Abstract
The DNA amplification performed by terminal protein-primed replication systems has not yet been developed for its general use to produce high amounts of DNA linked to terminal protein (TP). Here we present a method to amplify in vitro heterologous DNAs using the Φ29 DNA replication machinery and producing DNA with TP covalently attached to the 5′ end. The amplification requires four Φ29 proteins, DNA polymerase, TP, single-stranded DNA binding protein and double-stranded DNA binding protein (p6). The DNA to be amplified is inserted between two sequences that are the Φ29 DNA replication origins, consisting of 191 and 194 bp from the left and right ends of the phage genome, respectively. The replication origins do not need to have TP covalently attached beforehand to be functional in amplification and they can be joined to the DNA to be amplified by cloning or ligation. The facts that two functional origins were required at the ends of a linear template DNA and that the kinetics of DNA synthesis was very similar to that obtained using the TP-containing Φ29 genome as template support the proposal that genuine amplification is taking place. Amplification factors of 30-fold have been obtained. Possible applications of DNAs produced by this method are discussed.
Keywords: Φ29 DNA polymerase, origins of replication
Although the most common systems to start DNA replication are those in which a DNA or an RNA molecule is used as primer, DNA synthesis can also be started using a protein as primer. This type of system initiates DNA synthesis using as an acceptor of the first dNMP, the OH group of a specific serine, threonine, or tyrosine residue of a protein, instead of the 3′ OH group of a ribose or deoxyribose (1). This protein is generally named terminal protein (TP) because it becomes covalently attached at the 5′ termini of the DNA. The TP-primed DNA replication has been studied in a number of systems, such as bacteriophages Φ29, Nf, and GA-1 (2), PRD1 (3), and Cp-1 (4), linear plasmids from bacteria such as pSCL and pSLA2 (5), and in adenovirus (6). In addition, TP-containing DNAs have been identified in linear plasmids of mitochondria, yeast, and plants and in bacterial chromosomes (Streptomyces sp.) (see ref. 1 for a review). From all these, the bacteriophage Φ29 TP-DNA replication system has emerged as the best studied one (7). In vivo-based methods to generate linear DNAs with TP attached at the 5′ ends have been described for Streptomyces plasmids (8) and adenovirus (9). These methods take advantage of the observation that a DNA containing the correct terminal sequences but not having TP linked at the ends is capable, after transformation of an appropriate host and selection, to produce inside the cell the corresponding TP-DNA that is stably maintained. However, to date none of these systems has been used to produce TP-DNAs in high amounts appropriate for different molecular biology uses (see below). Regarding the in vitro approaches, as of today, the most efficient system has proved to be the Φ29 DNA replication one. To our knowledge, no other in vitro system produces TP-DNA amplification in high amounts using purified proteins (10).
The Φ29 genome replication system is a very efficient TP-DNA synthesizing machinery whether working in vitro or in vivo, provided that it is using as template the bacteriophage Φ29 DNA with TP covalently attached to its 5′ ends (11, 12). The Φ29 in vitro minimal amplification system of TP-DNA is based on four essential components, DNA polymerase, TP, double-stranded DNA binding protein p6, and single-stranded DNA binding protein (SSB) p5 (10). In the Φ29 DNA replication mechanism (13) (Fig. S1), the origins of replication are the 5′ TP-linked ends of the phage genome. These terminal regions have signals for the cooperative binding of protein p6, which stimulates the initiation of replication likely by opening the two strands of the DNA at the very ends of the genome (14). The heterodimer formed by TP and DNA polymerase (15) is recruited to the origin in a process enhanced by the 5′-linked TP (parental TP) (16). Then, the initiation of replication takes place in which the TP is used as a primer by the DNA polymerase followed by a transition stage when the polymerase synthesizes 10 nucleotides, dissociates from the TP, and switches to the DNA-primed elongation mode of replication until it reaches the end of the genome (17). The Φ29 DNA polymerase is endowed with a potent strand displacement activity and a high processivity that allows it to perform the 19-kb replication of the phage genome from a single binding event and without the need for any accessory factors (12). The Φ29 replication system has been reported to amplify the genome of the phage (TP-DNA) to microgram amounts starting from nanograms in a single-step isothermal reaction (10). It is also important to note that the amplification of the 19-kb phage DNA takes place with high fidelity yielding fully functional DNA (10).
Until now, no systematic attempt had been made to produce defined TP-linked heterologous DNAs in a fashion similar to the amplification of the phage Φ29 genome. Here we report an in vitro method to produce DNA with TP covalently attached to the 5′ end that is not dependent on replication in a host cell and has the potential to amplify DNAs of at least 18 kb of length. The method defines the minimal sequences and requirements that the origins should meet to be targets of amplification by the Φ29 replication system.
Results and Discussion
Design of a Plasmid Displaying Functional Ends of the Φ29 Genome After Linearization.
Previous results showed that the initiation of replication from templates having Φ29 DNA ends without TP was much less efficient than the initiation performed using the Φ29 TP-DNA as template (16). Notwithstanding, there were no experiments in which a comparison had been made between the Φ29 genome (TP-DNA) and other templates having two Φ29 DNA terminal regions without TP, with respect to in vitro amplification using the Φ29 replication machinery as described (10). For a template to be susceptible to amplification using the Φ29 system, it must have two fully functional Φ29 DNA ends that are correctly oriented. To test whether it was possible to obtain amplification starting from adequate DNA templates that do not have the parental TP covalently linked at the 5′ ends, the plasmid described in Fig. 1A was designed. This plasmid, named pETORPHI, contains the Φ29 DNA left and right ends up to base pairs 191 and 194, respectively (see below), a pUC-type replication origin, a kanamycin resistance marker, and two multicloning sites. The Φ29 DNA ends are cloned with the extremities joined, in inverted orientation, in such a way that a DraI restriction site is created (Fig. 1B), and this site is unique in the plasmid. After restriction of the plasmid with DraI, the two ends of the linearized plasmid are identical to the two ends of Φ29 DNA up to the mentioned positions (Fig 1C). The Φ29 DNA segments were chosen to span the minimal functional origins as characterized by in vitro experiments comprising the p6 nucleation sequences (18) and the last 12 base pairs from each end (19) which include the 6 bp inverted repeat (5′AAAGTA) with the AAA sequence of the very end required for the sliding-back mechanism of initiation of replication (20). Although the minimal origins are reported to be 68 bp for the left end and 125 bp for the right end (18), in our initial construct we lengthened the Φ29 DNA sequences up to positions 191 and 194 from the left and right ends, respectively, because those are the approximate distances at which the binding of p6 to the Φ29 genome ends starts to become weaker (18).
Fig. 1.
Construction of a plasmid that generates the Φ29 DNA replication origins upon linearization. (A) The pETORPHI plasmid contains fused fragments of 191 and 194 bp from the left and right ends, respectively, of the Φ29 genome. Left Ori and Right Ori stand for left and right Φ29 DNA origins, respectively. (B) The ends were synthesized with an inverted “end-to-end” orientation that generates a DraI site at the joining point as shown in the figure. (C) Linearization of the plasmid with DraI produces the DNA segment shown with the Φ29 ends in the correct orientation. MCS1 and MCS2 denote multiple cloning sites 1 and 2; the sequences of the MCSs are listed in SI Text.
Linearized pETORPHI Plasmid is Efficiently Replicated in Vitro by the Φ29 DNA Synthesis Machinery.
The ability of plasmid pETORPHI (5.8 kb) cut with DraI (from now on pETORPHI) to support TP-primed replication/amplification with the Φ29 replication system was tested. As shown in Fig. 2A, in the presence of p6, synthesis of TP-DNA took place as indicated by the accumulation of 32P-labeled material at the right position (19.2 kb); the reaction had reached a plateau starting with 25 ng of TP-DNA with little increase in the amount of final product when using 100 ng as template. When pETORPHI was used as template, in the presence of p6, there was also accumulation of correct-sized 5.8-kb product; the reaction was also at plateau-level using 25 ng of template. In the absence of p6, there was synthesis of TP-DNA, although, with 25 ng of template, the reaction did not reach the maximum level obtained with p6. On the other hand, when pETORPHI was used as template, DNA synthesis turned out to be almost totally dependent on the presence of p6. As a control, DNA synthesis directed from the two templates was completely dependent on TP (Fig. 2B), which means that all the DNA produced is initiated by TP-priming and thus it contains TP linked at the end.
Fig. 2.
The plasmid pETORPHI is an efficient template for DNA synthesis. (A) The assays were done as described in Materials and Methods using the amounts in nanograms of plasmid pETORPHI or Φ29 TP-DNA templates indicated in the figure. The absence or presence of p6 in the reactions is indicated. The size of the amplified bands is shown: 19.2 kb for Φ29 TP-DNA and 5.8 kb for pETORPHI cut with DraI. Lambda phage DNA cut with HindIII is shown as molecular weight marker (Mr). (B) The presence of TP is essential for amplification. The assay was performed as in A, but using p6 in all cases. The absence or presence of TP is indicated.
The Φ29 Replication System Produces True Amplification from Plasmid Templates.
To determine whether true amplification products were obtained, pETORPHI cut with DraI was further cut with either MluI or PvuI giving rise in each case to two DNA molecules of different size (see Fig. 1C). The two DNA fragments produced after restriction with MluI or PvuI could not be susceptible to exponential amplification because each DNA molecule would have only one of the Φ29 DNA origins and could only produce linear replication. Fig. 3 shows that, in any of the cases in which the two ends of the plasmid were separated, the amount of DNA synthesized decreased greatly (between 10- and 20-fold, depending on the cases, as quantitated by phosphoimager), compared with that obtained with the linearized pETORPHI. Even when the fragments produced after restriction with the two enzymes were mixed in the reaction, to check if they can hybridize or complement each other (lane pETORPHI Dra Mlu + Dra Pvu), the outcome was the same as the cases in which fragments cut with just one enzyme were used. As expected, the uncut plasmid gave no signal. The conclusion of these experiments is that the plasmid is undergoing true amplification utilizing the Φ29 DNA replication origins at both ends of the molecule.
Fig. 3.
The plasmid-templated amplification requires two Φ29 DNA ends in the same molecule. The pETORPHI plasmid was cut with the indicated enzymes under the conditions described in Materials and Methods. The amounts in ng of Φ29 TP-DNA and plasmid fragments utilized in the amplification reactions are shown. The bands corresponding to the plasmid cut with MluI or PvuI in addition to DraI are 4.6 and 4.4 kb, respectively.
To further ensure that the products generated by reactions using the linearized pETORPHI plasmid as template are bona fide amplification products, time-course amplification experiments were performed and the results obtained with TP-DNA or pETORPHI as templates were compared. Fig. 4A shows that for the two templates a similar pattern was obtained, with the maximum reached at 120 min (see Fig. S2 for quantitation of the kinetics). If the amounts of DNA synthesized are normalized, taking the values at 60 min as 100%, both templates, the Φ29 TP-DNA and pETORPHI, follow almost identical, nonlinear, amplification kinetics (Fig. 4B). Given that the Φ29 TP-DNA undergoes genuine amplification (10), the time-course results support the proposal that pETORPHI is also being amplified. The average amplification factors across experiments were 83 ± 19 SD for the phage genome and 30 ± 7 SD for the plasmid, starting from 25 ng of template in both cases, which corresponds to about 2 and 0.8 μg, respectively, of total DNA synthesized.
Fig. 4.
The amplification kinetics of pETORPHI is similar to that of Φ29 TP-DNA. (A) Reactions containing 25 ng of Φ29 TP-DNA or pETORPHI cut with DraI were allowed to proceed at 22 °C for the indicated times and then they were stopped by addition of SDS to 0.1% and EDTA to 10 mM. (B) The amounts of synthesized DNA, expressed as a percentage of the value reached at 60 min, was represented as a function of time for reactions with Φ29 TP-DNA (▴) or pETORPHI (●) as templates. The bars represent the standard deviations from three independent experiments.
Minimal Template Requirements of the System.
The minimal amount of template that can be used as input in the amplification reaction was determined. Fig. 5 shows that when the Φ29 TP-DNA is used as template the minimal amount that allows amplification is 10 ng and the maximal amount of product yielded is reached at 17.5 ng, indicating that the reaction is close to saturation at that point. When pETORPHI is used as template, the results are very similar, 17.5 ng being the amount of template needed to reach saturation. It should be noticed that the minimal amount of template that produces amplification does not correspond to the same initial number of molecules for pETORPHI and Φ29 TP-DNA, but to the same mass of DNA, which corresponds to 3.3 times more molecules of pETORPHI than those of TP-DNA.
Fig. 5.
Minimal amount of template required for amplification. The amplification reactions were set up with the indicated amounts in nanograms of Φ29 TP-DNA or pETORPHI plasmid cut with DraI. The pETORPHI68L (see Materials and Methods and Fig. S2) was cut with DraI and the experiment was performed as before with the indicated amounts of template (Right).
The minimal length of Φ29 DNA that supports p6-stimulated replication activity corresponds to the last 68 bp from the left end of the Φ29 genome (18). As explained above, this DNA includes the 6 bp inverted terminal repeat (5′ AAAGTA) and sequences from base pairs 35–58 from the left end that have been described as high affinity p6 binding sites (18, 21). For the right end of Φ29 DNA, the minimal origin is longer, 125 bp, and the p6 affinity sequences are not as precisely defined as those of the left end (18). Thus, as a first step to develop an amplification system based on a minimal origin, we tested whether a fragment of 68 bp from the left Φ29 DNA end is capable of supporting amplification. The 194-bp segment corresponding to the right end of the Φ29 genome from pETORPHI was replaced by the 68-bp minimal origin from the left end described above, to obtain pETORPHI68L (Fig. S3 and SI Text). The 68-bp fragment was cloned in inverted orientation as in pETORPHI allowing the generation of the Φ29 DNA ends upon linearization with DraI. As shown in Fig. 5 (Right) the amount of pETORPHI68L that serves as template for amplification is the same as that of the original pETORPHI; however, the final amount of product obtained with pETORPHI68L is lower (29 ± 8% SD with respect to pETORPHI).
Two Different Strategies to Produce Heterologous TP-DNAs.
We have shown that the plasmid pETORPHI, after linearization with the enzyme DraI, presents two functional origins of replication for the Φ29 minimal amplification machinery. The fact that the DraI site occurs very often in vectors and genomes with a medium to high AT content could constitute a problem if more DraI sites are present in the vector or in the DNA to be amplified. To overcome this problem, a procedure was designed to cut the pETORPHI plasmid in such a way that Φ29 DNA ends are generated using a restriction site different from that recognized by DraI. We chose the enzyme BaeI because its recognition site is much more rare in all kinds of DNAs as compared to the DraI site and it leaves ends compatible with the Φ29 DNA origins (Fig. 6A). Thus, a plasmid called pEYFPORBae was constructed (see SI Text) such that a BaeI restriction site was inserted at the junction between the Φ29 DNA ends from pETORPHI. BaeI cuts at two different points in each strand of the DNA, separated by 33 bp, having its recognition site in the middle and cutting the DNA independently of the sequence at the corresponding scission points. The cut with BaeI leaves 3′ extensions of five nucleotides at the resulting ends (Fig. 6A). Until now the effect of the 3′ extensions attached to the Φ29 DNA origins on the amplification efficiency of the Φ29 replication machinery had not been reported and, therefore, three different approaches for the amplification reaction were tried: (i) performing the reaction under standard conditions after BaeI treatment (lane pEYFPORBae); (ii) pretreatment of the BaeI-cut plasmid with the Φ29 DNA polymerase in the presence of dNTPs to eliminate the 3′ extensions, and then adding the TP and the rest of the components of the reaction (lane pEYFPORBae-Pretreat); and (iii) pretreatment with the Klenow polymerase and dNTPs, also to generate blunt ends, and then adding the components to set a standard Φ29 DNA amplification reaction (lane pEYFPORBae-Klenow). As shown in Fig. 6B, the DNA yield with approaches i and iii was 50% of that obtained with the original pETORPHI. The pretreatment with the Φ29 DNA polymerase produced a lower amount of DNA, perhaps due to the strong exonuclease activity of this enzyme. As a control, when pEYFPORBae cut with BaeI was further cut with SacII to separate the two origins, the amount of product obtained was greatly decreased (Fig. S4).
Fig. 6.
A pETORPHI derivative linearized with BaeI is efficient in amplification. (A) Scheme of BaeI recognition (underlined) and cutting sites (arrows) and position of Φ29 origins (capital letters) in pEYFPORBae. (B) Plasmid pEYFPORBae was prepared as described in Materials and Methods. Aliquots of 25 ng of pEYFPORBae cut with the enzyme BaeI (from now on pEYFPORBae) were included in the different reactions as templates. The amplification reactions were set up as usual (lane pEYFPORBae), or by pretreating the digested plasmid 10 min with Φ29 DNA polymerase and 100 μM dNTPs and then adding TP (pEYFPORBae-Pretreat), or pretreating the plasmid 10 min with the Klenow enzyme and 30 μM dNTPs, inactivating the enzyme, and then adding the rest of the components of the reaction (pEYFPORBae-Klenow). The size of the amplified bands is shown: 5.8 kb for pETORPHI cut with DraI and 5.3 kb for pEYFPORBae cut with BaeI.
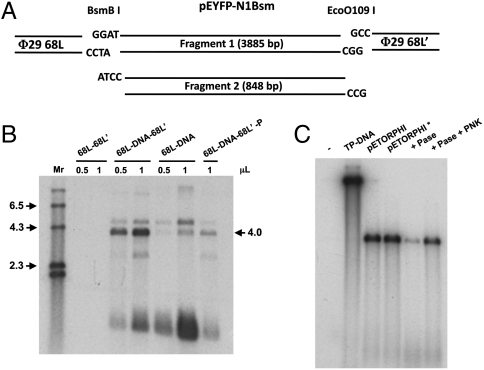
Having shown above that a fragment of 68 bp from the left end origin of Φ29 DNA is a functional amplification origin, we can design single-stranded oligonucleotides that, after hybridization, will generate the 68-bp fragment as dsDNA plus any extra sequence of interest, for example, single-stranded extensions to be ligated to foreign DNA ends obtained after digestion with restriction enzymes. By this procedure, one can generate Φ29 DNA “adapters” that join the Φ29 minimal origin to any linear DNA to be amplified with the Φ29 replication system. To test this approach, we introduced by site-directed mutagenesis a BsmBI site in plasmid pEYFP-N1 at position 4702. Then the pEYFP-N1Bsm plasmid was cut with the enzymes BsmBI and EcoO109I, which recognize unique sequences in that vector. We also used two sets of single-stranded synthetic oligonucleotides so that, after hybridization, each set would form the double-stranded 68-bp Φ29 minimal origin plus short single-stranded DNA extensions to ligate specifically to one of the two ends of pEYFP-N1Bsm obtained after restriction with BsmBI and with EcoO109I (Fig. 7A). The enzymes were chosen because they generate asymmetric ends, preventing self-ligation of the oligonucleotides bearing the half-site cohesive end and cross-ligation between the BsmBI and EcoO109I sites. In this way, each set of oligonucleotides, after hybridization, can ligate with the complementary restriction end of the plasmid but not with the other end, with the other set of oligonucleotides, or self-ligate. In addition, this approach makes it unnecessary to gel purify the DNA fragments or the ligation products. The use as template for amplification of the DNAs resulting from the ligase reaction would only allow true amplification from one of the ligation products, which is the 3,885-bp plasmid fragment (Fig. 7A) ligated to the two different 68-bp Φ29 DNA adapters. This product would generate a band of 4,021 bp corresponding to Φ29_68L-BsmBI-pEYFP-N1_fragment 1-EcoO109I-Φ29_68L′. The other fragment (fragment 2) would not be ligated to the adapters and thus would not be amplified. As shown in Fig. 7B, only the ligation containing all the correct components generates an amplified band of the right length (lane 68L-DNA-68L′), whereas reactions performed with ligation mixtures lacking one adapter (68L-DNA) gave clearly less signal, and control reactions containing only the adapters (68L-68L′) did not yield any amplified product. The band of higher molecular weight (4.6 kbp) that appears when 68L-DNA-68L′ or 68L-DNA are used as templates corresponds to plasmids cut with one enzyme and ligated to one type of adapter; these molecules should not serve as templates for amplification. Bands of lower molecular weight were detected, especially in the case of the DNA ligated with one adapter (68L-DNA). Low molecular weight DNA has been characterized as truncated molecules that become palindromic and so they acquire a duplicated replication origin (22). The results obtained show that it is possible to ligate Φ29 DNA minimal origins to a heterologous DNA to get TP-DNA of the right length amplified by the Φ29 replication system. Fig. 7B also shows that, when 5′ unphosphorylated oligonucleotides were used (lanes 68L-DNA-68L′-P), very low amplification was obtained. To confirm the importance of the 5′ phosphate group in amplification, the experiment shown in Fig. 7C was carried out. When pETORPHI cut with DraI was treated with Antarctic Phosphatase almost no amplification was obtained. Further treatment with T4 polynucleotide kinase restored the amplification close to the level obtained with pETORPHI or with pETORPHI treated as control.
Fig. 7.
Ligation of 68 bp from the Φ29 genome left end at the two ends of a heterologous DNA fragment supports amplification. (A) Scheme of DNA fragments present in the ligation reactions. The 68-bp Φ29 left end origins were assembled from single-stranded oligonucleotides bearing cohesive ends for the enzymes BsmBI (68L) or EcoO109I (68L′) as described in the text. The plasmid pEYFP-N1Bsm was cut with BsmBI and EcoO109I, generating the two fragments (1 and 2) indicated in the figure. Only fragment 1 has cohesive ends compatible with the Φ29 linkers. (B) Ligations were set up with either just one of the adapters plus the plasmid fragments 1 and 2 (see Materials and Methods) (lanes 68L-DNA), the two adapters 68L and 68L′, and the plasmid fragments (lanes 68L-DNA-68L′), or with the two types of adapters alone (68L-68L′). An aliquot of sample 68L-DNA-68L′ was left untreated with T4 polynucleotide kinase (PNK) (lanes 68L-DNA-68L′-P) (see Materials and Methods). Aliquots (0.5 or 1 μL) of the ligation mixtures were used as templates in the amplification reactions. (C) Aliquots of pETORPHI were either untreated (pETORPHI), treated with Antarctic Phosphatase (+ Pase), with PNK after the phosphatase (+Pase + PNK), or processed in the same way as the treated samples but without added enzymes (pETORPHI*). In all cases, the amount of pETORPHI in the reactions was 25 ng. The size of pETORPHI cut with DraI is 5.8 kb and the 68L-DNA-68L′ is 4.0 kb.
As mentioned before, no studies to develop in vitro TP-primed amplification of heterologous DNAs had been reported. Previous results showed that it was possible to initiate replication using dsDNAs that contain the ends of the genome without TP attached; however, the efficiency of the initiation/replication of these DNAs was much lower than that of the phage TP-DNA (20). In this work, we have shown that it is possible to obtain efficient amplification in vitro with the Φ29 DNA replication machinery using as template a linear DNA displaying Φ29 DNA replication origins, but not having the parental TP covalently linked at their ends. Two lines of evidence support the notion that the Φ29 system is producing true DNA amplification: One, the fact that the separation in two different molecules of the two Φ29 DNA origins greatly reduces the amount of product, and two, the kinetics of the reaction using pETORPHI as template is very similar to that obtained with TP-DNA as template. As shown in Fig. S5B, the DNA that can be inserted between the Φ29 origins for amplification is at least 18 kb. Furthermore, as mentioned above, the Φ29 DNA polymerase is very processive and over 70 kb of continuous replication has been obtained (12). Thus, the method we present here has the potential to perform very long amplifications.
Regarding the parental TP, in this work we have shown that it is not essential for amplification (Fig. 2A); however, it increases the amount of amplified product obtained and decreases the amount of template molecules needed. In addition, the amplification of pETORPHI was very dependent on the presence of p6, although that was not the case for TP-DNA under the experimental conditions used.
The minimal requirements for the in vitro amplification system in terms of proteins and DNA sequences are summarized in Fig. S6. The DNA sequences correspond to the Φ29 minimal 68 bp left origin. This 68-bp origin contains the last 12 bp from the Φ29 DNA end that are necessary for replication initiation and elongation, and it also contains a high-affinity p6 binding site corresponding to nucleotides 35 to 58 from the end. The phosphate group at position 5′ has also been shown to be a requirement for amplification. Our results also show that the p6 nucleation site is sufficient to obtain amplification in the presence of protein p6 and the minimal origin sequences, although a higher yield is obtained with the 194-bp origin.
The Φ29 system can efficiently generate amplified products from templates that have five nucleotides protruding at the 3′ end, as it is the case of plasmid pEYFPORBae linearized with BaeI. These 3′ end-protruding nucleotides were removed by the exonuclease activity of Φ29 DNA polymerase that would generate blunt ends identical to the genuine Φ29 replication origins. In order to expand the possibilities of the system, we have also shown that the 68-bp minimal replication origin can be attached to a foreign DNA by ligation and serve as amplification template.
The potential applications of a TP-primed amplification method based on the Φ29 replication system described here would be of two types. One, there would be the possibility to amplify in a defined way long linear DNAs, like viral genomes, chromosome fragments, or plasmids. Because the amplified DNAs have TP linked at the end, this TP can be removed, for example, by restriction digestion at the DNA ends, if the protein is unwanted for downstream applications. The other potential distinctive type of application of the method described in this work would be to generate in vitro high amounts of DNAs bound to TP for uses in which a hybrid protein–DNA molecule is required. One example of that would be the use of protein–DNA molecules as gene delivery vehicles, such that the DNA would be targeted to specific cells, ferried to subcellular compartments, and protected from exonucleases by the covalently linked protein in such a way that the TP is acting as a platform to which different targeting peptide modules are fused. In this sense, a previous report has shown that the expression of a CFP-TP fusion protein in vivo complements an infecting phage Φ29 mutant that cannot express the TP, indicating that fusion of proteins at the TP N terminus may not impair the TP function (23). Another use of the protein–DNA molecules could be based in the generation of enzyme-DNA aptamer hybrids that could exploit the advantages of both types of molecules. The enzyme would be fused to the TP that is utilized as a primer to amplify a short DNA that contains a sequence that folds into an aptamer-type structure. In the amplification process, multiple copies of the enzyme-TP would be linked to the aptamer-DNA synthesized. In the protein/DNA molecule (enzyme-TP/aptamer), the aptamer could recruit a given substrate on which the enzyme component would perform a specific reaction.
Regarding the biological activity of DNAs amplified with the TP-based Φ29 system, previous work showed that the quality of a TP-containing Φ29 DNA amplified in vitro in the presence of the minimal set of replication proteins (TP, DNA polymerase, SSB, and p6) is identical to the TP-containing DNA isolated from phage particles measured as the ability to produce viral progeny in a Bacillus subtilis protoplasts transfection assay (10). This supports the possible use of the system described in this paper to generate TP-DNAs with biological activity.
Concluding Remarks.
In summary, we describe a method to produce by amplification DNAs with TP covalently attached to the 5′ end using the in vitro Φ29 DNA replication system that includes four proteins and the DNA replication origins. Functional DNA origins do not need to have TP linked beforehand, can be between 194- and 68-bp long, can have protruding 3′ ends, and they require a phosphate group at the 5′ end. This method opens up possibilities in relation to amplification of DNA and the generation of hybrid protein–DNA molecules.
Materials and Methods
Nucleotides and DNAs.
Unlabeled nucleotides were purchased from General Electric Healthcare, and [α-32P]dATP [3,000 Ci/mmol (1 Ci = 37 GBq)] was obtained from Amersham Pharmacia. Oligonucleotides were obtained from Sigma-Genosys. Φ29 TP-DNA was prepared as described (24).
A DNA fragment consisting of DNA sequences corresponding to 191 and 194 bp from the left and right ends, respectively, of the Φ29 genome joined through the Φ29 DNA ends was obtained from Genescript cloned in plasmid pUC57 (pUC57ORPHI). The pETORPHI plasmid was constructed by cloning between sites SacI and HindIII of pET28b (Novagen) a DNA fragment excised from pUC57ORPHI using the same restriction enzymes. In this way the pETORPHI gets a unique DraI site, which is the one formed at the exact joint between the Φ29 origins of replication. Construction of plasmids pETORPHI68L, pETORPHIBae, and pEYFPORBae is described in SI Text. Digestion of the plasmids with the enzyme DraI was normally performed for 4 h at 37 °C in New England Biolabs Buffer 4 except for the cases where multiple digestions with enzymes DraI, MluI, and PvuI had to be made; in this case, New England Biolabs Buffer 3 plus 0.1 mg/mL of BSA were used. Digestion of plasmid pEYFPORBae with BaeI was done in New England Biolabs Buffer 4 supplemented with 0.1 mg/mL of BSA and 20 μM S-adenosylmethionine. After digestion with BaeI, the DNA was purified by Qiaquick Spin columns (Qiagen). All the enzymes were inactivated by incubating at 65 or 80 °C depending on the case. In all cases, the restriction cuts were confirmed by agarose gel electrophoresis. In the experiments that involved ligations, plasmid pEYFPBsm was cut with BsmBI and EcoO109I in New England Biolabs Buffer 3 for 8 h at 37 °C, purified by Qiaquick Spin columns (Qiagen), and then ligated in a 1∶4 proportion to the corresponding double-stranded hybridized oligonucleotides (oligonucleotides 68L + Comp68LBsm, 68L + Comp68LEco109; see SI Text, Oligonucleotides List). After inactivation of the ligase, the samples were treated with New England Biolabs T4 Polynucleotide Kinase, without changing buffer, for 30 min at 37 °C. After inactivation of the kinase, aliquots of the ligation reactions were used for amplification without any other treatment. Dephosphorylation was performed utilizing the Antarctic Phosphatase of New England Biolabs, adding its corresponding buffer and incubating at 37 °C for 15 min and then inactivating the enzyme by incubation for 20 min at 65 °C. After this treatment, the samples were ethanol precipitated with 20 μg of glycogen as carrier to eliminate the phosphatase buffer. After precipitation, the samples were resuspended in New England Biolabs Ligase buffer and used directly in the amplification experiments.
Protein Purification.
DNA polymerase and SSB were purified as described in Lázaro et al. (25) and Soengas et al. (26), respectively. TP and protein p6 were purified as described in SI Text.
Amplification Assay.
The incubation mixture contained, in 25 μL, Buffer MRI 1X [50 mM Tris·HCl (pH 7.5), 10 mM MgCl2, 5% glycerol, 1 mM DTT, 0.1 mg/mL BSA] plus ammonium sulfate to 20 mM final concentration, 100 μM each of the four dNTPs plus [α-32P]dATP (1 μCi), 7.5 μg Φ29 SSB, 1.5 μg Φ29 p6, 10 ng Φ29 DNA polymerase, and 20 ng Φ29 TP unless otherwise stated, and the indicated amounts of the corresponding type of DNA. Reactions were allowed to proceed for 2 h at 22 °C and stopped by addition of EDTA to 10 mM and SDS to 0.1% final concentrations, respectively. Then the samples were filtered through Sephadex G-50 columns equilibrated in 10 mM Tris·HCl (pH 7.5), 1 mM EDTA, and 0.1% SDS, and after that the samples were dried, resuspended in 50 μL of 0.5 M NaOH, and analyzed by electrophoresis in 0.7% agarose gels run in the presence of 30 mM NaOH and 1 mM EDTA. A typical result out of at least three independent experiments is shown. The radioactivity incorporated (cpm) was measured with an LKB Wallac 1219 Rackbeta liquid scintillation counter. Quantitation of gel bands was performed on BAS1000 phosphorimager equipment. The synthesized DNA was determined calculating the ratio of radioactivity incorporated into DNA with respect to total radioactivity present in the reaction and, using that ratio, the total mass of dNMPs incorporated into DNA was calculated by multiplying the initial mass of dNTPs present in the reaction by the ratio incorporated/total obtained from the radioactive nucleotide. Lambda phage DNA cut with HindIII, used as molecular weight marker, was labeled by incubation for 25 min at 25 °C with the Klenow enzyme and 50 μM each of dCTP, dGTP, and dTTP plus [α-32P]dATP (1 μCi) in Buffer EcoPol (New England Biolabs).
Supplementary Material
Acknowledgments.
We are grateful to José M. Lázaro for advice in protein p6 and TP purifications and for critical reading of the manuscript, and Laurentino Villar for technical assistance. This investigation has been aided by Grants BFU2008-00215/BMC and CONSOLIDER-INGENIO CSD2007-00015 from the Spanish Ministry of Science and Innovation (to M.S.), Grant S2009MAT-1507 from Madrid Autonomous Community (to M.S.), Grant PET2007-0160 from the Spanish Ministry of Education and Science (to M.d.V.), and by an institutional grant from Fundación Ramón Areces to the Centro de Biología Molecular “Severo Ochoa”. P.G. was recipient of a Collaboration Fellowship from the Spanish Ministry of Education.
Footnotes
Conflict of interest statement: A patent application related to this work has been filed for which M.M., P.G., M.d.V., and M.S. are inventors.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114397108/-/DCSupplemental.
References
- 1.Salas M. Protein-priming of DNA replication. Annu Rev Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- 2.Meijer WJ, Horcajadas JA, Salas M. Φ29 family of phages. Microbiol Mol Biol Rev. 2001;65:261–287. doi: 10.1128/MMBR.65.2.261-287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bamford DH, Caldentey J, Bamford JK. Bacteriophage PRD1: A broad host range DSDNA tectivirus with an internal membrane. Adv Virus Res. 1995;45:281–319. doi: 10.1016/s0065-3527(08)60064-0. [DOI] [PubMed] [Google Scholar]
- 4.Garcia P, Martin AC, Lopez R. Bacteriophages of Streptococcus pneumoniae: A molecular approach. Microb Drug Resist. 1997;3:165–176. doi: 10.1089/mdr.1997.3.165. [DOI] [PubMed] [Google Scholar]
- 5.Chang PC, Kim ES, Cohen SN. Streptomyces linear plasmids that contain a phage-like, centrally located, replication origin. Mol Microbiol. 1996;22:789–800. doi: 10.1046/j.1365-2958.1996.01526.x. [DOI] [PubMed] [Google Scholar]
- 6.Liu H, Naismith JH, Hay RT. Adenovirus DNA replication. Curr Top Microbiol Immunol. 2003;272:131–164. doi: 10.1007/978-3-662-05597-7_5. [DOI] [PubMed] [Google Scholar]
- 7.Salas M, de Vega M. Bacteriophage protein-primed DNA replication. In: Hefferon KL, editor. Recent Advances in DNA Virus Replication. Ithaca, NY: Research Signpost; 2006. pp. 259–288. [Google Scholar]
- 8.Shiffman D, Cohen SN. Reconstruction of a Streptomyces linear replicon from separately cloned DNA fragments: Existence of a cryptic origin of circular replication within the linear plasmid. Proc Natl Acad Sci USA. 1992;89:6129–6133. doi: 10.1073/pnas.89.13.6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crouzet J, et al. Recombinational construction in Escherichia coli of infectious adenoviral genomes. Proc Natl Acad Sci USA. 1997;94:1414–1419. doi: 10.1073/pnas.94.4.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanco L, Lázaro JM, de Vega M, Bonnin A, Salas M. Terminal protein-primed DNA amplification. Proc Natl Acad Sci USA. 1994;91:12198–12202. doi: 10.1073/pnas.91.25.12198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salas M, et al. Protein-nucleic acid interactions in bacteriophage Φ29 DNA replication. FEMS Microbiol Rev. 1995;17:73–82. doi: 10.1111/j.1574-6976.1995.tb00189.x. [DOI] [PubMed] [Google Scholar]
- 12.Blanco L, et al. Highly efficient DNA synthesis by the phage Φ29 DNA polymerase. Symmetrical mode of DNA replication. J Biol Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- 13.Salas M, de Vega M. Replication of bacterial viruses. In: Mahy BWJ, Regenmortel MHVv, editors. Encyclopedia of Virology. Oxford: Elsevier; 2008. pp. 339–406. [Google Scholar]
- 14.Serrano M, et al. Phage Φ29 protein p6: A viral histone-like protein. Biochimie. 1994;76:981–991. doi: 10.1016/0300-9084(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 15.Blanco L, et al. Effect of NH4+ ions on Φ29 DNA-protein p3 replication: Formation of a complex between the terminal protein and the DNA polymerase. J Virol. 1987;61:3983–3991. doi: 10.1128/jvi.61.12.3983-3991.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutiérrez J, et al. Signals in the Φ29 DNA-terminal protein template for the initiation of phage ϕ29 DNA replication. Virology. 1986;155:474–483. doi: 10.1016/0042-6822(86)90209-6. [DOI] [PubMed] [Google Scholar]
- 17.Méndez J, Blanco L, Salas M. Protein-primed DNA replication: A transition between two modes of priming by a unique DNA polymerase. EMBO J. 1997;16:2519–2527. doi: 10.1093/emboj/16.9.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serrano M, Gutiérrez J, Prieto I, Hermoso JM, Salas M. Signals at the bacteriophage Φ29 DNA replication origins required for protein p6 binding and activity. EMBO J. 1989;8:1879–1885. doi: 10.1002/j.1460-2075.1989.tb03584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutiérrez J, Garmendia C, Salas M. Characterization of the origins of replication of bacteriophage Φ29 DNA. Nucleic Acids Res. 1988;16:5895–5914. doi: 10.1093/nar/16.13.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Méndez J, Blanco L, Esteban JA, Bernad A, Salas M. Initiation of Φ29 DNA replication occurs at the second 3′ nucleotide of the linear template: A sliding-back mechanism for protein-primed DNA replication. Proc Natl Acad Sci USA. 1992;89:9579–9583. doi: 10.1073/pnas.89.20.9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serrano M, Gutiérrez C, Salas M, Hermoso JM. Superhelical path of the DNA in the nucleoprotein complex that activates the initiation of phage Φ29 DNA replication. J Mol Biol. 1993;230:248–259. doi: 10.1006/jmbi.1993.1140. [DOI] [PubMed] [Google Scholar]
- 22.Esteban JA, Blanco L, Villar L, Salas M. In vitro evolution of terminal protein-containing genomes. Proc Natl Acad Sci USA. 1997;94:2921–2926. doi: 10.1073/pnas.94.7.2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muñoz-Espin D, Holguera I, Ballesteros-Plaza D, Carballido-Lopez R, Salas M. Viral terminal protein directs early organization of phage DNA replication at the bacterial nucleoid. Proc Natl Acad Sci USA. 2010;107:16548–16553. doi: 10.1073/pnas.1010530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peñalva MA, Salas M. Initiation of phage Φ29 DNA replication in vitro: Formation of a covalent complex between the terminal protein, p3, and 5′-dAMP. Proc Natl Acad Sci USA. 1982;79:5522–5526. doi: 10.1073/pnas.79.18.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lázaro JM, Blanco L, Salas M. Purification of bacteriophage Φ29 DNA polymerase. Methods Enzymol. 1995;262:42–49. doi: 10.1016/0076-6879(95)62007-9. [DOI] [PubMed] [Google Scholar]
- 26.Soengas MS, Gutiérrez C, Salas M. Helix-destabilizing activity of Φ29 single-stranded DNA binding protein: Effect on the elongation rate during strand displacement DNA replication. J Mol Biol. 1995;253:517–529. doi: 10.1006/jmbi.1995.0570. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.