Abstract
The current gold standard for the culture of human pluripotent stem cells requires the use of a feeder layer of cells. Here, we develop a spatially defined culture system based on UV/ozone radiation modification of typical cell culture plastics to define a favorable surface environment for human pluripotent stem cell culture. Chemical and geometrical optimization of the surfaces enables control of early cell aggregation from fully dissociated cells, as predicted from a numerical model of cell migration, and results in significant increases in cell growth of undifferentiated cells. These chemically defined xeno-free substrates generate more than three times the number of cells than feeder-containing substrates per surface area. Further, reprogramming and typical gene-targeting protocols can be readily performed on these engineered surfaces. These substrates provide an attractive cell culture platform for the production of clinically relevant factor-free reprogrammed cells from patient tissue samples and facilitate the definition of standardized scale-up friendly methods for disease modeling and cell therapeutic applications.
Keywords: biomaterials, induced pluripotent stem cells, surface science, embryonic stem cells
Because human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs) have the potential to grow indefinitely in culture and to make any cell in the adult human body, they represent important resources for regenerative medicine (1) and for disease modeling efforts that seek to recreate a patient's disease pathogenesis in the laboratory (2, 3). hESCs and hiPSCs are typically grown on a “feeder” cell layer of mitotically inactivated mouse embryonic fibroblasts (mEFs) or on “feeder-free” culture systems composed of a variety of ECM/serum proteins coated onto tissue culture dishes (4–9) or synthetic materials (10–14). Nearly all these systems have been reported to promote hESC/hiPSC self-renewal when the cells are passaged in multicellular clumps and seeded at a suitably high cell density. Such characteristics make it laborious to generate large quantities of hESCs/hiPSCs when starting from small numbers of cells, especially from a single cell. However, genetic modification procedures of hESCs/hiPSCs involve growth from a single cell (15–17), and hiPSCs must be expanded from a few cells during reprogramming (18). Because the clinical application of stem cells may require as many as 1010 cells per patient (19) and disease modeling efforts typically require >106 cells to make a single differentiated cell type (20), robust methods of producing cells under conditions that accelerate proliferation could prove particularly valuable.
Recent work in developing feeder-free systems represents key progress in simplifying hESC/hiPSC production, but these methods still require components that are difficult to synthesize. A variety of substrates are used in these methods: synthetic polymers (11, 14), peptide-modified surfaces (10, 12), embryonic ECM laminin isoforms (7), fibronectin from ECM with a small molecule mixture (9), and various vitronectin proteins (4, 6). Some of these culture methods use animal products, which makes potential transplantation applications problematic (21). Most of these systems provide only modest gains in scaling-up hESC/hiPSC production because they still require seeding at a suitably high cell density and passaging through multicellular clumps. Even for the defined systems that support clonal growth (4, 9, 11), mass production of synthetic polymers, recombinant proteins, or small molecule mixtures may be a challenge, particularly when considering the number of cells needed for disease modeling and clinical application. Here, we develop a technology that notably provides expansion rates greater than the gold standard of feeder substrates; works with enzymatic passaging; and is compatible with current manipulations involving hESCs/hiPSCs, including gene modification and reprogramming. Importantly, in contrast to the previous reports that used polymers and/or peptides to coat the dish (10–12, 14), our UV/ozone (UV) treatment of common research materials is an attractive alternative because it is inexpensive and mass production of appropriately treated culture dishes for numerous applications should be straightforward.
Results
UV Treatment to Generate Optimal Surface Chemistry.
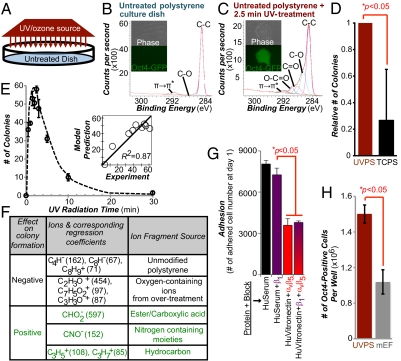
hESCs and hiPSCs aggregate or undergo apoptosis when passaged as single cells under conventional culture conditions (22, 23). We hypothesized that spatial control of the plating surface chemistry could influence early aggregation and could lead to improved cell propagation. Therefore, we sought to develop methods that would allow the rapid spatially controlled generation of chemically optimized surfaces for hESC/hiPSC propagation. We focused our efforts on polystyrene, the most commonly used plastic in cell culture. Recently, hundreds of polymers were evaluated to define the chemical and material properties of polymers that support the long-term culture of fully dissociated hESCs/hiPSCs (11). In these experiments, a distinct polymer surface chemistry was found to correlate with undifferentiated hESC growth, whereas indentation elastic modulus or roughness properties of biomaterials had less pronounced effects on growth (11). To generate optimized polymer surface chemistry [characterized by time of flight secondary ion mass spectrometry (ToF-SIMS) analysis by certain oxygen-containing ions and hydrocarbon ions (11)], virgin polystyrene was treated with short-wavelength UV (Fig. 1A). Polystyrene surfaces treated with different doses of UV were analyzed using ToF-SIMS (Fig. S1) to reveal mass spectral information from their topmost layers (∼10 Å) (24). Comparative analysis of these spectra (Figs. S2 and S3 A and B) confirmed that UV treatment yields chemically distinct surfaces from both virgin polystyrene and conventional tissue culture polystyrene (TCPS). Further, these results were consistent with surface elemental and functional composition resolved by X-ray photoelectron spectroscopy (XPS; Fig. 1 B and C). To test the effect of UV treatment on cell growth, we cultured fully dissociated transgenic Oct4-GFP–positive BG01 hESCs on the surfaces generated from different doses and precoated with fetal bovine serum (FBS) (Fig. 1E). hESCs robustly grew on moderately treated (1.5–3 min) polystyrene surfaces (Fig. 1C), whereas no cells attached to virgin polystyrene surfaces (Fig. 1B) and significantly fewer cells grew on TCPS (Fig. 1D).
Fig. 1.
More undifferentiated human pluripotent stem cells grow on chemically optimized substrates than on feeder-containing substrates. (A) Schematic diagram of UV treatment. XPS spectra of surface chemical functionality of an untreated “virgin” polystyrene culture dish (B) and after 2.5 min of UV treatment (C) are shown. Phase-contrast and fluorescence images of transgenic Oct4-GFP hESCs on these surfaces are shown. Bright green fluorescence on the UV-treated areas indicates strong expression of the pluripotency marker Oct4. (D) Relative number of hESC colonies on UVPS treated for 2.5 min vs. conventional TCPS on day 7 after cell seeding. (E) Colony formation on virgin polystyrene treated with various UV doses. The y axis lists the number of colonies formed on day 7. (Inset) PLS model prediction of the number of colonies plotted against the experimental results. For A–E, surfaces were precoated with 20% (vol/vol) bovine serum and cells were seeded in ROCK inhibitor for the first 8–12 h. (F) Using the PLS model of the ToF-SIMS data, surface ions with high positive or negative regression coefficients were identified as supporting or inhibiting hESC colony formation (expanded list is provided in Fig. S3C). (G) Number of adhered cells after 24 h of culture on UVPS coated with either human serum (HuSerum) or recombinant human vitronectin (HuVitronectin) and with the specified integrin-blocking antibody. Blocking with αvβ5 reduced adhesion, whereas blocking with β1 had minimal effect, either alone or in combination with αvβ5 blocking. (H) Number of undifferentiated Oct4-GFP–positive hESCs per well of a six-well plate at day 7 of culture on UVPS (red) and on standard mouse embryonic feeder (mEF)-containing substrates (gray). UVPS was precoated with vitronectin. All error bars indicate 95% confidence intervals (n = 3).
To identify specific functionalities generated by UV that were important for colony formation, a numerical model was developed using a multivariate chemometric technique (25). This technique, using partial least squares (PLS) regression, is particularly useful when analyzing high-dimensional datasets like ToF-SIMS spectra to identify salient features that correlate with another variable. In Fig. 1E, Inset, we correlated surface chemistry contained in the spectra of various polystyrene surfaces to the number of hESC colonies observed on the surfaces. Good agreement between the measured number of colonies and that predicted from the PLS model developed from the ToF-SIMS spectra was seen (R2 = 0.87). In the PLS model, each secondary ToF-SIMS ion from the UV-treated surfaces was listed with its regression coefficient, a quantitative measure of its contribution to hESC colony formation. Further, some secondary ions were identified to associate with certain classes of surface chemical functionality (Fig. 1F; an expanded list is provided in Fig. S3C). The assignments of surface chemical functionalities were also supported by XPS (Fig. 1 B and C) and are consistent with previous characterization studies involving UV treatment of virgin polystyrene (26). Consistent with the results from the polymer array (11), robust self-renewal of hESCs was supported by ester/carboxylic acid functionalities yielding oxygen-containing ions in the ToF-SIMS spectra (Fig. 1F and Fig. S3C). In particular, polystyrene surfaces treated with UV for 1.5–3 min produced high intensities of several secondary ions in the ToF-SIMS experiments that were identified to support hESC colony formation (e.g., C2H5O+ and C2H6N+ in Fig. S2).
Increased Number of hESCs/hiPSCs During Short-Term Culture.
At the optimal 2.5-min dose, UV-treated polystyrene (UVPS) provides an attractive platform for stem cell culture. hESCs could be readily passaged directly from standard mEF substrates to UVPS, and vice versa, without any significant cell death, indicating that hESCs do not require an adaptation period to be cultured on UVPS. When coated with human serum, UVPS supported clonal growth in serum-free mTeSR1 media (Stem Cell Technologies) at an efficiency of 30 ± 12%, which is comparable to previous acrylate-based polymer results (11), and cells were highly Oct4-GFP–positive after 7 d of culture (Fig. S4A). Media in these experiments were supplemented with rho-associated kinase (ROCK) inhibitor Y-27632 (Stemgent) for the first 8–12 h of culture to reduce initial apoptosis of completely dissociated hESCs (27). Similar to behavior seen on the best acrylate-based polymers (11), blocking the vitronectin-binding integrin αvβ5 resulted in a significant decrease in day 1 cell adhesion, whereas blocking a matrigel-binding (28) integrin β1 had no effect (Fig. 1G). When surfaces were coated with varying concentrations of a recombinant vitronectin solution in PBS, robust growth occurred above a threshold of 100 ng/cm2 (Fig. S4B). Because clonal cell seeding and extremely high dilution ratios during passaging can lead to the outgrowth of genetically abnormal hESCs (29, 30), we tested the ability of UVPS to support the culture of fully dissociated hESCs at the typical 1:3 dilution ratio used during conventional hESC passaging. At these relevant conditions, UVPS supported about three times more cells per area than traditional feeder-containing mEF substrates (Fig. 1H). These results indicate that the surface chemistry generated by optimal UV treatment, like the surface chemistry of acrylate polymer hits (11), outperforms mEF substrates, the gold standard for hESCs.
Spatial Patterning.
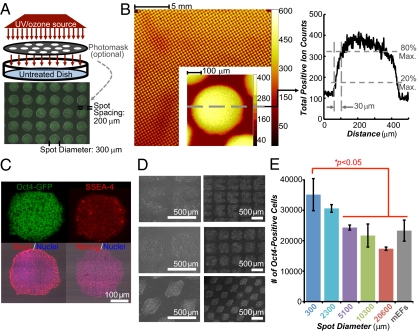
After 7 d of culture on UVPS, we observed the appearance of a few scattered large colonies (>1,200 μm in diameter) that were heterogeneous in pluripotency marker expression (Fig. S5A), indicating differentiation at the interior of the colony. To suppress the formation of these large colonies, we spatially controlled UV treatment by inserting a photomask between the polystyrene and radiation source (Fig. 2A). UV treatment generated a chemically heterogeneous surface as assayed by ToF-SIMS surface chemical analysis (Fig. 2B). Features down to ∼30 μm could be reliably generated by this masking technique (Fig. 2B). Spatially patterned UV treatment of polystyrene constrained cell migration and adhesion, such that hESCs attached and grew only on the areas of the dish that received UV treatment when precoated with FBS (Fig. 2A). Areas of the dish that were blocked from the UV treatment by the photomask did not support colony growth. Similar results were seen with five other human pluripotent stem cell lines, including three hiPSC lines (Fig. S5B), and when human serum or human vitronectin was used instead of FBS to coat the UV-patterned surfaces (Fig. S5 C and D).
Fig. 2.
Chemical and geometrical optimized polystyrene substrates. (A) Schematic diagram of UV treatment. Treatment can be spatially controlled by inserting a photomask between the UV source and the dish. An overlay of phase-contrast and fluorescent images of transgenic Oct4-GFP hESC cultures on a UV-patterned polystyrene substrate (300-μm spot diameter with 200-μm spacing, “UV-Pattern”) is shown. UVPS was precoated with FBS. (B) ToF-SIMS scan of a UVPS surface after patterning with a photomask with a spot diameter of 300 μm with 100-μm spacing. Color indicates the intensity of all positive ions. The profile indicates a resolution of 30 μm between 20% and 80% peak changes in ion intensity. Max., maximum. (C) Pluripotency marker immunostaining of cells on UV-Pattern described in A. (D) Patterning of hESCs or hiPSCs using other geometries. (E) Number of undifferentiated Oct4-GFP–positive cells in each well after 7 d of culture as assayed by flow cytometry on the constant area patterns (15,000 cells initially seeded per well). The cumulative UV-treated area per well is the same across all patterns (details are provided in Fig. S10A). Error bars indicate 95% confidence intervals (n = 3). Surfaces were precoated with 20% (vol/vol) bovine serum, and cells were seeded in ROCK inhibitor for the first 8–12 h.
Spatially patterned UV treatment could also generate robust substrates on a hydrogel-coated surface that supported robust colony formation and cell patterning (Fig. S6A). Like the polystyrene surfaces (Fig. 1E), an intermediate dose of UV on two other common research materials, polypropylene and hydrogel-coated surfaces, optimally supported colony formation from fully dissociated hESCs (Fig. S6B). Similar surface chemical functionalities supported colony growth on UVPS and polypropylene surfaces (Fig. 1F and Fig. S6D).
In contrast to UVPS without any spatial patterning, on the UV-patterned surfaces with 150- to 450-μm diameter spots, cells showed uniform staining of all pluripotency markers tested (Fig. 2C), and subsequent single-cell flow cytometric analysis indicated a small but significant decrease in the number of differentiating GFP-negative cells on a 300-μm spot pattern (Fig. S6E). Further, we detected poor spatial patterning when spot spacing was less than 200 μm, because cells could bridge the gaps between the UV-treated spots (Fig. S6F). Some of these bridged cells also exhibited low levels of pluripotency markers and exhibited differentiated cell morphology (Fig. S6F). Cells could be patterned in various geometries with sharp edges and curves (Fig. 2D) and could support more cells per well than traditional feeder-containing culture systems (Fig. S7 A and B). This behavior was consistent in two serum-free media formulations (Fig. S7C).
To gain insight into the spatial patterning parameters that control hESC behavior, we generated patterns of varying spot diameter but with the same cumulative UV-treated growth area per well. In this series of patterns, spot diameters varied from 300 to 20,600 μm (additional characteristics are presented in Fig. S10A). As before, we seeded fully dissociated hESCs on these patterns at a density typically used during routine cell culture. As the growth area became progressively partitioned into smaller and smaller spots, similar numbers of cells were observed initially 24 h after seeding (Fig. S8A), although more cells were found in large cell aggregates as spot diameters became larger (Fig. S8 B and C). After an additional 7 d of growth, cultures with the smaller spot diameters contained more undifferentiated cells (Fig. 2E) and proliferated faster on average, as indicated by a shorter doubling time (Fig. S8D).
Modeling Cell Movements on Patterned Substrates.
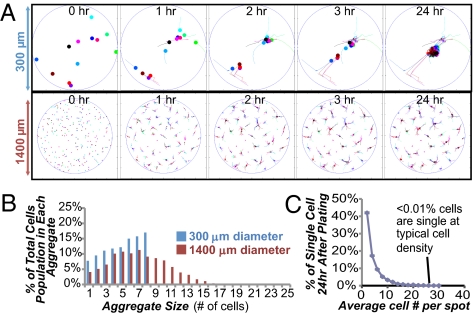
To probe why large cell aggregates preferentially form on larger spots, we modeled cell movements on patterned substrates through computer simulations (i.e., in silico) using the following assumptions. Cells were modeled to migrate on the surfaces through a random walk in the absence of neighboring cells, whereas in the presence of neighboring cells, single cells and cell aggregates preferentially migrate toward each other, as observed previously (31). Snapshots of cell migration behavior in silico on small and large spots (300 vs. 1,400 μm in diameter) indicate that the majority of cells form aggregates within 2–3 h postseeding (Fig. 3A and Fig. S9A). This time scale of aggregate formation was consistent with live imaging of cells in vitro on UV pattern (Movie S1). The number of cells in each aggregate after an additional 21 h of migration in silico was distributed differently between the small and large spot sizes (Fig. 3B). The 1,400-μm large spot pattern contained a long tail distribution of many aggregates of five cells or larger, whereas the 300-μm small spot pattern had a narrower distribution with more cells in smaller aggregates. Simulation results indicate that almost all the cells participate in colony formation, because the percentage of cells that are single is negligible (Fig. 3C) when seeded at a typical density during routine cell culture (60,000 cells per well in a 6-well plate). This result was further supported by the experimental data (Fig. S8C), and similar results were obtained from simulations of reduced cell migration in media with ROCK inhibitor (Fig. S9 B and C). In summary, both experimental and modeling results suggest that spots with a diameter of 300 μm control hESC colony formation by preventing the formation of large cell aggregates soon after seeding, which leads to lower average rates of growth (Fig. S8D).
Fig. 3.
Simulation of cell behavior on UV-patterned substrates. (A) Snapshots of hESCs/hiPSCs on spots with diameters of 300 and 1,400 μm during model simulation. The majority of cells aggregate within 3 h, as also seen during live imaging (Movie S1). (B) Distribution of cells in each aggregate as predicted from the cell migration model for two different patterns: 300- and 1,400-μm spot diameters (no ROCK inhibitor in media). (C) Percentage of cells that are not paired or in colonies (i.e., single) as a function of cell density. When seeded at a typical density during routine cell culture (60,000 cells per well in a 6-well plate), less than 0.01% of cells are single.
Long-Term Culture on Patterned Substrates.
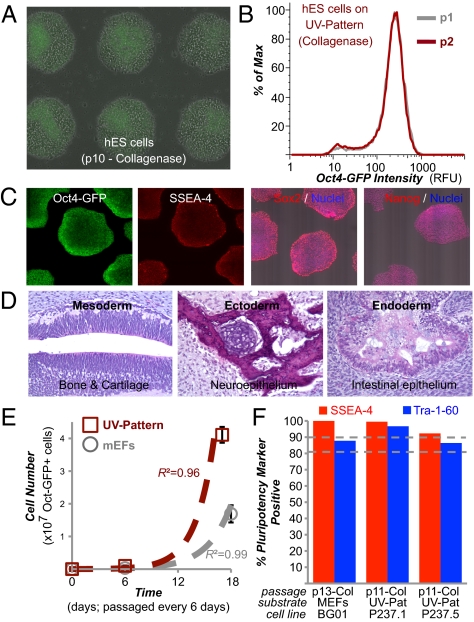
UV-patterned polystyrene was subsequently tested for its ability to support long-term culture. Cells after prolonged culture (>10 passages, 2 mo) on UV-patterned polystyrene using conventional collagenase or mechanical procedures were found to maintain an undifferentiated state at each passage (Fig. 4 A and B) and a normal karyotype (Fig. S10B). Further, cells after long-term culture robustly expressed all the pluripotency markers tested, including SSEA-4, Sox2, and Nanog (Fig. 4C). Derivatives of all three embryonic germ layers were seen in teratoma assays (Fig. 4D), demonstrating that hESCs cultured on the UV-patterned polystyrene maintain their full pluripotent potential. Routine cell culture with collagenase passaging yielded significant increases in the number of undifferentiated cells over mEFs that increased from passage to passage, indicating its utility for scale-up applications (Fig. 4E). Similar results were seen with two other hiPSC lines after >10 passages on UV-patterned polystyrene, where >80% of cells were positive for Tra-1-60 and >90% of cells were positive for SSEA-4 (Fig. 4F).
Fig. 4.
UV-patterned substrate supports standardized long-term culture. (A) Overlay of phase-contrast and fluorescent images of transgenic Oct4-GFP BG01 hESC cultures on “UV-Pattern” (as described in Fig. 2A) after 10 passages using collagenase dissociation. (B) Flow cytometry of cells after two consecutive passages on UV-Pattern. Max, maximum; p, passage; RFU, relative fluorescent unit. (C) Pluripotency marker immunostaining of cells on UV-Pattern. (D) Teratoma formation in immunodeficient mice by cells cultured on UV-Pattern. H&E staining was carried out on the teratoma, which contained tissues representing all three germ layers. (E) Number of undifferentiated Oct4-GFP–positive hESCs over three passages with accutase on UV-Pattern (red) and on standard mouse embryonic feeder (mEFs)-containing substrates (gray) when seeded with 24,000 cells per well of a six-well plate. Error bars indicate 95% confidence intervals (n = 3). High R2 coefficient of determination indicates good fit (R2 > 0.95) to exponential growth model (dashed lines). In A and E, the UV-Pattern was precoated with human serum. (F) Flow cytometry of cells for pluripotency markers SSEA-4 and Tra-1-60 after >10 consecutive passages on UV-Pattern for two different hiPSC lines, P237.1 and P237.5. Col, collagenase passaging. For the transgenic Oct4-GFP BG01 hESCs passaged on MEFs, only GFP-positive cells were analyzed for Tra-1-60 and SSEA-4 expression, allowing us to exclude MEFs from the analysis.
Clonal Growth During Gene Targeting and Reprogramming.
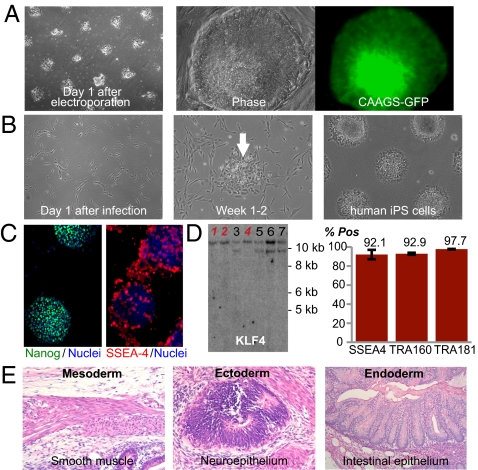
To examine whether these substrates could support clonal outgrowth of gene-targeted cells, hESCs on UV-patterned substrates were subjected to targeting of the AAVS1 locus using zinc-finger nuclease (ZFN)–mediated genome editing (Fig. 5A). Using previously described ZFN pairs and constructs (15), we targeted to the AAVS1 locus a donor plasmid expressing GFP under the control of the constitutively active CAGGS promoter. The targeting construct contained a splice acceptor-2A-puromycin selection cassette, and after using electroporation to introduce the constructs into hESCs, cells were grown in puromycin on UV-patterned polystyrene. After 2 wk of puromycin selection, we isolated clones that remained brightly GFP-positive after more than 2 mo of culture (Fig. 5A).
Fig. 5.
UV-patterned substrate supports reprogramming and gene modification of human pluripotent stem cells. (A) Phase-contrast images of BG01 hESCs on UV-patterned substrates (300-μm spot diameter with 400-μm spacing) after electroporation of CAAGS-GFP targeting and ZFN plasmids. Cells were seeded in ROCK inhibitor for the first 8–12 h after electroporation. A successfully targeted clone could be moved to mEFs and was highly GFP-positive after >2 mo of culture. (B) Phase-contrast and immunostained images of “patient-237” fibroblasts on UV-patterned polystyrene infected with a loxP flanked version of the pHAGE-STEMCCA vector, which is a Cre-excisable polycistronic vector encoding the reprogramming factors. The patterned surface was precoated with human serum, and the high serum content of the initial fibroblast media allows fibroblasts to adhere to the untreated areas of the dish. Over 4 wk, cells change morphology and form hiPSC colonies (arrow) on the substrates (10-cm dish, 300-μm spot diameter with 200-μm spacing) for 4 wk. (C) Pluripotency marker immunostaining of patient-237 hiPSC line on “UV-Pattern” (as described in Fig. 2A). (D) Southern blot analysis for Klf4 on genomic DNA from several patient-237 hiPSC lines after Cre-recombinase expression. Cell lines labeled in red indicate excision of reprogramming vector; these clones show loss of viral KLF4 bands (lower band) on Cre expression, indicating that vector-free hiPSCs can be clonally isolated postexcision. The bar graph on the right indicates % of cells positive for pluripotency markers SSEA-4, TRA-1-60, and TRA-1-81 as assayed by flow cytometry for a vector-free patient-237 hiPSC line after two passages on UV-Pattern as described in Fig. 2A. Error bars indicate 95% confidence intervals (n = 3). Pos, positive. (E) Teratoma formation in immunodeficient mice by vector-free hiPSCs reprogrammed and cultured on UV-Pattern. H&E staining was carried out on the teratoma, which contained tissues representing all three germ layers.
Given the ability of UV-patterned substrates to support hESC/hiPSC self-renewal with human serum, we also sought to reprogram human fibroblasts on UV-patterned substrates under xeno-free conditions. Fibroblasts established from two patient skin punch biopsies were infected with a Cre recombinase (Cre)-excisable vector carrying the reprogramming factors on UV-patterned polystyrene coated with human serum (Fig. 5B). After 4 wk of culture in serum-free mTeSR1 media, several colonies emerged (Fig. 5B) and hiPSC lines could be established that robustly expressed the pluripotency markers (Fig. 5C). Because residual expression of integrated copies of reprogramming factors can affect the gene expression and biological properties of hiPSCs (3, 32), the factors were excised after expressing Cre recombinase. When grown on UV-patterned polystyrene, >90% of the vector-free cells expressed several pluripotency markers (Fig. 5D). Also, the vector-free reprogrammed cells generated derivatives of all three embryonic germ layers in teratoma assays (Fig. 5E), demonstrating that the cells were fully pluripotent after reprogramming and excision of the reprogramming factors on UV-patterned substrates. Given that human serum can be readily replaced with recombinant human vitronectin, the UV-patterned substrates have strong potential to support the derivation and propagation of factor-free hiPSCs under fully defined xeno-free culture conditions.
Single-Cell Enzymatic Passaging.
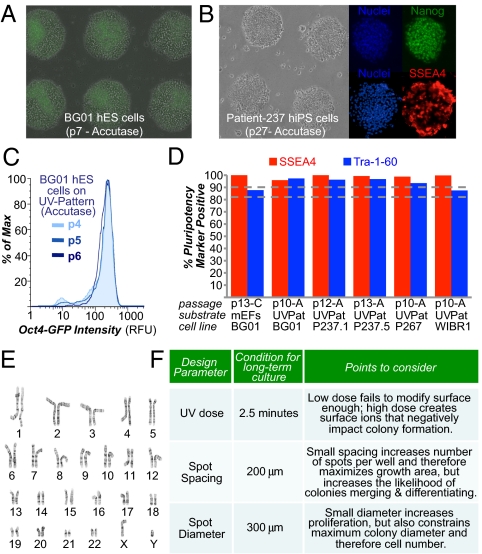
Lastly, UV-patterned substrates also supported the single-cell enzymatic passaging of hESCs with minimal use of small-molecule supplements. Current hESC/hiPSC passaging protocols use collagenase and/or mechanical dissociation to generate multicellular aggregates that vary in size, whereas dissociation into single cells using proteolytic enzymes (e.g., trypsin, accutase) greatly reduce this variability. Although dissociated hESCs/hiPSCs self-renew inefficiently, with the majority of single cells undergoing apoptosis (22, 23), we reasoned that the rapid establishment of multicellular aggregates on the patterned surfaces within 2–3 h after cell seeding could reduce the single-cell apoptosis and promote colony formation. Using accutase enzymatic dissociation to single cells, cultures could be consistently passaged at a 1:3 dilution every 5–6 d for both hESCs (Fig. 6A) and hiPSCs (Fig. 6B), wherein ROCK inhibitor was required only briefly for 8–12 h after dissociation. Cells robustly maintained expression of pluripotency markers consistently from passage to passage (e.g., Fig. 6C) and after long-term culture for five different cell lines (Fig. 6D). Karyotype analysis confirmed that all five cell lines had the correct number of chromosomes after >10 passages; in particular, patient-237 hiPSCs maintained a normal karyotype for more than 5 mo in culture (Fig. 6E). We note that using extremely harsh passaging conditions (e.g., using long incubations in trypsin, entirely omitting use of ROCK inhibitor postseeding) resulted in cultures with genetic abnormalities (Fig. S10B) as previously described (29, 30). We conclude that single-cell passaging on UV-patterned substrates can remove much of the variability in hESC/hiPSC culture generated by incomplete dissociation during mechanical or collagenase passaging, thus creating a standardized culturing system (e.g., Fig. 6C).
Fig. 6.
UV-patterned substrate supports single-cell passaging of human pluripotent stem cells. Images of BG01 hESC (A) and patient-237 hiPSC (B) cultures on “UV-Pattern” (as described in Fig. 2A) after 7 and 27 passages using single-cell accutase dissociation. The image of hESCs at passage (p) 7 in A contains an overlay of a fluorescent image indicating high expression of Oct4-GFP. Immunostaining of patient-237 hiPSCs at passage 27 indicates expression of the pluripotency marker Nanog (green) in all cell nuclei and high expression of SSEA-4 (red). Surfaces were precoated with 20% (vol/vol) bovine serum, and cells were seeded in ROCK inhibitor for the first 8–12 h. (C) Flow cytometry of BG01 hESCs with the Oct4-GFP reporter after three consecutive passages on UV-Pattern with accutase. (D) Flow cytometry of cells for pluripotency markers SSEA-4 and Tra-1-60, after >10 consecutive passages on UV-Pattern for five different cell lines. “A” indicates accutase-mediated passaging. For the transgenic Oct4-GFP BG01 cells passaged on mEFs, only GFP-positive cells were analyzed for Tra-1-60 and SSEA-4 expression. Therefore, mEFs were excluded from the analysis. (E) Normal 46XY karyotype was maintained for patient-237 hiPSCs propagated on UV-Pattern for more than 5 mo (27 passages). (F) Design parameters for developing a UV-treated culture system for human pluripotent stem cells.
Discussion
Through this work, we were able to generate substrates that replace or outperform mEF substrates, the gold standard in human pluripotent stem cell culture, in several key applications. The UV treatment itself without patterning creates a surface chemistry that supports at least threefold more cells per area (Fig. 1H and Fig. S7A) and creates at least twofold more colonies per cell seeded than conventional cell culture plastics (Fig. 1D). During routine cell culture, these differences grow exponentially from passage to passage and lead to even larger increases in undifferentiated cell number (Fig. 4E). Spatial patterning of the UV treatment can further increase the number of undifferentiated cells per growth area (Fig. 2E). By optimizing the surface treatment, we identified several parameters that were critical to the success of using UV treatment of polystyrene for hESC/hiPSC culture (Fig. 6F). The dose of UV was determined such that suitable amounts of carboxylic acid/ester and nitrogen-containing moieties were generated on the surface, whereas both under- and overexposure to UV led to undesirable surface chemistry (Fig. 1 E and F). Spatial patterning of UV treatment could constrain hESC/hiPSC colony size in two dimensions; however, hESCs/hiPSCs could only be consistently patterned into spots when spacing was greater than 100 μm (Fig. S6F). We consider several possibilities to explain these observations. Optimal spatial pattern of UV treatment may (i) establish favorable gradients of soluble factors guiding preferential migration toward other hESCs/hiPSCs (31), (ii) improve crosstalk between soluble factors and mechanical signal transduction (33), and (iii) facilitate the previously noted preference for hESCs/hiPSCs to be in cell-cell contact (22, 23, 34). Constraining cells to spots of small diameters prevented large multicellular aggregates from forming after seeding (Fig. S8 B and C), leading to higher levels of proliferation of undifferentiated cells (Fig. S8D). Small diameters, however, constrain the maximum colony size before cells are passaged. We found that a 300-μm spot diameter could support most applications of routine cell culture with hESCs/hiPSCs (Figs. 4–6), and similarly sized colonies were routinely observed on standard feeder substrates before they needed to be passaged (Fig. S10C). It should be possible to produce such plates in many different formats (e.g., multiwell plates, dishes) economically, because UV treatment does not require the relatively expensive gas handing and vacuum processing equipment used to manufacture standard tissue culture dishes.
Because use of the engineered substrates did not require any special steps or adaptation from cultures using existing feeder substrates, the surface treatments described here would likely integrate well with many existing protocols of manipulating human pluripotent stem cells. Further, the treated surfaces represent an important advance over the gold standard feeder substrates because they are fully defined synthetic substrates that enhance propagation of undifferentiated cells and support the long-term cell culture, clonal outgrowth of hESCs/hiPSCs, and reprogramming of human somatic cells. These surface-engineered substrates therefore have strong potential to replace feeder-containing substrates in almost any procedure envisioned with human pluripotent cells, enabling broad and rapid scale-up of these cells for both research and clinical applications.
Materials and Methods
A UV unit (Bioforce Nanoscience, Inc., USA) generated high-intensity light to treat surfaces. Before cell seeding, surfaces were coated for 15–30 min with human vitronectin, 20% human serum, or 20% (vol/vol) FBS. Further details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Q. Gao, P. Xu, D. Fu, R. Alagappan, P. Wisniewski, C. Araneo, T. Kiyomitsu, and I. Cheeseman for technical support and all members of the R.L. and R.J. laboratories for helpful discussions. R.J. is supported by the Howard Hughes Medical Institute and National Institutes of Health Grants R37-CA084198, RO1-CA087869, and RO1-HD045022. D.G.A., R.L., and Y.M. are supported by National Institutes of Health Grant DE016516. R.J. and J.M. are supported by the European Leukodystrophy Association. J.Y. is supported by Wellcome Trust Grant 085246, and K.S. is supported by the Society in Science: The Branco–Weiss Fellowship.
Footnotes
Conflict of interest statement: R.L., R.J., and D.G.A. are advisors to Stemgent, and R.L. and R.J. are cofounders of Fate Therapeutics.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114854108/-/DCSupplemental.
References
- 1.Nishikawa S-I, Goldstein RA, Nierras CR. The promise of human induced pluripotent stem cells for research and therapy. Nat Rev Mol Cell Biol. 2008;9:725–729. doi: 10.1038/nrm2466. [DOI] [PubMed] [Google Scholar]
- 2.Nelson TJ, Martinez-Fernandez A, Terzic A. Induced pluripotent stem cells: Developmental biology to regenerative medicine. Nat Rev Cardiol. 2010;7:700–710. doi: 10.1038/nrcardio.2010.159. [DOI] [PubMed] [Google Scholar]
- 3.Saha K, Jaenisch R. Technical challenges in using human induced pluripotent stem cells to model disease. Cell Stem Cell. 2009;5:584–595. doi: 10.1016/j.stem.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen G, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ludwig TE, et al. Feeder-independent culture of human embryonic stem cells. Nat Methods. 2006;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- 6.Braam SR, et al. Recombinant vitronectin is a functionally defined substrate that supports human embryonic stem cell self-renewal via alphavbeta5 integrin. Stem Cells. 2008;26:2257–2265. doi: 10.1634/stemcells.2008-0291. [DOI] [PubMed] [Google Scholar]
- 7.Rodin S, et al. Long-term self-renewal of human pluripotent stem cells on human recombinant laminin-511. Nat Biotechnol. 2010;28:611–615. doi: 10.1038/nbt.1620. [DOI] [PubMed] [Google Scholar]
- 8.Sun N, et al. Feeder-free derivation of induced pluripotent stem cells from adult human adipose stem cells. Proc Natl Acad Sci USA. 2009;106:15720–15725. doi: 10.1073/pnas.0908450106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsutsui H, et al. An optimized small molecule inhibitor cocktail supports long-term maintenance of human embryonic stem cells. Nat Commun. 2011;2:167. doi: 10.1038/ncomms1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klim JR, Li L, Wrighton PJ, Piekarczyk MS, Kiessling LL. A defined glycosaminoglycan-binding substratum for human pluripotent stem cells. Nat Methods. 2010;7:989–994. doi: 10.1038/nmeth.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mei Y, et al. Combinatorial development of biomaterials for clonal growth of human pluripotent stem cells. Nat Mater. 2010;9:768–778. doi: 10.1038/nmat2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melkoumian Z, et al. Synthetic peptide-acrylate surfaces for long-term self-renewal and cardiomyocyte differentiation of human embryonic stem cells. Nat Biotechnol. 2010;28:606–610. doi: 10.1038/nbt.1629. [DOI] [PubMed] [Google Scholar]
- 13.Gerecht S, et al. Hyaluronic acid hydrogel for controlled self-renewal and differentiation of human embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:11298–11303. doi: 10.1073/pnas.0703723104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Villa-Diaz LG, et al. Synthetic polymer coatings for long-term growth of human embryonic stem cells. Nat Biotechnol. 2010;28:581–583. doi: 10.1038/nbt.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hockemeyer D, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hockemeyer D, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nat Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
- 18.Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: Facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segers VFM, Lee RT. Stem-cell therapy for cardiac disease. Nature. 2008;451:937–942. doi: 10.1038/nature06800. [DOI] [PubMed] [Google Scholar]
- 20.Soldner F, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin MJ, Muotri A, Gage F, Varki A. Human embryonic stem cells express an immunogenic nonhuman sialic acid. Nat Med. 2005;11:228–232. doi: 10.1038/nm1181. [DOI] [PubMed] [Google Scholar]
- 22.Ohgushi M, et al. Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell. 2010;7:225–239. doi: 10.1016/j.stem.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 23.Chen G, Hou Z, Gulbranson DR, Thomson JA. Actin-myosin contractility is responsible for the reduced viability of dissociated human embryonic stem cells. Cell Stem Cell. 2010;7:240–248. doi: 10.1016/j.stem.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vickerman JC, Briggs D. ToF-SIMS: Surface Analysis by Mass Spectrometry. West Sussex, UK: IM Publications, Chichester; 2001. [Google Scholar]
- 25.Yang J, et al. Polymer surface functionalities that control human embryoid body cell adhesion revealed by high throughput surface characterization of combinatorial material microarrays. Biomaterials. 2010;31:8827–8838. doi: 10.1016/j.biomaterials.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell SA, et al. Cellular attachment and spatial control of cells using micro-patterned ultra-violet/ozone treatment in serum enriched media. Biomaterials. 2004;25:4079–4086. doi: 10.1016/j.biomaterials.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 27.Watanabe K, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- 28.Rowland TJ, et al. Roles of integrins in human induced pluripotent stem cell growth on Matrigel and vitronectin. Stem Cells Dev. 2010;19:1231–1240. doi: 10.1089/scd.2009.0328. [DOI] [PubMed] [Google Scholar]
- 29.Närvä E, et al. High-resolution DNA analysis of human embryonic stem cell lines reveals culture-induced copy number changes and loss of heterozygosity. Nat Biotechnol. 2010;28:371–377. doi: 10.1038/nbt.1615. [DOI] [PubMed] [Google Scholar]
- 30.Laurent LC, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L, et al. Individual cell movement, asymmetric colony expansion, rho-associated kinase, and E-cadherin impact the clonogenicity of human embryonic stem cells. Biophys J. 2010;98:2442–2451. doi: 10.1016/j.bpj.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soldner F, et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guilak F, et al. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell. 2009;5:17–26. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bauwens CL, et al. Control of human embryonic stem cell colony and aggregate size heterogeneity influences differentiation trajectories. Stem Cells. 2008;26:2300–2310. doi: 10.1634/stemcells.2008-0183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.