Abstract
Surveillance for drug-resistant parasites in human blood is a major effort in malaria control. Here we report contrasting antifolate resistance polymorphisms in Plasmodium falciparum when parasites in human blood were compared with parasites in Anopheles vector mosquitoes from sleeping huts in rural Zambia. DNA encoding P. falciparum dihydrofolate reductase (EC 1.5.1.3) was amplified by PCR with allele-specific restriction enzyme digestions. Markedly prevalent pyrimethamine-resistant mutants were evident in human P. falciparum infections—S108N (>90%), with N51I, C59R, and 108N+51I+59R triple mutants (30–80%). This resistance level may be from selection pressure due to decades of sulfadoxine/pyrimethamine use in the region. In contrast, cycloguanil-resistant mutants were detected in very low frequency in parasites from human blood samples—S108T (13%), with A16V and 108T+16V double mutants (∼4%). Surprisingly, pyrimethamine-resistant mutants were of very low prevalence (2–12%) in the midguts of Anopheles arabiensis vector mosquitoes, but cycloguanil-resistant mutants were highly prevalent—S108T (90%), with A16V and the 108T+16V double mutant (49–57%). Structural analysis of the dihydrofolate reductase by in silico modeling revealed a key difference in the enzyme within the NADPH binding pocket, predicting the S108N enzyme to have reduced stability but the S108T enzyme to have increased stability. We conclude that P. falciparum can bear highly host-specific drug-resistant polymorphisms, most likely reflecting different selective pressures found in humans and mosquitoes. Thus, it may be useful to sample both human and mosquito vector infections to accurately ascertain the epidemiological status of drug-resistant alleles.
Emergence of drug-resistant Plasmodium falciparum continues to pose a key obstacle to malaria control and elimination efforts (1, 2). It is widely recognized that P. falciparum has a daunting potential for developing resistance to any drug upon wider introduction, including the new combination regimens with or without artemisinin (3–7). Effective public health strategies for surveillance and containment of resistance are therefore paramount, and the World Health Organization has urged that drug efficacy be closely monitored (8).
An instrumental approach widely adopted to help control programs detect and thwart drug resistance is the molecular surveillance of drug resistance-conferring P. falciparum mutants. These molecular markers for resistance are best characterized for chloroquine (9) and antifolate drugs (10–12). Antifolates are extensively used across endemic countries in intermittent preventive therapy programs for malaria in pregnant women and in children (13, 14). Antifolates are also constituents of artemisinin combination therapy (ACT) regimens, such as proguanil–atovaquone–artesunate and artesunate–sulfadoxine/pyrimethamine.
Antifolate resistance by P. falciparum malaria is well recognized (15). Pyrimethamine resistance was first reported in rural Tanzania >50 y ago (16), suggesting the existence of populations of mutant organisms in the endemic region rather than rapid appearance of new mutations. Discovery of pyrimethamine-resistant mutations in parasite DNA encoding dihydrofolate reductase (DHFR) implicated amino acid substitutions at key residues—S108N, N51I, and C59R, as well as double or triple mutants (15, 17–19). Cycloguanil, the active metabolite of proguanil, is known to induce a different set of mutations in the gene encoding parasite DHFR—S108T and A16V, as well as the double mutant (15, 20, 21). During emergence of resistance, the mutation at DHFR amino acid codon 108 is known to arise (22), followed by mutations at the other positions in a stepwise course accompanied by increasing levels of resistance in the multiple mutants (11).
A combination of mutations S108N and I164L confers cross-resistance to pyrimethamine and cycloguanil, which is heightened when the C59R or N51I is also present. A parallel array of mutations in the P. falciparum dihydropteroate synthetase (DHPS) gene at codons 436, 437, 540, and 581 similarly confers resistance to DHPS inhibitors. Tracking such mutations has proved valuable for monitoring the emergence of antifolate drug resistance, as has been a corresponding chloroquine-resistant marker in the P. falciparum chloroquine-resistant transporter (Pfcrt) gene (23).
Current monitoring for P. falciparum drug-resistant alleles is mostly based on genotyping malaria infections in humans found positive by microscopic analysis of their blood. Infections in vector mosquitoes are rarely considered, despite the insects being the definitive host where parasite genetic recombination occurs. With evident field association between mosquito control and drug-resistant P. falciparum prevalence in humans (24, 25), we hypothesized that mosquitoes may play a role in influencing drug-resistant allele epidemiology. To better define the existence of drug-resistant alleles in native populations, we undertook genomic phenotyping of P. falciparum DNA in parasites from both humans and vector mosquitoes in an endemic region of southern Zambia.
Results
Our study of malaria in southern Zambia involved visits to rural households, where peripheral blood was drawn from the residents by finger prick and pyrethrum insecticide spray catches were performed in their sleeping rooms. The region in Choma District surrounding the settlement of Macha was known to exhibit seasonal malaria with thousands of cases of clinical malaria seen at the regional hospital. Sulfadoxine/pyrimethamine had been used in the area for decades. Insecticide-treated bed nets had been introduced 3 y earlier, as had ACT. A severe drought had occurred the previous year.
After giving informed consent, 2,779 human subjects were studied. The individuals ranged from 2 wk to 96 y of age, median age 12 y. Peripheral blood samples from all were examined by microscopy the same day. Only 169 individuals tested positive for malaria parasites (referred to as “blood smear-positive”). Most had low-grade asexual parasitemia, ranging from 38 to 245,000 parasites per microliter (geometric mean parasite density, 1,478; 95% CI, 1,060–2,061 parasites per microliter). Of these, 23 (14%) were pyrexic with axillary temperatures of ≥37.5 °C. Of the 2,610 individuals that tested negative by microscopy (referred to as “blood smear-negative”), 65 (2.5%) were pyrexic.
Insecticide spray catches yielded 796 malaria vector mosquitoes from human sleeping rooms. Nearly all (99%) were Anopheles arabiensis, and the remainder (1%) were Anopheles funestus. The mosquitoes were examined microscopically and individually dissected to separate the abdomen from the head and thorax section. Eighty-one (10%) were positive for P. falciparum in midguts and 64 (8%) in salivary glands, with only 4 (0.5%) infected in both body sections.
DHFR Mutant Parasites in Human Blood.
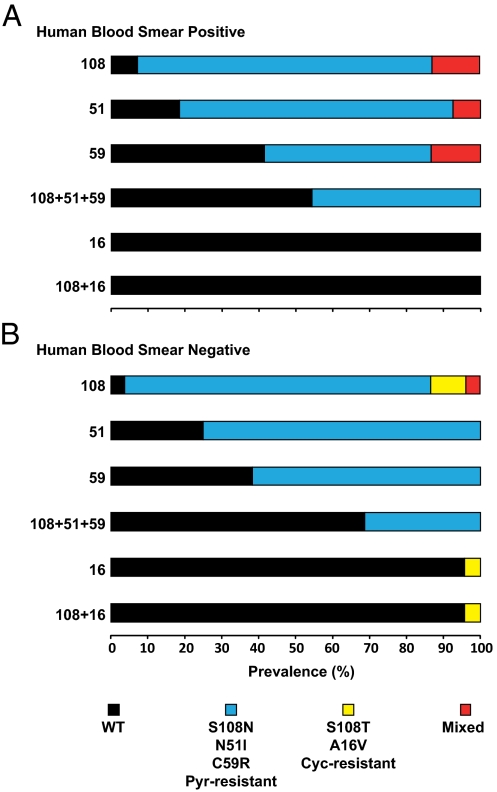
Because pyrimethamine had been used in the region for decades, resistance polymorphisms were expected in the P. falciparum gene encoding DHFR. From the 169 blood smear-positive individuals, a random subset of samples from 86 was PCR amplified. Genotypes for antifolate-resistant mutations (26) were obtained at one or more loci from all 86. From the 2,610 blood smear-negative individuals, a random subset of 475 samples was screened for submicroscopic parasitemia by the same nested PCR for detection of P. falciparum antifolate-resistant polymorphisms (26). Only 93 were PCR-positive, and 88 of these were successfully genotyped for antifolate-resistant mutations at one or more loci, whereas the other 5 had amplicon products too faint for analysis of restriction enzyme digestions (26). High levels of pyrimethamine-resistant mutations were observed in each group (Fig. 1 and Table 1). The sentinel mutation, S108N, was practically saturated (>90%). The other mutants, N51I, C59R, and the 108N+51I+59R triple mutant, were also prevalent (30–80%).
Fig. 1.
Prevalence of P. falciparum pyrimethamine- and cycloguanil-resistant DHFR mutants in human blood. Finger-prick samples were obtained from humans with microscopic parasitemia (blood film-positive; n = 86; A) and submicroscopic parasitemia (blood film-negative; n = 88; B). DNA was PCR amplified and analyzed for allele-specific polymorphisms by restriction digestion. y-axis numerals denote DHFR codons. Represented are WT, pyrimethamine-resistant mutants (S108N, N51I, C59R, and triple mutants), cycloguanil-resistant mutants (S108T, A16V, and double mutant), and mixed alleles (WT+S108T or S108N+S108T).
Table 1.
Determinations of P. falciparum DHFR resistant polymorphisms for parasites from blood smear-positive and -negative humans and from vector mosquito midguts and salivary glands
| Infection | S108N | N51I | C59R | S108N+N51I+C59R | A16V | I164L | 108T+16V |
| Human blood smear-positive | |||||||
| Mut | 67 | 60 | 37 | 36 | 0 | 0 | 0 |
| S108T | 0 | 0 | 0 | N/A | 0 | 0 | 0 |
| Mut + S108T | 0 | 0 | 0 | N/A | 0 | 0 | 0 |
| WT + mut | 11 | 6 | 11 | N/A | 0 | 0 | 0 |
| WT | 6 | 15 | 34 | 43 | 84 | 74 | 84 |
| Total | 84 | 81 | 82 | 79 | 84 | 74 | 84 |
| Human blood smear-negative | |||||||
| Mut | 43 | 33 | 29 | 10 | 2 | 0 | 2 |
| S108T | 5 | 0 | 0 | N/A | 0 | 0 | 0 |
| Mut + S108T | 2 | 0 | 0 | N/A | 0 | 0 | 0 |
| WT + mut | 0 | 0 | 0 | N/A | 0 | 0 | 0 |
| WT | 2 | 11 | 18 | 22 | 44 | 39 | 44 |
| Total | 52 | 44 | 47 | 32 | 46 | 39 | 46 |
| Mosquito midgut | |||||||
| Mut | 7 | 6 | 1 | 1 | 38 | 1* | 33 |
| S108T | 71 | 0 | 0 | N/A | 0 | 0 | 0 |
| Mut + S108T | 2 | 0 | 0 | N/A | 0 | 0 | 0 |
| WT + mut | 0 | 0 | 0 | N/A | 0 | 1 | 0 |
| WT | 1 | 43 | 62 | 40 | 29 | 44 | 34 |
| Total | 81 | 49 | 63 | 41 | 67 | 46 | 67 |
| Mosquito salivary gland | |||||||
| Mut | 27 | 7 | 11 | 4 | 2 | 0 | 1 |
| S108T | 26 | 0 | 0 | N/A | 0 | 0 | 0 |
| Mut + S108T | 0 | 0 | 0 | N/A | 0 | 0 | 0 |
| WT + mut | 1 | 0 | 0 | N/A | 0 | 0 | 0 |
| WT | 8 | 46 | 43 | 46 | 40 | 42 | 41 |
| Total | 62 | 53 | 54 | 50 | 42 | 42 | 42 |
Human blood smear-positive, n = 86; human blood smear-negative, n = 88; mosquito midgut, n = 81; mosquito salivary gland, n = 64. N/A, not applicable.
*Midgut, I164R.
Cycloguanil-resistant mutants were not expected because the progenitor molecule, proguanil, had not been routinely used in the region or adopted for the Zambian national malaria control program, despite clinical trials of the proguanil–atovaquone regimen (27). The sentinel cycloguanil-resistant mutant S108T was not detected in samples from blood smear-positive individuals (Fig. 1 and Table 1), but samples from blood smear-negative individuals revealed S108T, in a modest number (13%). The second mutant, A16V, and the S108T+A16V double mutants were not detected in samples from the blood smear-positive individuals, but they were found in a small percentage of samples (∼4%) from blood smear-negative individuals.
Because they were predominantly asymptomatic, 145 (83%) of the 174 individuals whose samples were genotyped were unaware of having taken antimalarials. Ten (6%) of the others had taken sulfadoxine/pyrimethamine within the past 2 mo. Proportions of pyrimethamine-resistant mutants were slightly higher in samples from these individuals, albeit not statistically significant, possibly due to the limited numbers. None of the individuals whose samples were typed had previously taken trimethoprim/sulfamethoxazole (Cotrimoxazole or Septrin).
DHFR Mutant Parasites in Vector Mosquitoes.
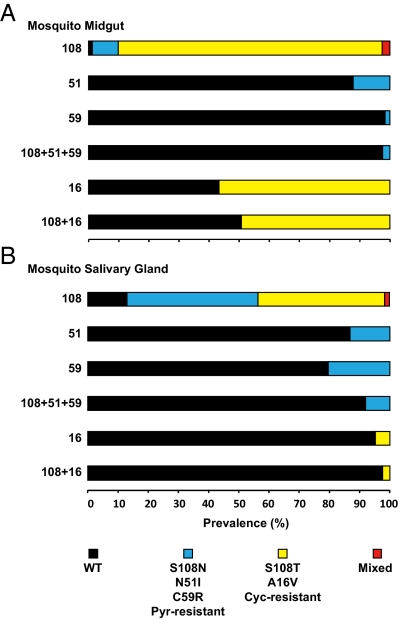
Despite high prevalence of mutant parasites in humans, prevalence of pyrimethamine-resistant mutant parasites was substantially diminished in midguts of Anopheles vector mosquitoes (Fig. 2 and Table1). This result was unexpected, because the mosquitoes had acquired parasites from humans as recently as 10 d previously. The likelihood of finding these mutants had dropped 100-fold in the mosquito phase; these differences were highly significant statistically (Table 2). Although parasites containing pyrimethamine-resistant mutations were least common in midguts, the sentinel mutation, S108N, was relatively more prevalent in salivary glands (>40%) than the other mutations (Fig. 2 and Table 1).
Fig. 2.
Prevalence of P. falciparum pyrimethamine- and cycloguanil-resistant DHFR mutants in 796 malaria vector mosquitoes. Spray catches were obtained. Midguts (A) and salivary gland (B) specimens were separated, and DNA was PCR amplified and analyzed for allele-specific polymorphisms by restriction digestion confirming midgut infections (n = 81) and mosquito gland infections (n = 62). y-axis numerals denote DHFR codons. Represented are WT, pyrimethamine-resistant mutants (S108N, N51I, C59R, and triple mutants), cycloguanil-resistant mutants (S108T, A16V, and double mutant), and mixed alleles (WT+S108T or designated mutant+S108T).
Table 2.
Odds for key P. falciparum DHFR resistant polymorphisms in human blood smear-positive samples compared with blood smear-negative samples and mosquito midguts and salivary glands
| Odds ratio (95% CI), human microscopy positives compared with |
|||
| Allele | Mosquito midgut | Mosquito salivary gland | Human submicroscopic infections |
| S108N | 101 (34.3–299.0); P < 0.001 | 15 (5.7–39.5); P < 0.001 | 1 (0.4–3.5); P = 0.753 |
| N51I | 32 (11.4–87.6); P < 0.001 | 29 (10.9–76.5); P < 0.001 | 1 (0.5–4.3); P = 0.684 |
| C59R | 88 (11.6–662.5); P < 0.001 | 6 (2.5–12.2); P < 0.001 | 1 (0.4–1.8); P = 0.724 |
| S108N+N51I+C59R triple mutant | 18 (1.9–172.2); P < 0.001 | 5 (1.2–22.7); P < 0.001 | 2 (0.6–5.8); P = 0.292 |
| S108T | —; P < 0.001 | —; P < 0.001 | —; P = 0.006 |
| A16V | —; P < 0.001 | —; P = 0.208 | —; P = 0.054 |
| S108T+A16V double mutant | —; P = 0.948 | —; P < 0.001 | —; P = 0.054 |
| I164L | —; P = 0.081 | — | — |
Mosquito midgut, n = 165; mosquito salivary gland, n = 146; human submicroscopic infections, n = 147. —, Odds ratio could not be calculated due to zero frequency in human infections.
Surprisingly high levels of cycloguanil-resistant mutants were observed in vector mosquitoes, especially in midgut samples (Fig. 2 and Table 1). Mosquito infections exhibited high prevalence of cycloguanil-resistant S108T, A16V, and the 108T+16V double mutant, which are currently believed to be rare or absent in natural P. falciparum infections of Africa and elsewhere (28). One midgut infection carried the I164L residue, which, as a multiple mutant with S108N, C59R, and/or N51I, confers high levels of resistance to both pyrimethamine and cycloguanil. Another midgut infection bore a unique I164R residue that has not been previously described for this locus. The composition of antifolate-resistant mutants in mosquito infections was independent of feeding status (Table 3).
Table 3.
Odds of P. falciparum DHFR resistant polymorphisms in mosquitoes with a blood meal compared with unfed mosquitoes
| Mutant | Odds ratio (95% CI) | P |
| S108N | 0.985 (0.371–2.616) | 0.975 |
| S108T | 0.971 (0.379–2.491) | 0.952 |
| N51I | 1.461 (0.295–7.227) | 0.924 |
| C59R | 2.824 (0.343–23.255) | 0.539 |
| A16V | 0.833 (0.274–2.535) | 0.748 |
| I164L* | — | — |
Mosquitoes with a blood meal, n = 105; unfed mosquitoes, n = 60.
*Statistics could not be computed because only one sample bore mutant.
DNA Sequence Confirmations.
Genotypes were confirmed by DNA sequencing. Amplicons from M3-F/ and F-M4 primers (26) were analyzed from subsets of human samples (six) and vector mosquito midgut (seven) and salivary gland samples (six). Polymorphisms in the gene encoding P. falciparum DHFR, as determined by PCR and allele-specific restriction enzyme digestions (Figs. 1 and 2 and Table 1), were confirmed by the DNA sequencing. The sequences flanking the sentinel mutation site at amino acid codon 108 are shown (Fig. 3).
Fig. 3.
DNA sequence alignments for P. falciparum DHFR. DNA flanking nucleotide 323 (amino acid codon 108) obtained from amplicons M3-F/ (M) and F-M4 (F) from human (H) and Anopheles mosquito (A) samples are plotted against standard clone 3D7 (GenBank accession no. AL844503; gene ID 812524). Nucleotide 323 encoding amino acid 108 is accentuated. AGC encodes WT S108; AAC encodes Pyr-resistant mutant S108N (blue); ACC encodes Cyc-resistant mutant S108T (yellow). Identical DNA sequences were obtained from both duplicate amplicons except one mosquito set (F19 A, M19 A) and one human set (F2 H, M2 H), which contained mixed infections. All results confirmed the PCR and restriction enzyme typing.
Structural Alterations of DHFR in Resistance Mutants.
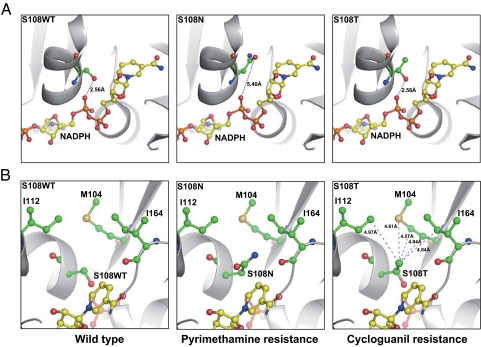
Atomic structures of P. falciparum DHFR illustrate the role of Ser-108 in the function of the wild-type (WT) enzyme and the consequences of S108N mutation [Protein Data Bank (PDB) ID codes 1J3I and 3JSU]. Essential for nucleotide biogenesis in all organisms, DHFR catalyzes reduction of dihydrofolate to tetrahydrofolate with oxidation of NADPH to NADP+.
Modeling with the WT structure as the template revealed major structural differences in the mutant enzymes. The binding pocket for NADPH within the WT DHFR contains an important 2.56-Å hydrogen bond linking the side chain of Ser-108 to the pyrophosphate moiety of NADPH (Fig. 4A Left). In contrast, the sentinel pyrimethamine-resistant mutant S108N projects a side chain that cannot form an effective hydrogen bond, because at 5.40 Å, the distance is too great (Fig. 4A Center). Similar to the WT enzyme, the cycloguanil mutant S108T maintains the 2.56-Å hydrogen bond with NADPH (Fig. 4A Right). The extra methyl group in S108T does not cause any steric collisions but forms van der Waals interactions with a nearby hydrophobic domain composed of Met-104, Ile-112, and Ile-164 (Fig. 4B Right).
Fig. 4.
Key structural variances at amino acid 108 in WT and resistant mutants of P. falciparum DHFR. Represented are crystal structures of WT DHFR (S108WT) and S108N mutant (PDB ID codes 1J3I and 3JSU). The structure of the S108T mutant was derived by molecular modeling. (A) Note a 2.56-Å hydrogen bond between the pyrophosphate moiety of NADPH and the hydroxyl group of the S108 WT or S108T residue; no effective hydrogen bond exists in the S108N mutant due to the 5.40-Å distance. (B) Note extra methyl group in the S108T mutant forms van der Waals interactions with a nearby hydrophobic area composed of Met-104, Ile-112, and Ile-164. The side chains of S108WT, S108T, S108N, Met-104, Ile-112, Ile-164, and NADPH are highlighted with sticks and spheres; the hydrogen bond is shown as the dashed line with the distance labeled, and the protein backbone is shown in a cartoon model.
Discussion
Our studies revealed contrasting drug-resistant polymorphisms in the P. falciparum gene encoding DHFR. The parasites in human blood demonstrated high levels of pyrimethamine-resistant mutants, whereas parasites in Anopheles vector mosquitoes exhibited high levels of cycloguanil-resistant mutants. This surprising contrast raises several questions. Are we using optimal allele detection methods? What is the role of antifolate drug resistance in the mosquito phase of malaria? What is the functional significance of the structural alterations of parasite DHFR? Are contrasting drug-resistant polymorphisms unique to a single malaria-endemic region, or is this a general phenomenon?
Our studies indicate that genotyping human blood smear-positive samples alone may underestimate the true profile of drug-resistant alleles. Furthermore, although cycloguanil-resistant mutants are currently believed to be rare or absent in natural P.falciparum infections in Africa (28–30), they were found to exist at high prevalence in the vector population. These mutants, S108T, A16V, and the double mutant S108T+A16V, are prevalent in parasites in mosquitoes, even though they are undetected by current surveillance based on typing human blood smear-positive samples. Rarer alleles, including I164L, and a unique allele, I164R, were also observed in vector mosquitoes. Mosquito midgut infections with P. falciparum were acquired from the human population within the previous 10 d, indicating that cycloguanil-resistant alleles were present in human parasitemia. Detection of low levels of S108T and A16V in samples from humans with blood smear-negative infections confirmed this hypothesis. It is likely that minority alleles in blood smear-positive infections escaped detection due to selective amplification of the dominant alleles during PCR. In Malawi, for example, presence of the rare I164L mutant was recently documented in human P. falciparum infections by using a heteroduplex tracking assay (30), but the allele was then subsequently detected in Kenya at low levels by regular PCR (31). Inclusion of human submicroscopic parasitemia and vector infections in epidemiological sampling may therefore facilitate effective tracking and curtailment of resistant alleles before they become public health problems. Although the possibility of de novo origins for mutants cannot be ruled out (32), this expanded sampling approach would remain useful.
Our results point to a pivotal role of mosquitoes in the epidemiology of drug-resistant P. falciparum. Apart from harboring minority alleles subpatent in humans, An. arabiensis clearly exerted a dramatic depletion (as much as 100-fold) on P. falciparum pyrimethamine-resistant alleles, despite prevailing sulfadoxine/pyrimethamine drug pressure in the area. This mosquito selection phenomenon on P. falciparum polymorphisms is distinct from Kublin's in vivo fitness burden in humans (33) and was evidently independent and opposing to existing drug selection.
We speculate that vector-selective constraints on parasite-resistant polymorphisms may reflect field links between mosquito control and suppression of P. falciparum drug resistance, as reported with sulfadoxine/pyrimethamine and with chloroquine (24, 25, 34). Increasing prevalence of P. falciparum DHFR WT alleles was observed in Tanzania after the use of insecticide-treated nets (24). In Zimbabwe, the introduction of indoor residual insecticide spraying (IRS) was associated with a fourfold reduction in the odds of chloroquine therapeutic failure (25), despite years of ongoing drug pressure. Cessation of the IRS was subsequently linked to a rebound in chloroquine resistance (25). Given the vector-selective pressure on parasite-resistant alleles observed in this study, it would be expected that mosquito control may affect the prevalence of drug resistance, and this link should be considered in control programs.
It is important to understand specific mechanisms by which the relative proportions of resistant alleles change between human and mosquito phases. One possibility might be the disparity in differentiation to gametocytes reported to occur between WT and resistant alleles (35). This difference may also relate to the curious switch in the proportions of S108T and S108N alleles seen between midgut and salivary gland infections. Drug-resistant alleles are known to be associated with altered biological fitness of the parasite (36, 37). It is therefore possible that survival differences under immune clearance within the human and mosquito phases, and presence or absence of drug selection, may play significant roles. Analogous differences were observed in isolation frequencies of Borrelia species from ticks and human patients from the same endemic region (38).
The P. falciparum polymorphisms found in humans were expected from selection pressure due to decades of sulfadoxine/pyrimethamine use. The underlying basis for P. falciparum polymorphisms in mosquitoes is not clear. Structural modeling revealed distinct differences between the pyrimethamine-resistant mutation, S108N, and the cycloguanil-resistant mutation S108T, and these changes may provide a molecular explanation for the host specificities. Overall, mutations causing pyrimethamine resistance were all nonconservative substitutions—replacement of a serine with an asparagine, an asparagine with an isoleucine, or a cysteine with an arginine. In contrast, mutations causing cycloguanil resistance were very conservative—replacement of a serine with a threonine or an alanine with a valine.
The Ser-108 residue plays a key role in DHFR catalysis and protein structure and may explain the contrasting antifolate-resistant polymorphisms reported here. Previous kinetic studies (39, 40) showed that S108N is a loss-of-function mutant, consistent with reduced fitness in the mosquito vector despite pyrimethamine-induced selection in humans (Figs. 1 and 2). In contrast, S108T is a gain-of-function mutant. The specific activity of S108T is approximately twice that of the WT, whereas Km values of this mutant for 7,8-dihydrofolate and NADPH are <25% of the WT (39). These data suggest that the S108T mutation enhances the DHFR activity, possibly conferring an advantage to the parasite, whereas within the midgut, P. falciparum parasites carrying this mutation may outgrow WT parasites while forming oocysts. After rupture of the oocysts, the emerging sporozoites are nonproliferating. Additionally, because sulfadoxine/pyrimethamine has long been used in this region of Africa, the advantage of S108T parasites may only apply to the mosquito cycle, and not the human cycle, because pyrimethamine imposes selective pressure favoring mutations such as S108N. In this study, S108T accounts for 90% of alleles in mosquito midgut infections, supporting the above hypothesis. Nevertheless, it does not explain why S108T prevalence is not elevated in human infections during absence of sulfadoxine/pyrimethamine selection pressure. Future studies will be needed to elucidate the candidate factors underpinning these allele patterns.
The human populations and mosquitoes surveyed in this study were all from one rural area in southern Zambia. Before general significance can be established, it will be essential to undertake similar surveys in other areas where malaria is endemic. Polymorphisms in P. falciparum genes encoding resistance to other antimalarial drugs should also be examined. If contrasting drug-resistant phenotypes are confirmed in other regions, it may be useful to examine both human and vector infections during epidemiological surveillance for antimalarial drug-resistant alleles.
Materials and Methods
See SI Materials and Methods for additional details.
Area and Population.
The study was conducted during the 2006–2007 peak malaria transmission season in 15 representative 25-km2 geographical grids near Macha in the Southern Province of Zambia. Willing individuals of all ages from the resident BaTonga communities were eligible to participate in the study.
Study Design and Sample Collection.
The study was a prospective cross-sectional design. P. falciparum malaria infections in sympatric contemporaneous human and mosquito populations were typed for DHFR-resistant alleles.
DNA Extractions.
Mosquito samples were subjected to a simplified Chelex protocol (41) for P. falciparum DNA extraction. Human blood samples spotted on filter paper were air-dried, and the Chelex protocol (12) was used to extract parasite DNA
Assays for Antifolate Drug-Resistant Alleles.
P. falciparum was genotyped by nested PCR and allele-specific restriction enzyme digestion (26) at DHFR amino acid codons 108, 51, 59, 16, and 164.
A random subset of six samples from human, seven mosquito midgut, and six mosquito salivary gland infections were subjected to independent sequence confirmation of P. falciparum amplicons at the Johns Hopkins DNA analysis core facility.
Structural Modeling.
The structure of S108T DHFR mutant was modeled with PyMol (www.pymol.org) by using the WT DHFR crystal structure (PDB ID code 1J3I) as the template. The quadruple DHFR mutant structure (PDB ID code 3JSU) was used as the S108N mutant.
Data Analysis.
Mantel–Haenzel's χ2 test and multivariate binary logistic models were used to analyze differences in composition of resistant alleles among human and mosquito phases. Odds of resistant mutants were determined from the logistic regression models.
Ethics.
The study was approved by both the national (University of Zambia) research ethics committee and the Johns Hopkins institutional review board.
Supplementary Material
Acknowledgments
We thank the residents, headmen, and chiefs of Mapanza, Muchila, Chikanta, and Macha for their support and participation; and Harry Hamapumbu, Petros Moono, Gift Moono, Patricia Muleya, and others from the Malaria Institute at Macha field and laboratory staff. This work was supported by the Johns Hopkins Malaria Research Institute, the Bill and Melinda Gates Foundation, and National Institutes of Health Grants U19 AI089680 (International Centers of Excellence for Malaria Research) and HL48268.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116162108/-/DCSupplemental.
References
- 1.Björkman A, Bhattarai A. Public health impact of drug resistant Plasmodium falciparum malaria. Acta Trop. 2005;94:163–169. doi: 10.1016/j.actatropica.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 2.Trape JF. The public health impact of chloroquine resistance in Africa. Am J Trop Med Hyg. 2001;64(1–2 Suppl):12–17. doi: 10.4269/ajtmh.2001.64.12. [DOI] [PubMed] [Google Scholar]
- 3.Crabb C. Plasmodium falciparum outwits Malarone, protector of travellers. Bull World Health Organ. 2003;81:382–383. [PMC free article] [PubMed] [Google Scholar]
- 4.Fivelman QL, Butcher GA, Adagu IS, Warhurst DC, Pasvol G. Malarone treatment failure and in vitro confirmation of resistance of Plasmodium falciparum isolate from Lagos, Nigeria. Malar J. 2002;1:1. doi: 10.1186/1475-2875-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noedl H, et al. Artemisinin Resistance in Cambodia 1 (ARC1) Study Consortium. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 6.Wongsrichanalai C, Meshnick SR. Declining artesunate-mefloquine efficacy against falciparum malaria on the Cambodia-Thailand border. Emerg Infect Dis. 2008;14:716–719. doi: 10.3201/eid1405.071601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noedl H, Socheat D, Satimai W. Artemisinin-resistant malaria in Asia. N Engl J Med. 2009;361:540–541. doi: 10.1056/NEJMc0900231. [DOI] [PubMed] [Google Scholar]
- 8.Laufer MK, Djimdé AA, Plowe CV. Monitoring and deterring drug-resistant malaria in the era of combination therapy. Am J Trop Med Hyg. 2007;77(6 Suppl):160–169. [PubMed] [Google Scholar]
- 9.Djimdé AA, et al. Molecular diagnosis of resistance to antimalarial drugs during epidemics and in war zones. J Infect Dis. 2004;190:853–855. doi: 10.1086/422758. [DOI] [PubMed] [Google Scholar]
- 10.Plowe CV. Monitoring antimalarial drug resistance: Making the most of the tools at hand. J Exp Biol. 2003;206:3745–3752. doi: 10.1242/jeb.00658. [DOI] [PubMed] [Google Scholar]
- 11.Plowe CV, et al. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine-sulfadoxine use and resistance. J Infect Dis. 1997;176:1590–1596. doi: 10.1086/514159. [DOI] [PubMed] [Google Scholar]
- 12.Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: Polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg. 1995;52:565–568. doi: 10.4269/ajtmh.1995.52.565. [DOI] [PubMed] [Google Scholar]
- 13.Andriantsoanirina V, et al. Origins of the recent emergence of Plasmodium falciparum pyrimethamine resistance alleles in Madagascar. Antimicrob Agents Chemother. 2010;54:2323–2329. doi: 10.1128/AAC.01511-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobbe R, et al. Follow-up survey of children who received sulfadoxine-pyrimethamine for intermittent preventive antimalarial treatment in infants. J Infect Dis. 2011;203:556–560. doi: 10.1093/infdis/jiq079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plowe CV. The evolution of drug-resistant malaria. Trans R Soc Trop Med Hyg. 2009;103(Suppl 1):S11–S14. doi: 10.1016/j.trstmh.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clyde DF, Shute GT. Resistance of Plasmodium falciparum in Tanganyika to pyrimethamine administered at weekly intervals. Trans R Soc Trop Med Hyg. 1957;51:505–513. doi: 10.1016/0035-9203(57)90039-1. [DOI] [PubMed] [Google Scholar]
- 17.Cowman AF, Morry MJ, Biggs BA, Cross GA, Foote SJ. Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1988;85:9109–9113. doi: 10.1073/pnas.85.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peterson DS, Walliker D, Wellems TE. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci USA. 1988;85:9114–9118. doi: 10.1073/pnas.85.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kublin JG, et al. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J Infect Dis. 2002;185:380–388. doi: 10.1086/338566. [DOI] [PubMed] [Google Scholar]
- 20.Foote SJ, Galatis D, Cowman AF. Amino acids in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum involved in cycloguanil resistance differ from those involved in pyrimethamine resistance. Proc Natl Acad Sci USA. 1990;87:3014–3017. doi: 10.1073/pnas.87.8.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peterson DS, Milhous WK, Wellems TE. Molecular basis of differential resistance to cycloguanil and pyrimethamine in Plasmodium falciparum malaria. Proc Natl Acad Sci USA. 1990;87:3018–3022. doi: 10.1073/pnas.87.8.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cortese JF, Caraballo A, Contreras CE, Plowe CV. Origin and dissemination of Plasmodium falciparum drug-resistance mutations in South America. J Infect Dis. 2002;186:999–1006. doi: 10.1086/342946. [DOI] [PubMed] [Google Scholar]
- 23.Djimdé A, et al. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med. 2001;344:257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- 24.Alifrangis M, et al. Increasing prevalence of wildtypes in the dihydrofolate reductase gene of Plasmodium falciparum in an area with high levels of sulfadoxine/pyrimethamine resistance after introduction of treated bed nets. Am J Trop Med Hyg. 2003;69:238–243. [PubMed] [Google Scholar]
- 25.Mharakurwa S, et al. Association of house spraying with suppressed levels of drug resistance in Zimbabwe. Malar J. 2004;3:35. doi: 10.1186/1475-2875-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duraisingh MT, Curtis J, Warhurst DC. Plasmodium falciparum: Detection of polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes by PCR and restriction digestion. Exp Parasitol. 1998;89:1–8. doi: 10.1006/expr.1998.4274. [DOI] [PubMed] [Google Scholar]
- 27.Mulenga M, et al. A randomised, double-blind, placebo-controlled trial of atovaquone-proguanil vs. sulphadoxine-pyrimethamine in the treatment of malarial anaemia in Zambian children. Trop Med Int Health. 2006;11:1643–1652. doi: 10.1111/j.1365-3156.2006.01726.x. [DOI] [PubMed] [Google Scholar]
- 28.Mawili-Mboumba DP, Ekala MT, Lekoulou F, Ntoumi F. Molecular analysis of DHFR and DHPS genes in P. falciparum clinical isolates from the Haut—Ogooué region in Gabon. Acta Trop. 2001;78:231–240. doi: 10.1016/s0001-706x(01)00084-5. [DOI] [PubMed] [Google Scholar]
- 29.Muehlen M, et al. Short communication: Prevalence of mutations associated with resistance to atovaquone and to the antifolate effect of proguanil in Plasmodium falciparum isolates from northern Ghana. Trop Med Int Health. 2004;9:361–363. doi: 10.1111/j.1365-3156.2004.01201.x. [DOI] [PubMed] [Google Scholar]
- 30.Juliano JJ, Trottman P, Mwapasa V, Meshnick SR. Detection of the dihydrofolate reductase-164L mutation in Plasmodium falciparum infections from Malawi by heteroduplex tracking assay. Am J Trop Med Hyg. 2008;78:892–894. [PMC free article] [PubMed] [Google Scholar]
- 31.Alker AP, et al. Mutations associated with sulfadoxine-pyrimethamine and chlorproguanil resistance in Plasmodium falciparum isolates from Blantyre, Malawi. Antimicrob Agents Chemother. 2005;49:3919–3921. doi: 10.1128/AAC.49.9.3919-3921.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rathod PK, McErlean T, Lee PC. Variations in frequencies of drug resistance in Plasmodium falciparum. Proc Natl Acad Sci USA. 1997;94:9389–9393. doi: 10.1073/pnas.94.17.9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kublin JG, et al. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis. 2003;187:1870–1875. doi: 10.1086/375419. [DOI] [PubMed] [Google Scholar]
- 34.Molyneux DH, Floyd K, Barnish G, Fèvre EM. Transmission control and drug resistance in malaria: A crucial interaction. Parasitol Today. 1999;15:238–240. doi: 10.1016/s0169-4758(99)01453-2. [DOI] [PubMed] [Google Scholar]
- 35.Sokhna CS, Trape JF, Robert V. Gametocytaemia in Senegalese children with uncomplicated falciparum malaria treated with chloroquine, amodiaquine or sulfadoxine + pyrimethamine. Parasite. 2001;8:243–250. doi: 10.1051/parasite/2001083243. [DOI] [PubMed] [Google Scholar]
- 36.Hayward R, Saliba KJ, Kirk K. pfmdr1 mutations associated with chloroquine resistance incur a fitness cost in Plasmodium falciparum. Mol Microbiol. 2005;55:1285–1295. doi: 10.1111/j.1365-2958.2004.04470.x. [DOI] [PubMed] [Google Scholar]
- 37.Osman ME, Mockenhaupt FP, Bienzle U, Elbashir MI, Giha HA. Field-based evidence for linkage of mutations associated with chloroquine (pfcrt/pfmdr1) and sulfadoxine-pyrimethamine (pfdhfr/pfdhps) resistance and for the fitness cost of multiple mutations in P. falciparum. Infect Genet Evol. 2007;7:52–59. doi: 10.1016/j.meegid.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 38.Picken RN, et al. Molecular characterization of Borrelia burgdorferi sensu lato from Slovenia revealing significant differences between tick and human isolates. Eur J Clin Microbiol Infect Dis. 1996;15:313–323. doi: 10.1007/BF01695664. [DOI] [PubMed] [Google Scholar]
- 39.Sano G, Morimatsu K, Horii T. Purification and characterization of dihydrofolate reductase of Plasmodium falciparum expressed by a synthetic gene in Escherichia coli. Mol Biochem Parasitol. 1994;63:265–273. doi: 10.1016/0166-6851(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 40.Sirawaraporn W, Prapunwattana P, Sirawaraporn R, Yuthavong Y, Santi DV. The dihydrofolate reductase domain of Plasmodium falciparum thymidylate synthase-dihydrofolate reductase. Gene synthesis, expression, and anti-folate-resistant mutants. J Biol Chem. 1993;268:21637–21644. [PubMed] [Google Scholar]
- 41.Musapa M, et al. A simple chelex protocol for DNA extraction from Anopheles spp. J Visualized Exp Immunol Infect. 2011 doi: 10.3791/3281. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.