Abstract
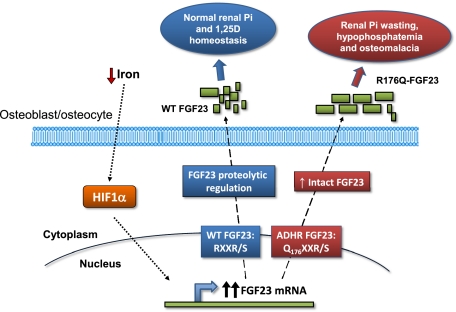
Autosomal dominant hypophosphatemic rickets (ADHR) is unique among the disorders involving Fibroblast growth factor 23 (FGF23) because individuals with R176Q/W and R179Q/W mutations in the FGF23 176RXXR179/S180 proteolytic cleavage motif can cycle from unaffected status to delayed onset of disease. This onset may occur in physiological states associated with iron deficiency, including puberty and pregnancy. To test the role of iron status in development of the ADHR phenotype, WT and R176Q-Fgf23 knock-in (ADHR) mice were placed on control or low-iron diets. Both the WT and ADHR mice receiving low-iron diet had significantly elevated bone Fgf23 mRNA. WT mice on a low-iron diet maintained normal serum intact Fgf23 and phosphate metabolism, with elevated serum C-terminal Fgf23 fragments. In contrast, the ADHR mice on the low-iron diet had elevated intact and C-terminal Fgf23 with hypophosphatemic osteomalacia. We used in vitro iron chelation to isolate the effects of iron deficiency on Fgf23 expression. We found that iron chelation in vitro resulted in a significant increase in Fgf23 mRNA that was dependent upon Mapk. Thus, unlike other syndromes of elevated FGF23, our findings support the concept that late-onset ADHR is the product of gene–environment interactions whereby the combined presence of an Fgf23-stabilizing mutation and iron deficiency can lead to ADHR.
Keywords: α-klotho, FGF-23, anemia, osteocyte
Autosomal dominant hypophosphatemic rickets (ADHR, Online Mendelian Inheritance in Man no. 193100) is characterized by low serum phosphate concentrations due to isolated renal phosphate wasting, inappropriately normal or low serum 1,25(OH)2 vitamin D (1,25D) concentrations, and rickets/osteomalacia and fracture (1). Heterozygous missense mutations in the fibroblast growth factor-23 (FGF23) gene cause ADHR (2). These mutations replace the arginine (R) residues at positions 176 or 179 with glutamine (Q) or tryptophan (W) within a 176RXXR179/S180 subtilisin-like proprotein convertase (SPC) site that separates the conserved FGF-like N-terminal domain from the variable C-terminal tail (2–4). Acting through the coreceptor α-Klotho (5) and a fibroblast growth factor receptor (FGFR) (5, 6), FGF23 reduces renal phosphate reabsorption through down-regulation of the sodium phosphate cotransporters NPT2a and NPT2c and suppresses kidney 1,25(OH)2 vitamin D production by inhibiting and increasing vitamin D 1α-hydroxylase (Cyp27b1) and 24-hydroxylase expression (Cyp24), respectively (7). Compared with WT Fgf23 protein, ADHR-mutant FGF23 shows increased but not complete resistance to SPC proteolytic cleavage (3, 4). When expressed in mammalian cells, the R176Q-, R179Q-, and R179W-FGF23 proteins are secreted primarily as the full-length (32-kDa) polypeptide, in contrast to the full-length and cleavage products (20 and 12 kDa) typically observed for WT FGF23 (3). This proteolytic event inactivates the mature FGF23 polypeptide, as full-length FGF23, but not N-terminal fragments (residues 25–179) or C-terminal fragments (residues 180–251), reduces serum phosphate concentrations when injected into rodents (4).
The ADHR biochemical and skeletal disease phenotypes parallel those of other disorders of excess FGF23 including X-linked hypophosphatemic rickets (XLH; loss-of-function mutations in phosphate-regulating gene with homologies to endopeptidases on the X chromosome, PHEX), autosomal recessive hypophosphatemic rickets (ARHR; loss-of-function mutations in dentin matrix protein 1, DMP1) (8, 9), and the acquired disorder, tumor-induced osteomalacia. ADHR was described first in a small family (10), and the subsequent analysis of an expanded ADHR kindred demonstrated that, unlike XLH and ARHR, ADHR displays incomplete penetrance and variable age of onset (1), with elevated circulating intact FGF23 concentrations that correlate with active disease status (11). ADHR contains two subgroups of affected individuals: One subgroup consists of patients who present during childhood with phosphate-wasting, rickets, and lower limb deformity; the second subgroup consists of phenotypic carrier individuals who are unaffected as children but later present clinically during adolescence or adulthood (1). Of significance, female ADHR carriers can have late-onset disease during puberty and following pregnancy, both physiological situations that are prone to iron deficiency (12), and male ADHR patients also have shown disease onset and regression over time (13). Further, reduced serum iron concentrations were correlated strongly with increased serum FGF23 in ADHR patients (14), and C-terminal FGF23 is negatively correlated with ferritin (15); however, the mechanisms underlying these observations are unknown.
Since late-onset ADHR can occur during physiological states in which iron deficiency is common and reduced serum iron has been associated with increased FGF23 concentrations in ADHR, the goals of the present studies were to test the hypothesis that an ADHR disease phenotype can be induced in a knock-in R176Q-Fgf23 mouse model by decreasing iron load, and to determine the molecular mechanisms underlying FGF23 expression in bone during iron deficiency. The studies described here have important therapeutic implications for rare and for more common disorders of phosphate metabolism involving FGF23 by providing a mechanistic basis for delayed onset of ADHR and revealing unique cross-over regulation between phosphate and iron metabolism.
Results
Murine Model Expressing an R176Q-Fgf23 ADHR Allele.
A mouse harboring an allele with an orthologous ADHR R176Q-Fgf23 mutation was developed using a knock-in strategy (Fig. S1 A and B). Following targeting of the R176Q-Fgf23 construct and establishment of germline transmission of the mutant allele (Fig. S1 A and B), RNA was isolated from heterozygous ADHR mice. Using intron-spanning primers to amplify Fgf23 mRNA by RT-PCR, appropriately-sized cDNAs were obtained and sequenced. The ADHR-Fgf23 mRNA showed correct splicing for the three exons, and the presence of the heterozygous knocked-in ADHR R176Q mutation [codon cgc (Arg; R) >cag (Glu; Q) for mouse] was confirmed (Fig. S1C). No other substitutions were found in the Fgf23 transcripts; thus these mice appropriately express mRNA encoding the ADHR R176Q-Fgf23 mutant allele under the native promoter and regulatory elements.
In Vivo Analysis of ADHR Onset and Progression: Mineral Metabolism.
Subjects with ADHR mutations are more likely to experience late-onset disease during physiological states associated with low iron status, therefore WT and ADHR mice were examined after receiving diet with normal iron content (45 mg/kg iron; control diet) or a low-iron diet (no added iron) from weaning (at 4 wk) for an additional 8 and 12 wk (n = 6–12 male and female mice per 8- and 12-wk cohorts). Similar diets and feeding periods have been used previously to reduce iron status in rodents (16, 17). Males and females were analyzed together, because there was no interaction between the primary variables and sex. To isolate the physiological effects of the mutant allele during iron deprivation, homozygous R176Q-Fgf23 ADHR mice were tested initially (see heterozygous ADHR results below). Although the male mice were heavier than female mice, the mice on both diets gained weight consistently over the treatment course (Fig. 1A).
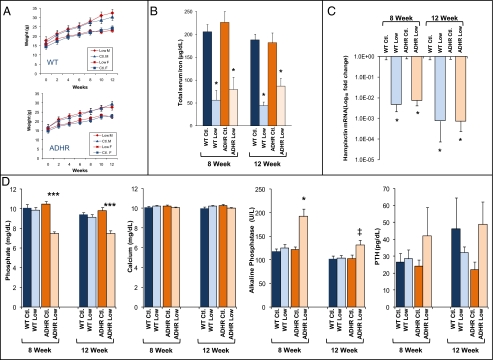
Fig. 1.
Mineral homeostasis in WT and ADHR mice. (A) Male (M) and female (F) WT and ADHR mice receiving control (Ctl; iron 45 mg/kg) and low-iron (Low; iron 0 mg/kg) diets gained weight continuously over the time course (n = 5–10 mice per group). (B) The low-iron diet regimen resulted in significantly reduced total serum iron in the WT and ADHR low-iron groups (*P < 0.01; n = at least 4 mice per group). (C) qPCR of liver RNA showed reduced Hamp mRNA concentrations at 8 and 12 wk in WT and ADHR mice provided low-iron diets (*P < 0.01; n = 5–10 mice per group). (D) Serum phosphate was reduced significantly at 8 and 12 wk of treatment in ADHR mice provided the low-iron diet compared with WT mice receiving control or low-iron diets and with ADHR mice provided the control diet. Serum calcium was unchanged. Serum alkaline phosphatase was elevated in ADHR mice on the low-iron diet. PTH was not different among groups (***P < 0.0001, **P < 0.001, *P < 0.01, ‡P < 0.05 vs. all groups; n = at least 12 mice per group).
To test the response of WT and ADHR mice to the low-iron diet, we examined serum iron concentrations as well as a gene that reliably adapts to iron status, the hepcidin antimicrobial peptide (Hamp), which inhibits iron absorption from the small intestine and is down-regulated during iron deficiency (18). Both WT and ADHR mice provided the low-iron diet for 8 and 12 wk had significantly reduced total serum iron concentrations compared with mice receiving the control diet (P < 0.01) (Fig. 1B). The mice receiving the low-iron diets also displayed an appropriate decrease in liver hepcidin mRNA (P < 0.01; Fig. 1C). Furthermore, WT and ADHR mice receiving the low-iron diet showed changes consistent with iron-deficient anemia in a complete blood cell count (CBC), including significantly reduced mean corpuscular volume, mean corpuscular hemoglobin, and increased red blood cell (RBC) distribution width, but no change in total RBC number (Table S1). The markers of iron status changed similarly in WT and ADHR mice, indicating that the Fgf23 R176Q-ADHR allele did not markedly alter the normal physiological responses to changes in dietary iron.
Next, to test the effects of iron deficiency in the context of the ADHR R176Q-Fgf23 allele, serum biochemistries were evaluated with regard to phosphate metabolism. At 8 wk serum phosphate concentrations did not differ in WT and ADHR mice receiving the control diet and WT mice fed the low-iron diet. In contrast to these groups, the ADHR mice provided the low-iron diet were significantly hypophosphatemic (P < 0.0001 for diet effect in ADHR and WT mice; Fig. 1D), and the hypophosphatemia persisted at 12 wk (P < 0.001) (Fig. 1D). Serum calcium levels did not differ among the groups (Fig. 1D). The ADHR mice receiving the low-iron diet had significant increases in alkaline phosphatase (P < 0.01 for diet effect in ADHR and WT mice; Fig. 1D), and alkaline phosphatase remained modestly elevated at 12 wk (P < 0.05; Fig. 1D). There were no significant differences in parathyroid hormone (PTH) at 8 and 12 wk across genotypes and diets (Fig. 1D). Serum creatinine was within the normal range (0.2–0.9 mg/dL) (8 wk: WT control/low-iron diet: 0.39 ± 0.04/0.4 ± 0.03; ADHR control/low-iron diet: 0.30 ± 0.2/0.29 ± 0.02; 12 wk: WT control/low-iron diet: 0.37 ± 0.04/0.39 ± 0.03; ADHR control/low-iron diet: 0.39 ± 0.05/0.42 ± 0.03). Thus, mice carrying R176Q-Fgf23 ADHR mutations and receiving the low-iron diet expressed a biochemical phenotype that paralleled the clinical manifestations of late-onset ADHR patients (1, 11).
Fgf23 Production and Processing in ADHR.
Fgf23 can be detected in the serum as intact, full-length Fgf23 or as circulating N- and C-terminal fragments that arise from proteolytic cleavage of the intact hormone at the 176RXXR179/S180 SPC site (3, 4). To test the regulation of circulating Fgf23 protein following the reduction of iron status in the ADHR mice, we used two serum assays, an “Intact Fgf23” ELISA that binds whole-molecule Fgf23 and a “C-terminal Fgf23” ELISA that recognizes the full-length protein and Fgf23 proteolytic fragments C-terminal to the Fgf23 176RXXR179/ S180 SPC site. To test for changes in serum Fgf23, subsets of WT and ADHR mice were assessed at 6 wk by tail bleed and after mice were killed at 8 and 12 wk.
Intact Fgf23.
Although there was a common age-dependent increase, serum intact Fgf23 did not differ by diet in WT mice (Fig. 2A). In contrast, at 6 wk the proportion of abnormally high intact Fgf23 (>161 pg/mL, 2.5 SD above the mean in the WT mice on the control diet) differed among the four groups (P < 0.05): Intact Fgf23 was abnormally high in 9 of 44 ADHR mice receiving the low-iron diet (cohort mean: 209.2 ± 59.1 pg/mL), in 2 of 31 ADHR mice on the control diet (cohort mean: 87.1 ± 7.9 pg/mL), and in 1 of 27 WT mice receiving the low-iron diet (cohort mean: 92.1 ± 7.8 pg/mL) but was not elevated in any of the 22 WT mice on the control diet (cohort mean: 78.7 ± 7 pg/mL). At 8 wk, some of the ADHR mice on the low-iron diet also manifested elevated intact Fgf23 (1,848, 1,770, 231, and 432 pg/mL; up to 22-fold increase vs. control), whereas no WT mice had values higher than twofold of the mean in these cohorts (n = 43). At 12 wk, serum intact Fgf23 levels were significantly lower in ADHR mice receiving the low-iron diet than in the other groups (P < 0.01), suggesting either down-regulation or altered Fgf23 processing during prolonged iron deficiency and hypophosphatemia (Fig. 2A).
Fig. 2.
Fgf23 following dietary intervention. (A) Intact Fgf23: WT mice receiving control or low-iron diets showed no differences between groups in intact Fgf23 at all time points. Fgf23 was elevated at 6 wk and reduced at 12 wk (*P < 0.01) in the ADHR mice on the low-iron diet. At 6 wk, the percentage of mice with intact Fgf23 concentrations >2.5 SD the WT control diet group mean is listed above the intact Fgf23 concentration for each cohort (‡P < 0.05 for ADHR mice on the low-iron diet v. all groups). (B) C-terminal Fgf23: C-terminal Fgf23 ELISA demonstrated significantly elevated Fgf23 in all low-iron groups (***P < 0.0001). (C) Immunoprecipitation and Western blot analysis of Fgf23 from bone lysates at 12 wk revealed that, compared with the largely intact Fgf23 protein produced by mice receiving the control diet, the low-iron diet resulted in increased immunoreactive Fgf23 proteolytic fragments in both WT and ADHR mice (arrows and bracket). Actin in the starting lysates was used as the loading control, and the negative control with no primary antibody (Neg. Ab) resulted in no immunoreactive protein. Molecular mass markers (M) are shown in kDa. (D) (Left) The intact/C-terminal Fgf23 concentrations showed a positive association in WT and ADHR mice receiving the control diet (P < 0.0001). (Right) For the low-iron diet intact did not change with C-terminal Fgf23 in WT mice, but ADHR mice had a positive correlation (P < 0.001 for difference in slopes). (E) Fgf23 transcripts were measured in total RNA from femur/tibia by qPCR. Transcript levels were not different between WT and ADHR mice receiving the control diet for 8 or 12 wk. In mice provided the low-iron diet, Fgf23 mRNA was significantly increased in WT and ADHR mice at 8 and 12 wk [**P < 0.001; ‡P < 0.05 vs. WT mice on the low-iron diet; n = 6–14 mice (mixed sexes) per group].
C-terminal Fgf23.
To test whether iron status was associated with Fgf23 proteolytic processing, we measured circulating Fgf23 using the C-terminal ELISA. C-terminal Fgf23 concentrations were three- to sixfold higher on low-iron diets than on control diets for both WT and ADHR mice (P < 0.0001; Fig. 2B). Immunoprecipitation of Fgf23 protein from bone lysates harvested at 12 wk qualitatively revealed that only intact 32-kD Fgf23 was detectable in the WT and ADHR mice receiving the control diets (Fig. 2C). In contrast, the WT and ADHR mice receiving the low-iron diets had increased immunoreactive Fgf23 and the notable appearance of lower-molecular-weight Fgf23 protein species (arrows and bracket in Fig. 2C).
We next examined the relationship between circulating intact and C-terminal Fgf23 concentrations at 8 wk. The WT and ADHR mice on the control diet showed a significant (P < 0.0001) positive relationship between serum intact and C-terminal Fgf23 (Fig. 2D, Left). Interestingly, the serum intact Fgf23 did not change with C-terminal Fgf23 in the WT mice receiving the low-iron diet (Fig. 2D, Right) but increased significantly in ADHR mice (P < 0.001 for difference in slopes) (Fig. 2D, Right). This relationship was consistent with a reduced ability of the ADHR mice to cleave the R176Q-Fgf23 protein species and recognition by the C-terminal ELISA of a greater proportion of circulating intact Fgf23 during iron deficiency.
In humans, the ADHR mutation presents as a dominant phenotype; therefore we also tested the impact of iron deficiency in heterozygous (Het) ADHR mice. The Het animals receiving the low-iron diet had increased C-terminal Fgf23 after 8 and 12 wk of treatment (P < 0.0001; Table S2), normocalcemia with modestly reduced serum phosphate (P < 0.01), and increased serum alkaline phosphatase (P < 0.01). Although Het mice carried both WT and ADHR alleles, intact Fgf23 was significantly reduced at 8 and 12 wk (suppressed 36% and 42%, respectively; P < 0.01), possibly in response to the prevailing hypophosphatemia. The biochemical manifestations were not as severe in Het mice as in homozygous R176Q-Fgf23 ADHR mice, suggesting a potential gene dosage effect of the mutant allele.
To test the mechanisms for the increased serum Fgf23 during iron deficiency, Fgf23 mRNA concentrations in femur were examined by quantitative PCR (qPCR). At 8 wk, WT and ADHR mice receiving the low-iron diet had significant increases in Fgf23 mRNA, by 3.2- and sixfold (P < 0.001), respectively (Fig. 2E). Fgf23 mRNA levels were increased further at 12 wk in WT mice on the low-iron diet (P < 0.001), and the elevation was even higher in ADHR mice on the low-iron diet (P < 0.05; Fig. 2E).
ADHR Skeletal Mineralization Phenotype.
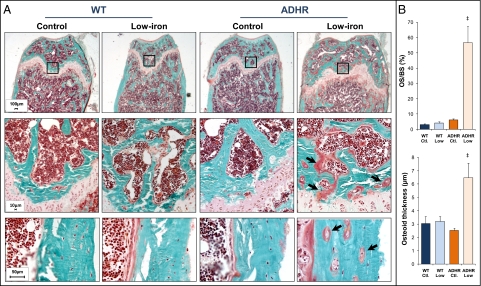
Although ADHR patients develop rickets and osteomalacia (1), the phenotype of the ADHR skeleton remains largely unknown. Histological analysis of the distal femur using Goldner's trichrome staining following 8 wk of treatment demonstrated that the WT mice on control and low-iron diets and ADHR mice on the control diet showed normal bone, as assessed by measures of osteoid surface and thickness (Fig. 3A), consistent with normophosphatemia in these groups. In contrast, ADHR mice on the low-iron diet had extensive osteomalacia in the cancellous bone (Fig. 3A). Upon analysis of cortical bone proximal to the metaphysis in ADHR mice on the low-iron diet, we detected heterogeneous populations of osteocytes: A portion of the cells appeared as osteocytic lesions, surrounded by appreciable areas of osteoid (arrows in Fig. 3A). These cells were adjacent to areas containing osteocytes that were indistinguishable from those in WT mice receiving either control or low-iron diets and in ADHR mice receiving the control diet (Fig. 3A). Quantification of the defective mineralization revealed that ADHR mice receiving the low-iron diet had a ninefold increase in the percent of osteoid surface/bone surface (OS/BS) compared with WT and ADHR mice provided the control diet and WT mice receiving the low-iron diet (WT control diet/WT low-iron diet/ADHR control diet: 6.17 ± 0.81%; ADHR low-iron diet: 56.62 ± 10.71%; P < 0.05 vs. all groups) (Fig. 3B), as well as increased osteoid thickness (P < 0.05) (Fig. 3B).
Fig. 3.
ADHR skeletal phenotype. (A) (Top) Goldner's staining of femur distal metaphysis revealed that WT mice receiving the control and low-iron diets and ADHR mice given the control diet had normal mineralization at 8 wk of treatment. ADHR mice receiving the low-iron diet had marked osteomalacia (3× magnification). (Middle) Enlarged images (20× magnification) of boxed areas in the top row demonstrated areas of intense osteomalacia in the ADHR low-iron group (arrows). (Bottom) Analysis of cortical bone demonstrated areas of osteoid surrounding osteocytes (arrows) that were adjacent to normal-appearing cells in the ADHR mice provided the low-iron diet (20× magnification). (B) Quantitative histomorphometric analyses demonstrated significantly increased osteoid surface/bone surface percent (OS/BS%) as well as increased osteoid thickness (‡P < 0.05 vs. all groups; n = 3–6 mice per group).
Renal Alterations in ADHR.
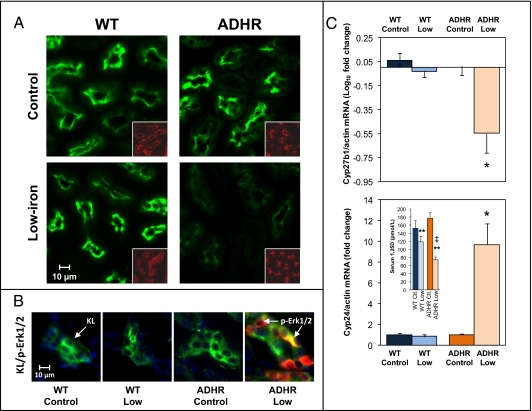
To understand the molecular events that resulted in the hypophosphatemic phenotype observed in the ADHR mice, expression of genes regulated by Fgf23 was examined at 8 wk. There were no differences in the expression of the type IIa sodium-phosphate cotransporter (Npt2a) protein between WT and ADHR mice receiving the control diet and WT mice on the low-iron diet. However, a marked reduction in Npt2a expression was observed in ADHR mice on the low-iron diet (Fig. 4A). Na/H exchange regulatory factor-1 (Nherf1), which delineates the apical membrane, showed similar expression across diets and genotypes (Fig. 4A, Inset). We previously determined that, when delivered in vivo, Fgf23 initiates phospho-Erk1/2 (p-Erk1/2) signaling within the kidney distal convoluted tubule, the primary site of renal α-Klotho expression (19, 20). Using similar analysis of fixed kidney sections from control diet and low-iron diet cohorts, only the ADHR mice receiving the low-iron diet showed strong p-Erk1/2 staining that localized with Klotho (Fig. 4B).
Fig. 4.
Molecular manifestations of ADHR in the kidney. (A) Immunofluorescent analyses on fixed kidney sections revealed that at 8 wk of dietary treatment the ADHR mice on the low-iron diet (Lower Right; n = 4–6 mice per group) had detectable but reduced Npt2a (green) protein compared with WT mice receiving the low-iron diet (Lower Left; n = 4–6 mice per group). The apical membrane control Nherf1 (red; Insets) was similar in sections from the same animals. (B) WT mice provided control or low-iron diets and ADHR mice receiving the control diet showed no kidney p-Erk1/2 but were positive for Klotho (KL) (green), whereas ADHR mice on the low-iron diet showed increased p-Erk1/2 activity (red) that localized with Klotho. (C) qPCR analyses demonstrated suppression of Cyp27b1 (Upper) and increased expression of Cyp24 (Lower) mRNAs in the ADHR low-iron cohort compared with the other groups. For Cyp27b1, the horizontal line indicates the normalized mean of WT mice on the control diet (*P < 0.01; n = at least 10 mice per group). (Lower Inset) Serum 1,25D concentrations at 8 wk of treatment (**P < 0.001 vs. mice on the control diet; ‡P < 0.05 vs. WT mice on the low-iron diet; n = 6–9 mice per group).
Fgf23 is known to reduce circulating 1,25D concentrations through transcriptional repression of the kidney anabolic enzyme vitamin D 1-α-hydroxylase (Cyp27b1) and elevation of the catabolic vitamin D 24-hydroxylase (Cyp24) (7). At 8 wk Cyp27b1 mRNA was 72.5 ± 8.9% reduced in ADHR mice receiving the low-iron diet (P < 0.01; Fig. 4C, Upper). Cyp24 mRNA was increased by 10.8 ± 2.13-fold in ADHR mice on the low-iron diet (P < 0.01) Fig. 4C, Lower). Levels of Cyp27b1 and Cyp24 were not significantly decreased or increased, respectively, in WT mice on either diet (Fig. 4C). Mice on the low-iron diet had reduced serum 1,25D (ADHR: 58%, WT: 23%; P < 0.001), and the reduction was significantly greater in ADHR mice (P < 0.05 vs. WT low-iron diet) (Fig. 4C; Lower Inset).
Isolation of Fgf23 Regulation in Vitro.
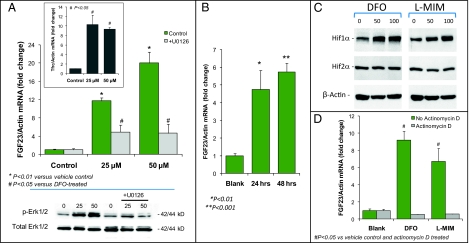
The osteoblastic cell line UMR-106 expresses transferrin receptor-1 (TfRc1) and maintains saturable transferrin binding (21, 22). To test for direct regulation of Fgf23 during iron deficiency in bone cells, UMR-106 cells were treated with the therapeutic iron chelator deferoxamine (DFO) for 48 h. As expected, TfRc1 mRNA was increased 10-fold by DFO treatment, confirming the development of low cellular iron (P < 0.05; Fig. 5A, Inset). Fgf23 mRNA was increased significantly, 12- and 20-fold, with 25 and 50 μM DFO treatment, respectively (P < 0.01 each vs. vehicle control; Fig. 5A Upper); these results are consistent with our findings on Fgf23 mRNA during iron deficiency in vivo (Fig. 2E). DFO has been shown previously to increase Mapk activity (23). To test the role of Mapk in the increase of Fgf23 production during iron deficiency, UMR-106 cells were pretreated with the Mek1 inhibitor U0126 (10 μM). Mitogen-activated protein kinase kinase-1 (MEK1) inhibition blocked the DFO-dependent rise in Fgf23 mRNA by more than 70% (U0126 data are shown in Fig. 5A; P < 0.05). DFO (25 and 50 μM, 48 h) significantly increased p-Erk1/2, and U0126 blunted this effect in UMR-106 cells (Fig. 5A, Lower). To examine the molecular mechanisms for the elevated Fgf23 mRNA under low-iron conditions, cells were treated with l-mimosine (l-MIM; 50 μM), a specific egg-laying-defective nine (EGLN) prolyl hydroxylase inhibitor that activates the hypoxia-induced factor (HIF) transcription factors (24), key metabolic sensors that control genes in response to iron deficiency in bone and other tissues (25). Importantly, l-MIM produced a significant, time-dependent increase in Fgf23 mRNA (P < 0.01; Fig. 5B), and both DFO and l-MIM treatment resulted in stabilization of HIF1α protein but not HIF2α in UMR-106 cells (Fig. 5C). To test whether the DFO- and l-MIM–dependent elevations in Fgf23 mRNA were caused by increased transcription, cells were pretreated with actinomycin D (0.1 μg/mL; 30 min), followed by DFO or l-MIM for 24 h. Actinomycin D ablated Fgf23 transcriptional activity with these agents (P < 0.05; Fig. 5D).
Fig. 5.
Regulation of Fgf23 in vitro. (A) (Upper) UMR-106 cells were treated with 25 and 50 μM DFO, resulting at 48 h in dose-dependent increases in Fgf23 mRNA of 12- and 20-fold, respectively (*P < 0.01 vs. vehicle). The Mek inhibitor U0126 (10 μM) blunted the increase in Fgf23 mRNA by more than 70% (#P < 0.05 vs. DFO treatment). (Inset) As a positive control for iron reduction in the cultures by chelation, TfRc1 mRNA was increased over the same treatment course (#P < 0.05 vs. vehicle). (Lower) Western blot analyses of UMR-106 cell lysates demonstrated that p-Erk1/2 was increased at 48 h following 25- or 50-μM DFO treatment. p-Erk1/2 activity was inhibited by U0126 (10 μM); the control total-Erk1/2 was similar across treatments. (B) The EGLN inhibitor l-MIM (50 μM) significantly increased Fgf23 mRNA (*P < 0.01; **P < 0.001). (C) DFO and l-MIM (50 or 100 μM) were associated with stabilized HIF1α but not HIF2α protein. (D) Transcriptional activation of Fgf23 was examined with actinomycin D (0.1 μg/mL) before DFO or l-MIM treatment (24 h). Actinomycin D treatment abolished the Fgf23 mRNA increases (#P < 0.05 vs. both vehicle control and actinomycin D treatment).
In summary, these studies determined that Fgf23 mRNA is stimulated robustly in bone during iron deficiency in mice. WT mice can counteract this response by proteolytically cleaving excess hormone at a secondary regulatory step to maintain stable serum intact Fgf23 and normal serum phosphate. Our results show that this proteolytic step is compromised in ADHR mice carrying the R176Q-Fgf23 mutation, leading to increased serum intact Fgf23, to alterations in genes controlling phosphate reabsorption and 1,25D production, and to hypophosphatemic bone disease. Collectively, these findings support the notion that the Fgf23 176RXXR179/S180 SPC site is central to controlling normal blood concentrations of bioactive hormone. Furthermore, this work demonstrates that, in the context of an ADHR R176Q-Fgf23 mutation in mice, the induction of Fgf23 mRNA transcription during iron deficiency has a stronger biological effect than the combined suppressive actions of hypophosphatemia and low 1,25D; these results are consistent with the inability of ADHR patients to reduce serum FGF23 during late-onset, active disease.
Discussion
Here we provide evidence that late-onset ADHR, unlike other disorders associated with elevated FGF23, is the product of gene–environment interactions, whereby the presence of an ADHR mutation and iron deficiency cause the disease phenotype. Decreased serum total iron concentrations have been associated with elevated FGF23 in ADHR patients (14), and our data support the concept that during fluctuations in serum iron, normal individuals can control excess FGF23 through proteolytic processing via a secondary regulation that maintains the concentration of serum intact FGF23. This identified regulatory step is likely compromised in ADHR patients because of the mutations within the FGF23 R176XXR179/S180 SPC cleavage site, which disrupt normal proteolytic control of intact Fgf23 protein. When the biological iron load was reduced through dietary intervention in ADHR mice (Fig. 1 and Table S1), these animals demonstrated the hallmark biochemical alterations associated with increased intact Fgf23 secretion, including hypophosphatemia and inappropriately reduced serum 1,25D concentrations, increased serum alkaline phosphatase, and no significant differences in serum PTH (Fig. 1). These biochemistries are similar to observations from other mouse models associated with increased circulating Fgf23, including the Hyp mouse (26) and the ARHR model, the Dmp1-null mouse (8). In contrast to these models, which show a fully penetrant phenotype from birth, similar to XLH and ARHR patients (8, 9, 27), the ADHR disease phenotype associated with the R176Q-Fgf23 allele was induced through a stimulus of iron deficiency. Several observations support the notion that the molecular etiology of the active ADHR disease state is not solely the result of differences in the ability of WT or ADHR mice to control iron status. In this regard, the serum iron concentrations were similar in both WT and ADHR mice receiving the control or low-iron diets at 8 and 12 wk (Fig. 1), and we observed the appropriate biological responses to iron deficiency in terms of genes that regulate iron homeostasis and the development of iron deficiency anemia for both genotypes (Fig. 1 and Table S1).
Excess FGF23 is associated with hypophosphatemia in ADHR and other hypophosphatemic patients (11, 28–30) and in murine models (8, 26, 31). We detected a significant increase in serum intact Fgf23 in the ADHR mice receiving the low-iron diet at 6 wk and in a proportion of the ADHR mice at 8 wk. The normal or increased levels of intact Fgf23 at 8 wk can be considered “inappropriate,” given the degree of hypophosphatemia (Fig. 1), consistent with the stimulation of Fgf23 by a process external to the classical Fgf23–phosphate endocrine axis, namely iron deficiency. In WT mice provided the low-iron diet, as shown by a combination of ELISAs that recognize intact and C-terminal Fgf23 (Fig. 2), we determined that these animals produced increased C-terminal fragments to maintain normal serum intact Fgf23 and serum phosphate (Fig. 1) in the face of elevated Fgf23 mRNA (Fig. 2E). The inherently different physical-chemical natures of the two assays and our current lack of knowledge of the exact Fgf23 C-terminal fragment composition and turnover in blood make it difficult to calculate the absolute amount of C-terminal FGF23. We therefore examined matched controls for each diet, time point, and genotype to provide the appropriate relative comparisons within the intact and C-terminal ELISA results; taken together, these data strongly support the stabilizing influence of the ADHR mutations in vivo. In addition, Western blot analyses showed increased Fgf23 immunoreactive protein, as well as lower-molecular-weight species in bone from the iron-deficient mice (Fig. 2C). These findings are in accord with a cellular mechanism of first producing intact Fgf23 and then subsequent proteolytic cleavage. Whether these lower-molecular-mass species represent forms of Fgf23 with differing O-glycosylation (4) and proteolytic cleavage fragments (including additional endoproteolytic cleavage sites beyond the known 176RXXR179/S180 SPC site) or combinations of these potential regulatory processes remains to be determined.
Previous studies indicated that injection of the Fgf23 N- and C-terminal proteolytic fragments had no effect on serum phosphate concentrations, whereas delivery of whole-molecule Fgf23 was capable of reducing serum phosphate (4). In agreement with this prior work, although serum C-terminal fragments were increased significantly in the WT and ADHR animals receiving the low-iron diet (Fig. 2), we detected no difference in the serum phosphate levels (Fig. 1D) or bone osteomalacia (Fig. 3) in WT animals receiving the control or low-iron diets or in ADHR animals provided control diets. At this time, we cannot rule out the possibility that R176Q-Fgf23 undergoes additional processing in vivo at domains other than the 176RXXR179/S180 SPC site or inactivation by other mechanisms during chronic Fgf23 mRNA up-regulation. It is likely that combinations of these effects may be present in some ADHR patients during disease onset and progression and might reflect the variable metabolic bone and biochemical phenotypes associated with this disorder (1). Although we determined that the R176XXR179/S180 SPC site in Fgf23 is necessary for regulating circulating intact Fgf23 concentrations (at least under low-iron conditions), the sensing mechanisms that communicate the biological conditions under which Fgf23 should be cleaved actively remain to be determined. The SPC enzyme responsible for FGF23 processing also is currently unknown; however, SPC2 and its cofactor 7B2 recently have been associated with FGF23 proteolysis (32). The fact that ADHR mice were hypophosphatemic with normal or suppressed serum intact Fgf23 following a low-iron diet (Fig. 2) could indicate that the combined actions of Fgf23 proteolysis and potential renal compensatory mechanisms at the level of its signaling complex cannot completely abolish Fgf23-dependent bioactivity. Whether this reduction in intact Fgf23 in the face of elevated Fgf23 mRNA is specific for iron-deficient states remains to be determined. Of significance, these observations in ADHR mice parallel those in a subset of hypophosphatemic ADHR patients who had inappropriately normal serum FGF23 (11). Further, the influence of other factors, including sex steroids, on iron deficiency-dependent Fgf23 expression and processing are unknown.
Mice up-regulate Npt2a (33) and Cyp27b1 and down-regulate Cyp24 (34) during diet-induced hypophosphatemia. In line with the known in vivo effects of Fgf23, we detected a marked decrease of Npt2a in ADHR mice on the low-iron diet (Fig. 4A) (31), with suppressed Cyp27b1 and elevated Cyp24 (Fig. 4C) corresponding to the inappropriately low 1,25D (Fig. 4C, Inset). We also detected a modest decrease in serum 1,25D in WT mice provided the low-iron diet at 8 wk (Fig. 4C, Inset). In WT mice this decrease coincided with a slight increase in serum intact Fgf23 at the same time point (Fig. 2A). Given that, when injected in vivo, Fgf23 affects 1,25D production more potently than serum phosphate production (35), this decrease could indicate a partial ”leakiness” of intact Fgf23 proteolytic processing in the face of significantly elevated Fgf23 mRNA (Fig. 2E). Consistent with the hypophosphatemia in the ADHR mice receiving the low-iron diet, these mice presented with marked osteomalacia and osteocytic lesions (Fig. 3), which also are apparent in bone from Hyp mice and XLH patients (36).
The endocrine relationships between serum iron, phosphate, 1,25D, and Fgf23 are likely complex. Of note, medications such as infusions of select iron-containing compounds increase serum FGF23 concentrations, with patients developing hypophosphatemia and decreased serum 1,25D (37–39). In ADHR patients, however, low serum iron was correlated with elevated serum FGF23 concentrations (14), and mice fed a low-iron diet up-regulate serum Fgf23 (Fig. 2B) and bone Fgf23 mRNA (Fig. 2E), a result that appears converse to the aforementioned case reports involving iron infusion. The physiological reasons for this divergence are currently unknown; however, it is noteworthy that in the clinical reports both saccharated ferric oxide and iron-polymaltose infusion caused an elevation of FGF23, but dextrin-iron infusions did not (39–41). Thus, the difference in the kinetics of iron release from these pharmacological preparations or a rapid increase of iron may affect FGF23 expression differently than the long-term, physiologic regulation of iron in humans during normal variations in puberty or pregnancy or than the changes in dietary iron intake over weeks in mice during alterations in dietary iron exposure. Collectively, our work suggests that the ADHR phenotype may be controlled by administration of some forms of iron.
Our in vitro results demonstrated that decreasing free iron by DFO chelation and the activation of the HIF1α transcription factor using the EGLN inhibitor l-MIM (Fig. 5) (24) resulted in increased Fgf23 mRNA (Fig. 5). Further, Fgf23 transcription following DFO and l-MIM treatment was abolished by actinomycin D, indicating that Fgf23 was up-regulated by these agents through enhanced transcription (Fig. 5). In support of HIF activation in vivo, hepcidin, a direct target of HIF activity (42), was suppressed markedly by a low-iron diet in both WT and ADHR mice (Fig. 1). These findings support a model whereby iron deficiency results in stimulation of HIF1α, leading to increased Fgf23 transcription; however, whether HIFs directly or indirectly control Fgf23 remains to be determined. The physiological basis for the control of Fgf23 by iron deficiency through HIF regulation is unclear currently, although there are known biological associations between iron and phosphate homeostasis. In this regard, iron is contained within the hydrophilic core of the carrier protein ferritin as ferric hydroxide-phosphate complexes. Kinetic studies show that the ferritin iron-loading process is 10-fold more efficient in the presence of phosphate ions than in the presence of other anions such as nitrate and carbonate (43); therefore maintaining overall phosphate balance through Fgf23 potentially could influence iron storage. Certainly, additional studies will be required to test and expand our understanding of these specific biological interactions.
In sum, our findings indicate that ADHR is a disease produced through gene–environment interactions. We demonstrated that Fgf23 mRNA is up-regulated in bone in mice during iron deficiency, and in the context of an ADHR R176Q-Fgf23 allele this biological situation leads to elevated intact Fgf23 and to hypophosphatemic bone disease. Further, although elevated in WT mice during iron deficiency, native Fgf23 protein is proteolytically cleaved to maintain stable serum intact Fgf23 and phosphate. These results provide a mechanistic link between iron and phosphate homeostasis that explains the clinical phenotype of ADHR and may reveal therapeutic targets for both rare and more common disorders of phosphate handling and biomineralization.
Materials and Methods
Animal Studies.
Animal studies were performed according to the Institutional Animal Care and Use Committee (IACUC) for Indiana University and comply with National Institutes of Health guidelines for the use of animals. All ES cell procedures and initial test breeding for ADHR knock-in animal production were undertaken with IACUC approval by the Indiana University Transgenic Facility according to their standard protocols.
Gene Targeting to Create the R176Q-Fgf23 ADHR Allele.
To derive a mouse model with control of an R176Q-Fgf23 ADHR mutant allele under the native Fgf23 5′ promoter and regulatory elements, a knock-in strategy was used. A targeting construct for homologous recombination at the Fgf23 locus was developed by isolating an 11-kb fragment of the murine 129/SvJ strain Fgf23 gene containing exons 2 and 3 as an Xba/SacI fragment. This Fgf23 129-strain clone (a generous gift of Beate Lanske, Harvard University, Cambridge, MA) was used to make an Fgf23-null mouse (44) and was subcloned into pBluescript(SK+) (pBSK). The neomycin resistance gene (Neor) flanked by LoxP sites was removed from the pBSK-PGKneoLox2 vector (the generous gift of Fred Dick, University of Ottawa, Ottawa) using an EcoRV-SmaI digest (New England Biolabs, Inc.) and was ligated in the forward direction into a Bstz17I (New England Biolabs, Inc) site in Fgf23 intron 2 (Fig. S1A).
The R176Q-Fgf23 ADHR mutation (from nt 528cgc530–528cag530) and a silent SacI site 3′ to the R176Q mutation (from nt 627gagctG632–627gagctC632) were directed to the AatII-SbfI fragment of Fgf23 exon 3 using PCR with complementary primers containing the mutated nucleotides in the center position and the high-fidelity polymerase PfU (Agilent Technologies). The silent SacI site provided the ability to test for recombination of the ADHR allele (Fig. S1A). The coding region of the targeting construct was sequenced to ensure no cloning gaps or mutations in the intron–exon junctions. Once sequenced, the construct also was submitted to in vitro Cre recombination using the 294-Cre Escherichia coli (stably integrated Cre; Clone #47071; American Type Tissue Collection) which resulted in successful removal of the floxed Neor cassette. Transfection of the construct into 129/SvJ-strain ES cells was performed by the Indiana University Transgenic Facility staff according to standard electroporation protocols. Recombinant cells were selected in G418, and DNA lysates (Gentra Systems) from resistant colonies were screened by Southern blot analysis using SacI digests and a 3′ external probe as well as a Neor probe (Fig. S1A), according to standard protocols. The ES cells showed correct homologous recombination for the R176Q-Fgf23 allele within an expected 1–2% efficiency (Fig. S1A).
After ES cell implantation and derivation of 129/SvJ/C57Bl6 chimeric mice, chimeric male mice with high ES cell contribution were backcrossed to C57BL/6 females; germ line transmission was predicted by coat color and confirmed by Southern blot analysis of genomic tail-clip DNA (Fig. S1A). The heterozygous ADHR mice carrying the Neor cassette (Fig. S1A) then were mated to the ubiquitously expressed EIIa-Cre transgenic mice (Jackson Laboratory) to remove the floxed Neor gene from Fgf23 intron 2 in vivo. The deletion of the Neor gene through this mating was confirmed by the fact that the PCR products spanning the intronic site carrying Neor migrate more slowly by electrophoresis than the same genomic position in WT DNA because of the presence of the remaining LoxP site (Fig. S1B) and also was confirmed by DNA sequencing. Genotyping was performed with PCR primers that span the LoxP site remaining from Cre recombination of the Neor gene and primers that span the ADHR-R176Q Fgf23 mutation (Fig. S1B). All studies were performed on WT and ADHR generation N10 mice backcrossed to C57Bl6.
Rodent Diets.
The experimental diets were obtained from Research Diets, Inc. The control diet was the standard AIN-76A base diet with the S10001 mineral mix containing iron at a concentration of 45 mg Fe/kg (carbonyl iron). The iron-deficient diet was the AIN-76A base diet and S10001 mineral mix with the iron removed (∼0.1 ppm iron final concentration). All diets contained customary 0.55% phosphorus final concentration, with low trace element casein (Avicel PH-101 Cellulose). Diets and water were introduced at weaning (age 4 wk) and were provided ad libitum throughout the study.
Serum Biochemistries.
Blood samples were collected by tail bleed according to approved protocols or by cardiac puncture when mice were killed. Routine serum biochemistries were determined by standard automated methods: Calcium (Ca), phosphate (Pi), alkaline phosphatase (AP), creatinine (Cr), and total serum iron (Fe) were measured using a COBAS MIRA Plus Chemistry Analyzer (Roche Diagnostics). Serum 1,25(OH)2 vitamin D was measured using an enzyme immunoassay (Immunodiagnostic Systems), and PTH was measured with a mouse intact PTH Kit (Immutopics International), according to the manufacturers’ instructions. A mouse CBC was performed on whole blood using a Mascot HemaVet950FS automated processor.
Serum Fgf23 Measurements.
Serum intact Fgf23 concentrations were assessed using a commercial ELISA according to the manufacturer's protocol (Kainos Laboratories Inc.). This two-site, monoclonal antibody ELISA has been shown previously to recognize rodent Fgf23. Serum Fgf23 also was measured using a rodent-specific C-terminal Fgf23 ELISA with capture and detection antibodies that bind to rodent Fgf23 protein 3′ C-terminal to the 176RXXR179/S180 SPC site, according to the manufacturer's specifications (Immutopics International). For the rodent-specific C-terminal Fgf23 kit, anti-peptide polyclonal antibodies were derived as follows: The capture antibodies recognize epitopes within mouse Fgf23 residues 186–206, and the detection antibodies recognize epitopes within mouse FGF23 residues 225–244.
Histology and Immunofluorescence.
After animals were killed, the kidneys and femur were removed and dissected free of connective tissue. Kidneys were prepared as previously described (19). Sections were probed with anti-mouse Npt2a (1:300; generous gift of Heini Murer, Jürg Biber, and Nati Hernando of the Institute of Physiology, University Zürich, Zurich), anti-mouse Klotho (1:5; Santa Cruz Biotechnology), anti–p-ERK1/2 (1:50; Cell Signaling Technology), or anti-Nherf1 (1:50; Sigma) followed by incubation with anti-rabbit and anti-rat (1:200) fluorescent secondary antibodies (Alexa Fluor; Invitrogen, Inc.). Slides were mounted in the presence of DAPI (Vector Laboratories) and were imaged with the same setting across groups using a Leica DM5000B fluorescent microscope (Leica Microsystems, Inc.) with a SPOT camera and analysis software (Diagnostic Instruments).
Femurs were removed and fixed in 4% paraformaldehyde. The distal femurs were embedded in methyl methacrylate, and midsagittal (4-μm) sections of cancellous bone from the distal femur were cut using a microtome (2050 Supercut; Reichert-Jung). The sections were treated with Goldner's stain according to established protocols and were examined blinded to mouse diet and genotype. Histomorphometric measures were obtained at the distal femur metaphysis using standard protocols (45) and a semiautomatic analysis system (Bioquant OSTEO; Bioquant Image Analysis Co.) attached to a Nikon microscope. A defined region of interest (∼0.5 mm from the growth plate and encompassing 3–4 mm2) was used to determine bone surface, osteoid surface, and osteoid thickness. The terminology and units used are those recommended by the Histomorphometry Nomenclature Committee of the American Society for Bone and Mineral Research (46).
RNA Preparation and Quantitative RT-PCR.
Kidney, liver, and bone were harvested and homogenized in 1 mL of TRIzol reagent (Invitrogen, Inc.) according to the manufacturer's protocol using a TissueTearor rotor-stator (Biospec Products, Inc.). Total RNA from UMR-106 cellular lysates was prepared using the RNeasy Kit (Qiagen, Inc.). RNA samples were tested with intron-spanning primers specific for mouse vitamin D 24-hydroxylase (Cyp24), mouse vitamin D 1-α-hydroxylase (Cyp27b1), mouse and rat Fgf23, mouse Hamp, and rat TfRc1. Mouse or rat β-actin was used as an internal control. All qPCR primers and probes were purchased as preoptimized reagents (Applied Biosystems, Inc.) or were produced in-house, and sequences are available upon request.
The TaqMan One-Step RT-PCR kit was used to perform qPCR. PCR conditions for all experiments were 30 min at 48 °C, 10 min at 95 °C, followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. The data were collected and analyzed by the 7500 Real Time PCR system and software (Applied Biosystems). All primer sets were tested for specific amplification of mRNA by parallel analyses of controls that included omitting RT or template and resulted in no fluorescent signal detection. Each RNA sample was analyzed in at least triplicate, and each in vitro experiment was performed independently at least three times. The 2-ΔΔCT method described by Livak and Schmittgen (47) was used to analyze the data.
Cell Culture.
UMR-106 cells (American Type Culture Collection) were cultured in d-MEM/F-12 (Invitrogen) supplemented with 10% FBS (HyClone; Thermo-Fisher, Inc.) and 25 mM penicillin-streptomycin at 37 °C and 5% CO2. For bioactivity assays, cells (1.5 × 105) were seeded in 24-well plates. Cells were iron depleted with the iron chelator DFO (25–50 μM) for 24–48 h. These DFO concentrations are within a range known to have biological effects in UMR-106 cells (21, 22). Cells also were pretreated with actinomycin D as specified. To test Mapk signaling, cells were pretreated with the MEK inhibitor U0126 (10 μM) 1 h before DFO addition. The EGLN inhibitor l-mimosine (25–100 μM) was used to activate HIFs.
Western Blot Analysis.
UMR-106 cells were lysed with 100 μL 1× Lysis buffer (Cell Signaling Technologies, Inc.) with 1 μg/mL 4-(2-aminomethyl)benzenesulfonyl fluoride hydrochloride (AEBSF) protease inhibitor (Sigma-Aldrich, Inc.). Cell lysate protein concentrations were determined with the Better Bradford Kit (Thermo-Fisher Scientific) according to the manufacturer's instructions. Western blot analysis was performed as previously described (48) with 50 μg UMR-106 cellular lysates. The blots were incubated with primary antibodies [anti–p-ERK1/2 (Cell Signaling Technologies, Inc.); anti–total-ERK (Promega); anti-HIF1α (NB100-134), and anti-HIF2α (NB100-122) (Novus Biologicals)] and then were incubated with the appropriate secondary antibody at 1:3,000 [anti–rabbit-HRP (Bio-Rad, Inc.)] at 1:3,000. Detection was performed using the ECL Plus Western Blotting Detection Reagents (Amersham-GE Healthcare) and X-Omat film (Eastman-Kodak Co.).
Fgf23 Immunoprecipitation.
Femurs and tibias from treated mice were cleaned of connective tissue and homogenized in 1 mL 1× lysis buffer (Cell Signaling Technologies, Inc.) with 1 μg/mL AEBSF. The solid particles were removed by centrifugation at 15,000 × g at 4 °C, and the supernatant was stored at −80 °C until use. Fgf23 was precipitated from 1,600 mg/mL of bone lysate with 125 ng of the goat anti-mouse Fgf23 capture and detection antibodies from the C-terminal mouse Fgf23 ELISA Kit (see above; purified antibodies were generous gifts of Immutopics International, Inc.) for 4 h at room temperature. The IgG complexes were captured using Protein A&G resin binding (Pierce, Inc.) and washed three times in 1× lysis buffer. The precipitants were solubilized in 2× Leammli sample buffer (Bio-Rad, Inc.) at 95 °C before gel electrophoresis and Western blotting, which were performed as outlined above. The Western blots of precipitated Fgf23 were incubated with the detection antibody from the mouse C-terminal Fgf23 ELISA, a biotin-conjugated anti-mouse Fgf23 (2 μg/mL; generous gift of Immutopics International), followed by incubation with avidin-HRP (1:1,000; Bio-Rad, Inc.) and ECL detection. Anti-actin (1:1,000; Santa Cruz Biotechnology, Inc.) was used as the internal lysate control for gel loading.
Statistical Analysis.
All analyses were performed separately for male and female mice, and if there was no interaction between the primary predictor variable and sex, the data were combined, and sex was included as a covariate when appropriate. Subsequent analyses were performed for each outcome of interest including serum biochemistries and expression levels of key genes, log-transformed to normalize the distribution if necessary. Two-way ANOVA was used to test for genotype–diet interaction at each time point on each outcome, assuming unequal within-group variances for expression data. Repeated-measures ANOVA was used to test for between-group differences in serial FGF23 measurements. Significance interactions were followed by pairwise comparisons between diets within genotype using t tests except for FGF23 measurements with extreme bimodal distributions at weeks 6 and 8. In the latter case the outcome was dichotomized into abnormally high (>2.5 SD above the mean of the WT mice on control diet) or not abnormally high, and Fisher's exact test was used to test for differences in proportions between groups. Data are presented as SEM.
Supplementary Material
Acknowledgments
We thank Dr. Fred Dick, University of Ottawa, for consultation during targeting vector analysis; Anthony Acton, Jr., Ronald McClintock, and Dr. Keith Condon for assistance with sample processing; Dr. Xin Zhang for microscope use; and Jeff Lavigne and Richard Zahradnik from Immutopics International, Inc. for generously providing Fgf23 antibody reagents and C-terminal Fgf23 ELISA kits. This work was supported by National Institutes of Health Grants DK063934 and an American Recovery and Reinvestment Act supplement under the same grant number (to K.E.W.), DK054111 (to J.C.F.), R21 HL092524 (to R.J.C.), HL104867 (C.B.G.), and NS050227 (to R.V.). Support also was provided by the Indiana Genomics Initiative of Indiana University, which is supported in part by the Lilly Endowment, Inc.; by a National Kidney Foundation postdoctoral fellowship (E.G.F.); by the Indiana University Medical Student Affairs Program in Academic Medicine. This project also was supported, in part, by the Indiana Clinical and Translational Sciences Institute, which is funded in part by Grant RR025761 from the National Institutes of Health, National Center for Research Resources, Clinical and Translational Sciences Award.
Footnotes
Conflict of interest statement: K.E.W. receives royalties for licensing the FGF23 gene to Kyowa Hakko Kirin Co. Ltd., and M.P. is involved in a clinical trial with Kyowa Hakko Kirin Co. Ltd.
This article is a PNAS Direct Submission.
See Author Summary on page 18587.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110905108/-/DCSupplemental.
References
- 1.Econs MJ, McEnery PT. Autosomal dominant hypophosphatemic rickets/osteomalacia: Clinical characterization of a novel renal phosphate-wasting disorder. J Clin Endocrinol Metab. 1997;82:674–681. doi: 10.1210/jcem.82.2.3765. [DOI] [PubMed] [Google Scholar]
- 2.ADHR Consortium Autosomal dominant hypophosphataemic rickets is associated with mutations in FGF23. Nat Genet. 2000;26:345–348. doi: 10.1038/81664. [DOI] [PubMed] [Google Scholar]
- 3.White KE, et al. Autosomal-dominant hypophosphatemic rickets (ADHR) mutations stabilize FGF-23. Kidney Int. 2001;60:2079–2086. doi: 10.1046/j.1523-1755.2001.00064.x. [DOI] [PubMed] [Google Scholar]
- 4.Shimada T, et al. Mutant FGF-23 responsible for autosomal dominant hypophosphatemic rickets is resistant to proteolytic cleavage and causes hypophosphatemia in vivo. Endocrinology. 2002;143:3179–3182. doi: 10.1210/endo.143.8.8795. [DOI] [PubMed] [Google Scholar]
- 5.Urakawa I, et al. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature. 2006;444:770–774. doi: 10.1038/nature05315. [DOI] [PubMed] [Google Scholar]
- 6.Kurosu H, et al. Regulation of fibroblast growth factor-23 signaling by klotho. J Biol Chem. 2006;281:6120–6123. doi: 10.1074/jbc.C500457200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimada T, et al. Cloning and characterization of FGF23 as a causative factor of tumor-induced osteomalacia. Proc Natl Acad Sci USA. 2001;98:6500–6505. doi: 10.1073/pnas.101545198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng JQ, et al. Loss of DMP1 causes rickets and osteomalacia and identifies a role for osteocytes in mineral metabolism. Nat Genet. 2006;38:1310–1315. doi: 10.1038/ng1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorenz-Depiereux B, et al. DMP1 mutations in autosomal recessive hypophosphatemia implicate a bone matrix protein in the regulation of phosphate homeostasis. Nat Genet. 2006;38:1248–1250. doi: 10.1038/ng1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bianchine JW, Stambler AA, Harrison HE. Familial hypophosphatemic rickets showing autosomal dominant inheritance. Birth Defects Orig Artic Ser. 1971;7:287–295. [PubMed] [Google Scholar]
- 11.Imel EA, Hui SL, Econs MJ. FGF23 concentrations vary with disease status in autosomal dominant hypophosphatemic rickets. J Bone Miner Res. 2007;22:520–526. doi: 10.1359/jbmr.070107. [DOI] [PubMed] [Google Scholar]
- 12.Breymann C, Honegger C, Holzgreve W, Surbek D. Diagnosis and treatment of iron-deficiency anaemia during pregnancy and postpartum. Arch Gynecol Obstet. 2010;282:577–580. doi: 10.1007/s00404-010-1532-z. [DOI] [PubMed] [Google Scholar]
- 13.Gribaa M, et al. An autosomal dominant hypophosphatemic rickets phenotype in a Tunisian family caused by a new FGF23 missense mutation. J Bone Miner Metab. 2010;28:111–115. doi: 10.1007/s00774-009-0111-5. [DOI] [PubMed] [Google Scholar]
- 14.Imel EA, et al. Iron modifies plasma FGF23 differently in autosomal dominant hypophosphatemic rickets and healthy humans. J Clin Endocrinol Metab. 2011 doi: 10.1210/jc.2011-1239. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durham BH, Joseph F, Bailey LM, Fraser WD. The association of circulating ferritin with serum concentrations of fibroblast growth factor-23 measured by three commercial assays. Ann Clin Biochem. 2007;44:463–466. doi: 10.1258/000456307781646102. [DOI] [PubMed] [Google Scholar]
- 16.Ohira Y, Gill SL. Effects of dietary iron deficiency on muscle fiber characteristics and whole-body distribution of hemoglobin in mice. J Nutr. 1983;113:1811–1818. doi: 10.1093/jn/113.9.1811. [DOI] [PubMed] [Google Scholar]
- 17.Tompkins GR, O'Dell NL, Bryson IT, Pennington CB. The effects of dietary ferric iron and iron deprivation on the bacterial composition of the mouse intestine. Curr Microbiol. 2001;43:38–42. doi: 10.1007/s002840010257. [DOI] [PubMed] [Google Scholar]
- 18.Nemeth E, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 19.Farrow EG, Davis SI, Summers LJ, White KE. Initial FGF23-mediated signaling occurs in the distal convoluted tubule. J Am Soc Nephrol. 2009;20:955–960. doi: 10.1681/ASN.2008070783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farrow E, et al. Altered renal FGF23-mediated activity involving MAPK and Wnt: Effects of the Hyp mutation. J Endocrinol. 2010;207:67–75. doi: 10.1677/JOE-10-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasai K, Hori MT, Goodman WG. Characterization of the transferrin receptor in UMR-106-01 osteoblast-like cells. Endocrinology. 1990;126:1742–1749. doi: 10.1210/endo-126-3-1742. [DOI] [PubMed] [Google Scholar]
- 22.Kasai K, Hori MT, Goodman WG. Transferrin enhances the antiproliferative effect of aluminum on osteoblast-like cells. Am J Physiol. 1991;260:E537–E543. doi: 10.1152/ajpendo.1991.260.4.E537. [DOI] [PubMed] [Google Scholar]
- 23.Kim BS, et al. Involvement of p38 MAP kinase during iron chelator-mediated apoptotic cell death. Cell Immunol. 2002;220:96–106. doi: 10.1016/s0008-8749(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 24.Warnecke C, et al. Activation of the hypoxia-inducible factor-pathway and stimulation of angiogenesis by application of prolyl hydroxylase inhibitors. FASEB J. 2003;17:1186–1188. doi: 10.1096/fj.02-1062fje. [DOI] [PubMed] [Google Scholar]
- 25.Semenza GL, et al. Structural and functional analysis of hypoxia-inducible factor 1. Kidney Int. 1997;51:553–555. doi: 10.1038/ki.1997.77. [DOI] [PubMed] [Google Scholar]
- 26.Liu S, et al. Pathogenic role of Fgf23 in Hyp mice. Am J Physiol Endocrinol Metab. 2006;291:E38–E49. doi: 10.1152/ajpendo.00008.2006. [DOI] [PubMed] [Google Scholar]
- 27.Tenenhouse HS, Econs MJ. Mendelian Hypophosphatemias. In: Valle D, editor. The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill; 2001. pp. 1–9. [Google Scholar]
- 28.Jonsson KB, et al. Fibroblast growth factor 23 in oncogenic osteomalacia and X-linked hypophosphatemia. N Engl J Med. 2003;348:1656–1663. doi: 10.1056/NEJMoa020881. [DOI] [PubMed] [Google Scholar]
- 29.Weber TJ, Liu S, Indridason OS, Quarles LD. Serum FGF23 levels in normal and disordered phosphorus homeostasis. J Bone Miner Res. 2003;18:1227–1234. doi: 10.1359/jbmr.2003.18.7.1227. [DOI] [PubMed] [Google Scholar]
- 30.Yamazaki Y, et al. Increased circulatory level of biologically active full-length FGF-23 in patients with hypophosphatemic rickets/osteomalacia. J Clin Endocrinol Metab. 2002;87:4957–4960. doi: 10.1210/jc.2002-021105. [DOI] [PubMed] [Google Scholar]
- 31.Larsson T, et al. Transgenic mice expressing fibroblast growth factor 23 under the control of the alpha1(I) collagen promoter exhibit growth retardation, osteomalacia, and disturbed phosphate homeostasis. Endocrinology. 2004;145:3087–3094. doi: 10.1210/en.2003-1768. [DOI] [PubMed] [Google Scholar]
- 32.Yuan B, Meudt J, Blank R, Feng J, Drezner M. Hexa-D-arginine reversal of osteoblast 7B2 dysregulation in Hyp-mice normalizes the HYP biochemical phenotype. J Bone Miner Res 25. 2010. Available at http://www.asbmr.org/Meetings/AnnualMeeting/AbstractDetail.aspx?aid=c5c1b1c6-9d98-4ec8-bafa-833ec5088097. Accessed October 3, 2011.
- 33.Tenenhouse HS. Regulation of phosphorus homeostasis by the type iia na/phosphate cotransporter. Annu Rev Nutr. 2005;25:197–214. doi: 10.1146/annurev.nutr.25.050304.092642. [DOI] [PubMed] [Google Scholar]
- 34.Portale AA, Halloran BP, Morris RC., Jr Physiologic regulation of the serum concentration of 1,25-dihydroxyvitamin D by phosphorus in normal men. J Clin Invest. 1989;83:1494–1499. doi: 10.1172/JCI114043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimada T, et al. FGF-23 is a potent regulator of vitamin D metabolism and phosphate homeostasis. J Bone Miner Res. 2004;19:429–435. doi: 10.1359/JBMR.0301264. [DOI] [PubMed] [Google Scholar]
- 36.Marie PJ, Glorieux FH. Relation between hypomineralized periosteocytic lesions and bone mineralization in vitamin D-resistant rickets. Calcif Tissue Int. 1983;35:443–448. doi: 10.1007/BF02405074. [DOI] [PubMed] [Google Scholar]
- 37.Sato K, et al. Saccharated ferric oxide (SFO)-induced osteomalacia: In vitro inhibition by SFO of bone formation and 1,25-dihydroxy-vitamin D production in renal tubules. Bone. 1997;21:57–64. doi: 10.1016/s8756-3282(97)00084-7. [DOI] [PubMed] [Google Scholar]
- 38.Sato K, Shiraki M. Saccharated ferric oxide-induced osteomalacia in Japan: Iron-induced osteopathy due to nephropathy. Endocr J. 1998;45:431–439. doi: 10.1507/endocrj.45.431. [DOI] [PubMed] [Google Scholar]
- 39.Schouten BJ, Hunt PJ, Livesey JH, Frampton CM, oule SG. FGF23 elevation and hypophosphataemia following intravenous iron polymaltose—a prospective study. J Clin Endocrinol Metab. 2009;94:2332–2337. doi: 10.1210/jc.2008-2396. [DOI] [PubMed] [Google Scholar]
- 40.Schouten BJ, Doogue MP, Soule SG, Hunt PJ. Iron polymaltose-induced FGF23 elevation complicated by hypophosphataemic osteomalacia. Ann Clin Biochem. 2009;46:167–169. doi: 10.1258/acb.2008.008151. [DOI] [PubMed] [Google Scholar]
- 41.Shimizu Y, et al. Hypophosphatemia induced by intravenous administration of saccharated ferric oxide: Another form of FGF23-related hypophosphatemia. Bone. 2009;45:814–816. doi: 10.1016/j.bone.2009.06.017. [DOI] [PubMed] [Google Scholar]
- 42.Peyssonnaux C, et al. Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs) J Clin Invest. 2007;117:1926–1932. doi: 10.1172/JCI31370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cutler C, Bravo A, Ray AD, Watt RK. Iron loading into ferritin can be stimulated or inhibited by the presence of cations and anions: A specific role for phosphate. J Inorg Biochem. 2005;99:2270–2275. doi: 10.1016/j.jinorgbio.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 44.Sitara D, et al. Homozygous ablation of fibroblast growth factor-23 results in hyperphosphatemia and impaired skeletogenesis, and reverses hypophosphatemia in Phex-deficient mice. Matrix Biol. 2004;23:421–432. doi: 10.1016/j.matbio.2004.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allen MR, et al. Morphological assessment of basic multicellular unit resorption parameters in dogs shows additional mechanisms of bisphosphonate effects on bone. Calcif Tissue Int. 2010;86:67–71. doi: 10.1007/s00223-009-9315-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parfitt AM, et al. Report of the ASBMR Histomorphometry Nomenclature Committee Bone histomorphometry: Standardization of nomenclature, symbols, and units. J Bone Miner Res. 1987;2:595–610. doi: 10.1002/jbmr.5650020617. [DOI] [PubMed] [Google Scholar]
- 47.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 48.Larsson T, et al. Fibroblast growth factor-23 mutants causing familial tumoral calcinosis are differentially processed. Endocrinology. 2005;146:3883–3891. doi: 10.1210/en.2005-0431. [DOI] [PubMed] [Google Scholar]