDR gene expression pattern (35). An earlier study using the NCI-60 panel showed that 40 of 58 cell lines analyzed contained mutant p53 (39 of them being homozygous for p53 mutation), whereas 18 cell lines contained wild-type p53 (36). Further analyses revealed that 70% of the cancer cell lines containing mutant p53 showed high p53 protein expression (36).
The data we present represent an average of all of the different cell types in a cancer with the exception of metastatic melanoma. In this work, the metastatic melanoma samples were laser microdissected from tissue sections yielding, based on their histology, a clonal population of tumor cells; yet no correlation was found between the gene expression profiles of clinical and in vitro melanoma samples. Nonetheless, cell lines may capture one cancer cell type among many in a heterogeneous population and further analyses at the single cell level might provide insight to resolve this issue.
Tumor heterogeneity is an important factor in the development of drug resistance, because chemotherapy can select a drug-resistant subpopulation, leading to the failure of the anti-cancer treatment. In our previous work, we demonstrated that a breast cancer cell line (MCF-7) under long-term drug selection generated cells with cancer stem cell characteristics (37). We further characterized the gene expression profiles of multidrug resistance genes by using the TLDA in these cells sorted for CD44+/CD24− (stem-like cells) and CD44+/CD24+, and found that CD44+/CD24− cells had activated p53 and p21 pathways and decreased expression of many antiapoptotic genes. Moreover, the CD44+/CD24− cells are similar to seven of the nine cultured cancer cell types studied in this work in which p53 pathway activation is the highest-ranked network. These patterns of gene expression are therefore more similar among cultured cells and different from the patterns seen “on average” in the clinical cancer samples. Are these subpopulations of cancer stem cells or other cells within clinical cancers that mimic the expression patterns in the cultured cells (either stem cell subpopulations or whole populations)? To study this possibility, we prepared a CD133-positive subpopulation of human breast cancer cells isolated directly from surgical samples. The gene expression of this subpopulation of stem-like cells was not distinguishable from that of the breast cancer as a whole, and it was different from that of either stem-cell–like populations found among MCF-7 drug-resistant cells, or the entire population of MCF-7 cells or other cultured breast cancer cells (Fig. S5). Other investigators, such as Li et al., found that chemotherapy increased the number of stem-like cells within breast cancer biopsies (38). These studies suggest that drug selection either in vitro or in vivo can select for a population of highly resistant cancer cells, some of which include a stem-like population.
Although cultured cells can be used to study many aspects of cancer biology and response of cells to drugs, this study clearly shows that results derived from established cancer cell lines used for the study of either the mechanism or modulation of clinical drug resistance in cancer should be interpreted with caution. Investigators using these cell lines should be aware of the associated caveats and temper their extrapolations so as not to infer direct applicability to clinical medicine. This study emphasizes the necessity for new in vitro cancer models and the use of primary tumor models in which gene expression can be manipulated and small molecules tested in a setting that more closely mimics the in vivo cancer microenvironment so as to avoid radical changes in gene expression profiles brought on by extended periods of cell culture.
Materials and Methods
Cancer Cell Lines.
In vitro models.
Total RNA from 59 of the 60 cancer cell lines that make up the NCI-60 panel was prepared and provided by Developmental Therapeutics Program (DTP; http://www.dtp.nci.nih.gov/branches/btb/ivclsp.html). Total RNA for the ME:MDA-N line was unavailable at DTP.
The ovarian cancer cell line OVCAR-5 was grown in both monolayer and 3D (3D scaffold) models. In monolayer culture, the cells were maintained in RPMI 1640 supplemented with l-glutamine, penicillin, streptomycin, and 10% FCS. The 3D methods assessed included Algimatrix and Geltrex (Invitrogen), and the hanging drop and low attachment plate methods (Fisher Scientific). For the first two methods, the cells were maintained and harvested according to the manufacturer's instructions. According to the hanging drop method, 750 cells were seeded in a 30-μL drop spotted on the up-turned inner surface of the lid of a 100-mm tissue culture dish, in which 10 mL of PBS were added. In the low attachment plate method, cells were maintained in RPMI 1640 with 4.5 g/L glucose supplemented with l-glutamine, penicillin, streptomycin, and 10% FCS.
Two cisplatin-resistant (CP-r) cell lines and their parental cell lines were profiled: The human epidermoid carcinoma cell line KB-3-1 and its independent CP-r derivative, KB-CP.5, were selected in a single step at 0.5 μg of cisplatin/mL of medium (25, 26). The KB-CP20 cell line, and the human liver carcinoma cell line BEL-7404 and its CP-r derivative 7404-CP20, were selected by stepwise increases to 20 μg of cisplatin/mL of medium, as described (25, 26). All of the CP-r cells were maintained in the presence of cisplatin, but cisplatin was removed from the medium 3 d before preparation of RNA. All cell lines were grown as monolayer cultures at 37 °C in 5% CO2, using Dulbecco's modified Eagle medium with 4.5 g/L glucose supplemented with l-glutamine, penicillin, streptomycin, and 10% FCS. All of the cells were incubated under standard culture conditions (5% CO2 and 37 °C).
Total RNA from 10 additional cisplatin-resistant cell lines was received from the Cleveland Clinic, including the HOSE (39), PEO1 (40, 41), PEO4 (40, 41), A2780 (40), A2780/CP (40), C30 (40), CP70 (40), C200 (40), PAT-7 (42), and OC-2 (43) cell lines.
In vivo models.
All studies were conducted in an Association for Assessment and Accreditation of Laboratory Animal Care International accredited facility, in compliance with the US Public Health Service guidelines for the care and use of animals in research. The ovarian cancer cell line OVCAR-5 was grown by 2D and 3D methods, harvested, and used to generate xenograft models. The immunodeficient beige/nude/scid mice were challenged by s.c. and i.p. implantation of a cell pellet.
Tumor Samples.
The collection of tumors for research and specifically molecular analysis was first approved by the institutional review board of each of the four participating cancer treatment centers, and written informed consent was obtained from the patients. Three hundred eighty MDR-linked gene profiles from 80 specimens of ovarian untreated primary serous carcinoma and 32 ascites were used from studies performed in our laboratory (27).
Nine glioblastoma samples including four primary tumors, three recurrent tumors, and two metastatic samples were obtained from Georgetown University. Seven paired samples (normal and tumor) of colorectal cancer were obtained from the Laboratory of Human Carcinogenesis (National Cancer Institute). Leukemia (AML and T-ALL) samples were received from the Karolinska Institute and the National Cancer Institute Clinical Center. The AML cohort was comprised of 15 samples taken at diagnosis, whereas the T-ALL cohort was comprised of 12 untreated samples and 11 heavily pretreated samples. Nine metastatic melanoma samples were received from the National Cancer Institute Clinical Center. Eleven breast tissue samples, seven tumors, and four normal tissues were received from the Cooperative Human Tissue Network.
Preparation of Total RNA, Reverse Transcription, and TLDA Processing.
These were performed as previously published (27).
Data Import and Preprocessing.
Data from TLDAs was collected for 80 untreated ovarian primary serous carcinomas, 32 ascites, 36 in vitro and in vivo models of ovarian cancer, and 59 cell lines from the NCI-60 set of cancer cell lines. Samples from different origins were matched by using gene names. The median expression of each sample was subtracted from all gene expressions for that sample. The expressions from replicate probes were averaged together. The dataset is available at Gene Expression Omnibus (accession no. GSE30034 at www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE30034).
TLDA Data Processing.
TLDA cards were analyzed with RQ Manager Software, and the median was normalized as described (23). We then multiplied the gene expression values by −1 for ease of interpretation. By changing the sign of the normalized Ct values, high positive values of the new gene expression value represent high gene expression (above the median), whereas low negative values represent low gene expression (below the median).
Determination of Classification Accuracy.
To elucidate the accuracy of classification for the cancer types, an algorithm was set up by specifying the P value cutoff threshold and then excluding one sample. The P values for each gene were computed by using ANOVA; this P value signifies how much they are differentially expressed in the different cancer types. The P values were adjusted by using the Benjamini Hochberg procedure for multiple comparison correction. The genes with P values less than the specified threshold were used to create a diagonal linear discriminant classifier and the training sample. The excluded sample was classified, and this classifier was repeated for all samples excluded. The accuracy of the classifier was computed, indicating the proportion of times the predicted cancer class was the correct one. This procedure was repeated for all P value thresholds.
Classification of Cancer Types.
The resulting normalized values were analyzed by BRB-ArrayTools. (Biometric Research Branch, National Cancer Institute). Genes were excluded in the analysis if <20% of expression data had at least a 1.5-fold change in either direction, compared with the median gene expression, bringing the total number of genes to 354. After performing the multivariate permutation test, 185 genes were identified to be differentially expressed between the tissue types based on P < 0.05 and FDP < 10%. The parametric P values for the F-test statistic and the geometric means for each tissue type derived from the log base 2 transformed expression values were found. Hierarchical clustering analysis was then performed by using a correlation distance metric with average linkage. Multidimensional scaling was also carried out. After the ANOVA, post hoc Tukey–Kramer tests were completed to determine the specific cancer tissue types in which each gene is differentially expressed. The Tukey–Kramer tests were done by using SAS v9.
Pathway Analysis.
The genes differentially expressed in the tissue types were then analyzed by Ingenuity Pathways Analysis software (Ingenuity Systems) to determine which biological relationships exist between the genes present in each list. The reference set for this analysis was the Ingenuity Knowledge Base (genes only), and the network analysis was set to direct relationships. The human data sources including human cell line and tissue were used to run the analysis. A numerical value was determined by the software to rank networks according to relevance to the genes in the input dataset. This score takes into account the number of focus genes in the network and the size of the network to approximate relevance of the network to the original list of focus genes. These calculations are based on the hypergeometric distribution calculated via the computationally efficient Fisher's exact test for 2 × 2 contingency tables. Path Designer software was used to generate pathway images. Two pathways, for ovarian cancer and melanoma, were further enhanced by using SIMUSITE (BioPhase Systems). The correlations were drawn between gene expression profiles generated by using TLDAs and the growth inhibitory profiles of 1,429 candidate anticancer drugs tested against the NCI-60 panel (44). The drug index was calculated by using a module of SIMUSITE where the raw cell survival values were converted and integrated with the TLDA values of 380 genes. The pathways were further specified by merging with MDR-specific proteins. The relationships between the proteins were authenticated by the proprietary database generated from the public literature. SIMUSITE's analytical and data rendering libraries were interwoven with the open-source applications/databases available from the Cancer Biomedical Informatics Grid, National Cancer Institute; https://cabig.nci.nih.gov.
Comparison of the Cell Lines and Clinical Samples Analyzed.
Hierarchical clustering was performed by using the average linkage algorithm and 1-Pearson correlation as the distance measure. The differences between the in vitro and in vivo samples persist when using Euclidean distance clustering (Fig. S6 and Fig. S7). Actually, the clustering done by using Pearson correlation will not change under any linear normalization schemes (i.e., normalization that consists of subtracting the same quantity from the expression of all genes for each sample or multiplying or dividing the same quantity from the expression of all genes for each sample). Specifically, the correlations between samples remain the same and, thus, the clustering will be the same under any of the following normalization schemes: (i) no normalization, (ii) median normalization, and (iii) median normalization followed by converting all samples to the same variance (e.g., converting to z scores). If a nonlinear normalization (such as loess or quantile normalization) is done, then the correlations can potentially change and the clustering can be different. However, these kinds of normalization are not required for TLDA data (45).
The differences between samples of different origins were more directly assessed by a class comparison between samples of primary tumor, ascites, ovarian cancer models, and normal ovarian tissue. This was done for each gene by using an ANOVA between the expressions in the three sample groups.
Supplementary Material
Acknowledgments
We thank the Developmental Therapeutics Program for the total RNA from the NCI-60 cell panel; George Leiman for editorial assistance; and L. Stenke, M. Lindberg, M. Björkholm, J. Sjoberg, K. Viktorsson, R. Lewensohn (Karolinska Institutet), S. A. Rosenberg [Tumor Immunology Section Branch, National Cancer Institute (NCI), National Institutes of Health (NIH)], T. Waldman (Lombardi Comprehensive Cancer Center, Georgetown University), C. Harris, A. Schetter (Laboratory of Human Carcinogenesis, NCI, NIH), J. Keller (Laboratory of Cancer Prevention, NCI, NIH), P. Liu (Genetics and Molecular Biology Branch, National Human Genome Research Institute, NIH), L. Wolff (Laboratory of Cellular Oncology, NCI, NIH), T. Fojo, and L. Mickley Huff (Medical Oncology Branch, NCI, NIH) for generously providing clinical cancer samples. The BRB-ArrayTools were developed by Richard Simon and colleagues (Biometric Research Branch, National Cancer Institute). This research was supported by the Intramural Research Program of the NIH, Center for Cancer Research, NCI.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE30034).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111840108/-/DCSupplemental.
References
- 1.Rhodes DR, et al. Molecular concepts analysis links tumors, pathways, mechanisms, and drugs. Neoplasia. 2007;9:443–454. doi: 10.1593/neo.07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simon R. Lost in translation: Problems and pitfalls in translating laboratory observations to clinical utility. Eur J Cancer. 2008;44:2707–2713. doi: 10.1016/j.ejca.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gillet JP, Gottesman MM. Advances in the molecular detection of ABC transporters involved in multidrug resistance in cancer. Curr Pharm Biotechnol. 2011;12:686–692. doi: 10.2174/138920111795163931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borrell B. How accurate are cancer cell lines? Nature. 2010;463:858. doi: 10.1038/463858a. [DOI] [PubMed] [Google Scholar]
- 5.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma SV, Haber DA, Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer. 2010;10:241–253. doi: 10.1038/nrc2820. [DOI] [PubMed] [Google Scholar]
- 7.Borst P, Wessels L. Do predictive signatures really predict response to cancer chemotherapy? Cell Cycle. 2010;9:4836–4840. doi: 10.4161/cc.9.24.14326. [DOI] [PubMed] [Google Scholar]
- 8.Lukk M, et al. A global map of human gene expression. Nat Biotechnol. 2010;28:322–324. doi: 10.1038/nbt0410-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 10.Quackenbush J. Microarray analysis and tumor classification. N Engl J Med. 2006;354:2463–2472. doi: 10.1056/NEJMra042342. [DOI] [PubMed] [Google Scholar]
- 11.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 12.Scherf U, et al. A gene expression database for the molecular pharmacology of cancer. Nat Genet. 2000;24:236–244. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- 13.Gillet JP, Gottesman MM. Mechanisms of multidrug resistance in cancer. In: Zhou J, editor. MultiDrug Resistance in Cancer. Totowa, NJ: Humana; 2010. pp. 47–76. [DOI] [PubMed] [Google Scholar]
- 14.Gillet JP, Efferth T, Remacle J. Chemotherapy-induced resistance by ATP-binding cassette transporter genes. Biochim Biophys Acta. 2007;1775:237–262. doi: 10.1016/j.bbcan.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Szakács G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 16.Gillet JP, et al. Microarray-based detection of multidrug resistance in human tumor cells by expression profiling of ATP-binding cassette transporter genes. Cancer Res. 2004;64:8987–8993. doi: 10.1158/0008-5472.CAN-04-1978. [DOI] [PubMed] [Google Scholar]
- 17.Szakács G, et al. Predicting drug sensitivity and resistance: Profiling ABC transporter genes in cancer cells. Cancer Cell. 2004;6:129–137. doi: 10.1016/j.ccr.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 18.Efferth T, et al. Expression profiling of ATP-binding cassette transporters in childhood T-cell acute lymphoblastic leukemia. Mol Cancer Ther. 2006;5:1986–1994. doi: 10.1158/1535-7163.MCT-06-0086. [DOI] [PubMed] [Google Scholar]
- 19.Chapuy B, et al. Intracellular ABC transporter A3 confers multidrug resistance in leukemia cells by lysosomal drug sequestration. Leukemia. 2008;22:1576–1586. doi: 10.1038/leu.2008.103. [DOI] [PubMed] [Google Scholar]
- 20.Chapuy B, et al. ABC transporter A3 facilitates lysosomal sequestration of imatinib and modulates susceptibility of chronic myeloid leukemia cell lines to this drug. Haematologica. 2009;94:1528–1536. doi: 10.3324/haematol.2009.008631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinbach D, et al. ABCA3 as a possible cause of drug resistance in childhood acute myeloid leukemia. Clin Cancer Res. 2006;12:4357–4363. doi: 10.1158/1078-0432.CCR-05-2587. [DOI] [PubMed] [Google Scholar]
- 22.Langmann T, Mauerer R, Schmitz G. Human ATP-binding cassette transporter TaqMan low-density array: Analysis of macrophage differentiation and foam cell formation. Clin Chem. 2006;52:310–313. doi: 10.1373/clinchem.2005.059774. [DOI] [PubMed] [Google Scholar]
- 23.Orina JN, et al. Evaluation of current methods used to analyze the expression profiles of ATP-binding cassette transporters yields an improved drug-discovery database. Mol Cancer Ther. 2009;8:2057–2066. doi: 10.1158/1535-7163.MCT-09-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gillet JP, de Longueville F, Remacle J. DualChip microarray as a new tool in cancer research. Expert Rev Mol Diagn. 2006;6:295–306. doi: 10.1586/14737159.6.3.295. [DOI] [PubMed] [Google Scholar]
- 25.Liang XJ, Shen DW, Garfield S, Gottesman MM. Mislocalization of membrane proteins associated with multidrug resistance in cisplatin-resistant cancer cell lines. Cancer Res. 2003;63:5909–5916. [PubMed] [Google Scholar]
- 26.Shen D, Pastan I, Gottesman MM. Cross-resistance to methotrexate and metals in human cisplatin-resistant cell lines results from a pleiotropic defect in accumulation of these compounds associated with reduced plasma membrane binding proteins. Cancer Res. 1998;58:268–275. [PubMed] [Google Scholar]
- 27.Gillet JP, et al. Clinical relevance of multidrug resistance gene expression in ovarian serous carcinoma effusions. Mol Pharm. 2011 doi: 10.1021/mp200240a. 10.1021/mp200240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garson K, Shaw TJ, Clark KV, Yao DS, Vanderhyden BC. Models of ovarian cancer—are we there yet? Mol Cell Endocrinol. 2005;239:15–26. doi: 10.1016/j.mce.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 29.Connolly DC. Animal models of ovarian cancer. Cancer Treat Res. 2009;149:353–391. doi: 10.1007/978-0-387-98094-2_17. [DOI] [PubMed] [Google Scholar]
- 30.Bast RC, Jr, Hennessy B, Mills GB. The biology of ovarian cancer: New opportunities for translation. Nat Rev Cancer. 2009;9:415–428. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konstantinopoulos PA, Spentzos D, Cannistra SA. Gene-expression profiling in epithelial ovarian cancer. Nat Clin Pract Oncol. 2008;5:577–587. doi: 10.1038/ncponc1178. [DOI] [PubMed] [Google Scholar]
- 32.Gómez-Raposo C, Mendiola M, Barriuso J, Hardisson D, Redondo A. Molecular characterization of ovarian cancer by gene-expression profiling. Gynecol Oncol. 2010;118:88–92. doi: 10.1016/j.ygyno.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 33.Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol. 2008;9:702–712. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- 34.Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol. 2010;2:a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Connor PM, et al. Characterization of the p53 tumor suppressor pathway in cell lines of the National Cancer Institute anticancer drug screen and correlations with the growth-inhibitory potency of 123 anticancer agents. Cancer Res. 1997;57:4285–4300. [PubMed] [Google Scholar]
- 37.Calcagno AM, et al. Prolonged drug selection of breast cancer cells and enrichment of cancer stem cell characteristics. J Natl Cancer Inst. 2010;102:1637–1652. doi: 10.1093/jnci/djq361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100:672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 39.Maeda T, et al. Establishment of an immortalised human ovarian surface -epithelial cell line without chromosomal instability. Br J Cancer. 2005;93:116–123. doi: 10.1038/sj.bjc.6602662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson SW, Laub PB, Beesley JS, Ozols RF, Hamilton TC. Increased platinum-DNA damage tolerance is associated with cisplatin resistance and cross-resistance to various chemotherapeutic agents in unrelated human ovarian cancer cell lines. Cancer Res. 1997;57:850–856. [PubMed] [Google Scholar]
- 41.Wolf CR, et al. Cellular heterogeneity and drug resistance in two ovarian adenocarcinoma cell lines derived from a single patient. Int J Cancer. 1987;39:695–702. doi: 10.1002/ijc.2910390607. [DOI] [PubMed] [Google Scholar]
- 42.Lee FY, et al. BMS-247550: A novel epothilone analog with a mode of action similar to paclitaxel but possessing superior antitumor efficacy. Clin Cancer Res. 2001;7:1429–1437. [PubMed] [Google Scholar]
- 43.Uyar D, et al. Apoptotic pathways of epothilone BMS 310705. Gynecol Oncol. 2003;91:173–178. doi: 10.1016/s0090-8258(03)00481-5. [DOI] [PubMed] [Google Scholar]
- 44.Staunton JE, et al. Chemosensitivity prediction by transcriptional profiling. Proc Natl Acad Sci USA. 2001;98:10787–10792. doi: 10.1073/pnas.191368598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang B, et al. Systematic evaluation of three microRNA profiling platforms: Microarray, beads array, and quantitative real-time PCR array. PLoS ONE. 2011;6:e17167. doi: 10.1371/journal.pone.0017167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.