Abstract
Batrachochytrium dendrobatidis (Bd) is a globally ubiquitous fungal infection that has emerged to become a primary driver of amphibian biodiversity loss. Despite widespread effort to understand the emergence of this panzootic, the origins of the infection, its patterns of global spread, and principle mode of evolution remain largely unknown. Using comparative population genomics, we discovered three deeply diverged lineages of Bd associated with amphibians. Two of these lineages were found in multiple continents and are associated with known introductions by the amphibian trade. We found that isolates belonging to one clade, the global panzootic lineage (BdGPL) have emerged across at least five continents during the 20th century and are associated with the onset of epizootics in North America, Central America, the Caribbean, Australia, and Europe. The two newly identified divergent lineages, Cape lineage (BdCAPE) and Swiss lineage (BdCH), were found to differ in morphological traits when compared against one another and BdGPL, and we show that BdGPL is hypervirulent. BdGPL uniquely bears the hallmarks of genomic recombination, manifested as extensive intergenomic phylogenetic conflict and patchily distributed heterozygosity. We postulate that contact between previously genetically isolated allopatric populations of Bd may have allowed recombination to occur, resulting in the generation, spread, and invasion of the hypervirulent BdGPL leading to contemporary disease-driven losses in amphibian biodiversity.
Keywords: chytridiomycosis, infectious disease, extinction, epidemiology
Emerging fungal diseases present a growing threat to the biodiversity of free-ranging animal species (1, 2). In recent years, a single species within a basal clade of fungi little recognized for their pathogenicity, the Chytridiomycota, has gained substantial notoriety owing to its impact on global amphibian biodiversity (1). Batrachochytrium dendrobatidis (Bd) is known to have driven the local extinction (extirpation) of up to 40% of species in affected communities (3), and to have spread rapidly through diverse environments (1). Despite widespread research efforts, the geographic origin of this emerging infection and its subsequent patterns of global spread remain largely unknown (2, 4, 5). For example, the hypothesis that Bd originated in Africa and spread via the global trade in Xenopus spp. during the first half of the 20th century (6) is disputed by the detection of lower genetic diversity in African isolates of Bd compared with isolates from North America (4). This observation, however, was based on only five African isolates collected from two sites in the South African Cape, compared with 29 US isolates collected from multiple sites across the United States. The genetically depauperate nature of African Bd has been further challenged by the discovery of isolates of African descent exhibiting pronounced differences in morphology and virulence (5, 7).
A separate marker-based study conducted by Morehouse et al. (8) using 10 loci from 35 isolates found very low levels of polymorphism (five variable positions) and fixed heterozygous sites, suggesting a primarily clonal mode of reproduction, although with some evidence for spatially localized genetic recombination (9). These results, and other marker-based studies (4, 6), support the novel pathogen hypothesis, by suggesting that Bd is a recently emerged pathogen (10). Although these results describe the population structure of Bd at a coarse scale, patchily sampled genomes combined with a chronic lack of genetic diversity at the sequenced loci have prevented a reliable inference of Bd's evolutionary history. Recently, new whole-genome typing methods have greatly increased our ability to decipher genealogies by enabling unbiased sampling of the entire genome, thus increasing our power to date the coalescence of lineages and to identify recombination events. Here, we compare the whole genomes of 20 isolates of Bd to examine the recent evolutionary history of Bd and its patterns of global genome diversity.
Results
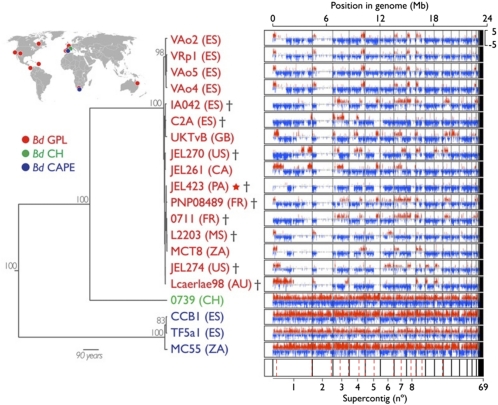
Using ABI's SOLiD system (sequencing by oligonucleotide ligation and detection), we achieved high-density coverage (mean 9.5× deep) of the 24-Mb genome for 20 globally distributed (Europe, North and Central America, South Africa, Australia) isolates of Bd from 11 amphibian host species. Eight isolates were from regions where epizootics have been documented (Table 1). We aligned the reads to the genome sequence of isolate JEL423 (http://www.broadinstitute.org/; 11) and searched for discrepancies using a depth-dependent binomial method (SI Appendix, Figs. S1 and S2), finding in total 51,915 nonredundant homozygous SNPs and 87,121 nonredundant heterozygous positions (SI Appendix, Figs. S3 and S4). Of these, 21% of the homozygous SNP positions and 19% of the heterozygous positions (22 Kb total) were covered ≥4 reads in all 20 samples and were used for phylogenetic analysis. Sixteen of the 20 samples, including the reference strain JEL423, were >99.9% genetically identical and fell within a single highly supported clade (Fig. 1 and SI Appendix, Fig. S5 and Table S1). This “global panzootic lineage” (BdGPL) includes all previously genotyped isolates of Bd and all of the isolates in our panel that are associated with regional epizootics, recovered from five continents (4).
Table 1.
The samples used and details of alignments
| Collection site | Amphibian host | Year | Collector | Culture reference | Reads aligned (millions) | Depth (X) |
| America, Colorado | B. boreas | 1999 | JEL | JEL274 | 4.7 | 6.0 |
| America, PtReyes | R. catesbeiana | 1999 | JEL | JEL270 | 9.4 | 12.0 |
| Australia, Rockhampton | L. caerulea | 1999 | LB | Lcaerulae98 | 10.1 | 12.9 |
| Canada, Quebec | R. catesbeiana | 1999 | JEL | JEL261 | 7.0 | 9.0 |
| England, Kent | L. vulgaris | 2007 | MF | U.K.TvB | 8.3 | 10.6 |
| France, Ansabere | A. obstetricans | 2007 | MF | 0711/1 | 6.3 | 8.1 |
| France, Lac Arlet | A. obstetricans | 2008 | MF | PNP08489 | 6.0 | 7.7 |
| Mallorca, Cocó de sa Bova | A. muletensis | 2007 | MF | CCB1 | 10.5 | 13.4 |
| Mallorca, Torrent des Ferrerets | A. muletensis | 2007 | MF | TF5a1 | 4.3 | 5.4 |
| Montserrat | L. fallax | 2009 | TG | L2203 | 5.8 | 7.5 |
| Panama, Guabal | P. lemur | 2004 | JEL | JEL423 | 10.5 | 13.5 |
| South Africa | A. fuscigula | 2009 | CW | MCT8 | 5.5 | 7.0 |
| South Africa | H. natalensis | 2009 | CW | MC55 | 9.3 | 11.9 |
| Spain, Ibon Acherito | A. obstetricans | 2004 | TG | IA042 | 6.9 | 8.8 |
| Spain, Penalara | A. obstetricans | 2002 | MF | C2A | 11.6 | 14.8 |
| Spain, Valencia | A. obstetricans | 2008 | MF | VAo2 | 6.3 | 8.0 |
| Spain, Valencia | A. obstetricans | 2008 | MF | VAo4 | 8.0 | 10.3 |
| Spain, Valencia | A. obstetricans | 2008 | MF | VAo5 | 7.4 | 9.5 |
| Spain, Valencia | R. perezi | 2008 | MF | VRp1 | 6.8 | 8.7 |
| Switzerland | A. obstetricans | 2007 | TG | 739 | 6.2 | 8.0 |
Bd isolates and locations that were resequenced. The first four columns provide information for the recommended naming scheme outlined by Berger et al. (18). The number of reads (millions) aligning to the Bd JEL423 genome assembly and the corresponding depth of coverage. Amphibian hosts include Afrana fuscigula (Cape river frog), Alytes muletensis (Mallorcan midwife toad), Alytes obstetricans (common midwife toad), Bufo boreas (Western toad), Hadromophryne natalensis (natal ghost frog), Litoria caerulea (green tree frog), Lissotriton vulgaris (smooth newt), Litoria fallax (Eastern dwarf tree frog), Peltophryne lemur (Puerto Rican toad), Rana catesbeiana (American bullfrog), Rana perezi (Iberian green frog). JEL, Joyce Longcore; LB, Lee Berger; MF, Matthew Fisher; TG, Trent Garner; and CW, Che Weldon.
Fig. 1.
Phylogenetic analysis of the 20 resequenced Bd mitochondrial genomes demonstrates three divergent lineages. The locations of the isolates belonging to the different lineages are shown using the same colors as in the phylogeny. Each genome is represented to the Right of the phylogeny. A nonoverlapping sliding window of SNPs minus heterozygous positions across the genome illustrates regions where heterozygosity predominates (blue) and where homozygosity predominates (red), illustrating the hallmark loss-of-heterozygosity events in the pan-global BdGPL lineage. The block below the 20 genomes denotes the supercontigs with black lines and the GARD recombination breakpoints are shown in red dotted lines. The star signifies the reference genome JEL423; crosses represent isolates that have been recovered from epizootics.
The remaining four newly sampled isolates form two novel, deeply divergent highly supported lineages. The “Cape lineage” (BdCAPE) includes two isolated from the island of Mallorca, and one from the Cape Province, South Africa. This clade supports the hypothesis of Walker et al. (5) that spillover of Bd from captive Cape clawed frogs (Xenopus gilli) into Mallorcan midwife toads (Alytes muletensis) led to the introduction of Bd onto the island through a captive breeding and reintroduction program for this endangered species. The discovery of BdCAPE as a separate lineage to previously genotyped isolates demonstrates that multiple emergences of amphibian chytridiomycosis have occurred, as well as confirming that the trade of amphibians is directly responsible for at least one of these emergences. A third novel lineage, “Swiss lineage,” (BdCH) is composed of a single isolate derived from a common midwife toad (Alytes obstetricans) from a pond near the village of Gamlikon, Switzerland. Further sampling is necessary to establish whether BdCH is a European-endemic isolate or more broadly distributed.
To ascertain whether genomic features were associated with these lineages, we searched for evolutionary differences by looking for mutation biases, copy number variation (CNV), and genes undergoing diversifying selection in the BdCAPE and BdCH lineages (SI Appendix, Figs. S6–S13) compared with BdGPL. We found that most (89%) of the polymorphisms in complementary determining sequence (CDS) regions caused a transition (A:T↔G:C) with a mean Ts/Tv of 8.03 with no lineage-specific biases. Synonymous mutations accounted for most of the polymorphisms in the CDS (ratio of 2.21 to nonsynonymous) concordant with the fact that most transitional mutations at twofold-degenerate sites are synonymous. Using the DoS measure of selection proposed by Stoletski and Eyre-Walker (12) and dN/dS ratios (13), we identified 114 genes in the BdCAPE and BdCH lineages that were significantly different from neutral expectations or had a dN/dS > 1 (SI Appendix, Fig. S9). Most of these genes had no significant blastp matches in the National Center for Biotechnology Information (NCBI) nr database and were otherwise indistinguishable from the rest of the transcriptome. We also detected no evidence for deviations in CNV between lineages using average read depth over each gene as a proxy.
Although no obvious interlineage variation was uncovered by these analyses, clear differences emerged when we examined the locations of the polymorphic sites within the genome. We identified a highly uneven distribution of homozygous SNPs and heterozygous positions across the 16 BdGPL isolates (Fig. 1), which could be observed using a range of window sizes (SI Appendix, Fig. S14). Furthermore, this pattern was found to be unique to the BdGPL (SI Appendix, Fig. S15) and was relatively similar between isolates. For example, we observed a region of low heterozygosity spanning supercontig 2 that was common to all isolates within BdGPL. To investigate whether this clustered distribution was due to recombination events, we tested whether the observed pattern was explained by multiple different evolutionary histories using the GARD (genetic algorithm for recombination detection) method (14). We identified 26 recombination breakpoints across the nuclear genome, 14 of which remained significant after Kishino–Hasegawa (KH) testing (SI Appendix, Fig. S16 and Table S3). Seven of these breakpoints were found within chromosomes and seven were found occurring between chromosomes. This suggests that the independent assortment of individual chromosomes expected under a model of frequent meiosis has not eroded the congruent phylogenetic signal between chromosomes, suggesting that meiosis is rare. Taken together, these data support the hypothesis of a single hybrid origin of BdGPL via an ancestral meiosis as proposed by James et al. (4).
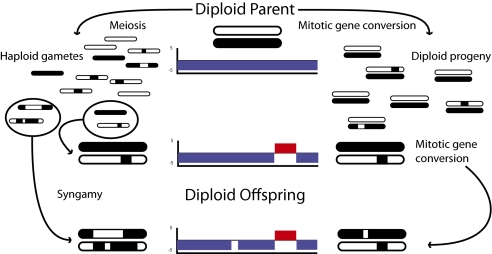
To further characterize the differences between the 15 significantly different recombination segments, we reconstructed separate topologies for each segment, but sharing the same model of evolution, using Bayesian phylogenetic inference techniques implemented in BEAST v.1.6.1 (15) (SI Appendix, Fig. S16). The three distinct lineages were recovered in all 15 segment trees, thereby ruling out recombination between them. In contrast, the topologies among recombination segments within BdGPL were markedly different from one another. This pattern can be explained by two alternative biological explanations: (i) different segments have different genealogies reflecting meiotic events or (ii) a highly heterozygous ancestor has since undergone recurrent mitotic gene conversion events at different places in the genome (Fig. 2). Although we cannot formally reject either hypothesis, rare meiotic recombination is the more parsimonious explanation for the observed pattern. In particular, four of our sequenced isolates (VRp1, VAo4, VAo2, and VAo5) originate from the same pond (near Valencia, Spain) at the same time point. The mitochondrial tree (Fig. 1), which is arguably expected to reflect the species tree best given its nonrecombining nature, gives high posterior support for this group being monophyletic. It therefore seems likely that the Valencia isolates originated from a single ancestor introduced to the pond. However, only 10 of the 15 gene trees (shown in SI Appendix, Fig. S20) give posterior support for this group being monophyletic, with consistently different groupings observed for the other cases. For this pattern to be compatible with a model of gene conversion, as described by hypothesis 2, we would need to assume that the population that colonized the pond near Valencia had levels of diversity comparable to BdGPL as a whole, and that this diversity was lost through rapid gene conversion only after the local colonization event. Distinguishing between these two hypotheses awaits the experimental determination of relative rates of meiotic versus mitotic recombination.
Fig. 2.
Two possible mechanisms of achieving the uneven distribution of heterozygous and homozygous SNPs throughout the Bd genome. Each black and white bar represents a haplotype identified in a parental isolate, and the charts in the Middle represent the plots illustrating regions where heterozygosity predominates (blue) and where homozygosity predominates (red for homozygous SNPs and empty for homozygous identical to reference). On the Left of the diagram, meiosis generates recombinant haploid genomes that then are united via syngamy into new diploid offspring with patchy heterozygosity. Meiosis involving fusion of diploid gametes could result in similar patterns if chromosomal segregation remains independent. On the Right of the diagram, mitotic gene conversion generates patches of homozygous sites via homologous DNA repair in diploid progeny.
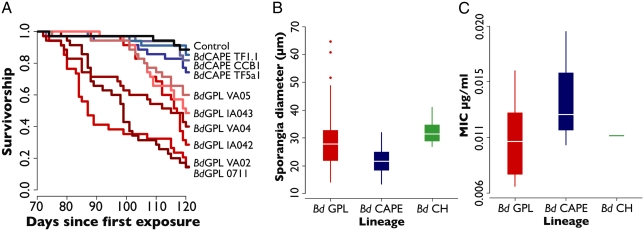
To ascertain whether Bd virulence recapitulated phylogeny, we experimentally exposed common toad (Bufo bufo) tadpoles to repeated, high-concentration doses of Bd using multiple isolates from the two lineages with replication (BdGPL and BdCAPE) and from two clusters within BdGPL (Valencia and the Pyrenees). Toads exposed to BdCAPE experienced significantly reduced infection and mortality compared with those exposed to BdGPL (Fig. 3A and SI Appendix, Fig. S17). Significant variation in virulence was also detected among isolates within the BdGPL (Fig. 1). Environmental conditions were standardized and animals were kept in isolation from each other precluding any effect of environmental forcing. The possibility does exist that the observed variation among treatments where BdGPL isolates were used could be due to variation in host susceptibility, as mass at metamorphosis and infection dynamics of individual toadlets both played a role in determining mortality. Nevertheless, any effect of host, or the effects of variation among isolates from BdGPL on virulence, is minor compared with the observed effect of lineage on postmetamorphic mortality.
Fig. 3.
Kaplan–Meier survival curves illustrating postmetamorphic survival of animals exposed to BdGPL isolates (red) and BdCAPE lineage isolates (blue). (A) BdGPL isolates were significantly different from both BdCAPE lineage isolates and the negative controls. (B) Significant differences in sporangia size between the three lineages. (C) No significant difference was observed between the three lineages in minimum inhibitory concentration values.
The discovery of BdCH came too late for this lineage to be included in our in vivo experimental framework. However, extensive population surveillance of Bd infected and uninfected populations across Switzerland has demonstrated a lack of association between infection status and population decline (http://www.bd-maps.net/maps); if BdCH is representative of the lineages infecting other areas of Switzerland, then this is circumstantial evidence that BdCH, like BdCAPE, is hypovirulent. Previous studies of morphological variation among Bd isolates have shown that phenotypic profiles are linked to the virulence of isolates (7) and that isolates belonging to BdCAPE exhibit smaller sporangial sizes compared with isolates belonging to BdGPL. We repeated this study using a greater number of BdGPL isolates from other continents and including BdCH. These data (Fig. 3B and SI Appendix, Figs. S18 and S19) show that BdCAPE does exhibit significantly smaller sporangia and increased hyphal length relative to BdGPL; however, these characters are not diagnostic for the two lineages. BdCH exhibits larger sporangia than the other two lineages, but no difference in hyphal length. Together, these data show that genetic differentiation between lineages of Bd has resulted in significant morphological variation. However, we did not detect differences in sensitivity to itraconazole among lineages (Fig. 3C and SI Appendix, Fig. S20 and Table S2).
Surveillance data shows that Bd is increasing its global range, and rapid emergence across geographic regions has been documented in the United States (16), Central America (17), and Australia (10). Eight of our sequenced isolates are associated with rapid population/community extirpations in diverse biomes across the New World, Europe, and Australia (3), exemplified by the rapid and virtual extirpation of the critically endangered mountain chicken frog Leptodactylus fallax in Montserrat by BdGPL isolate L2203. The widespread occurrence of BdGPL across five continents suggests that the emergence of this lineage is responsible for these Bd-driven collapses in diversity. To date the emergence of this lineage, we took advantage of the fact that the samples were collected over a period of 10 y and used this to inform a strict clock across the phylogenies of the 15 breakpoint segments (SI Appendix, Fig. S20). The median dates for the emergence of the global hypervirulent lineage range from 35 to 257 y before present (ybp) (SI Appendix, Table S4). Amphibian declines were first noted in the 1970s simultaneously in the United States (Sierra Nevadas), Central America, and Australia (18, 19); therefore, our dating suggests that the globalization of BdGPL is compatible with its spread within the amphibian trade.
Discussion
One of the more puzzling aspects of the emergence of amphibian chytridiomycosis has been that, whereas epizootics have been widely observed, many susceptible amphibian communities apparently coexist alongside Bd with no evidence of disease. Such coexistence has been attributed to the context-dependent nature of susceptibility to disease, and whereas this is undoubtedly an important factor (20), our data show that Bd genotype is also an important epidemiological determinant. Here, we found that there is a much greater diversity of Bd than was previously recognized, and that multiple lineages are being vectored between continents by the trade of amphibians (BdGPL and BdCAPE). We have characterized hypervirulence in BdGPL, suggesting that the emergence and spread of chytridiomycosis largely owes to the globalization of this recently emerged recombinant lineage (21).
Research has recently suggested the existence of a Bd lineage that is associated with the Japanese giant salamander, Andrias japonicus (22). This lineage, defined by sequencing a short fragment of the ribosomal DNA, is dissimilar to the rDNA sequences of BdGPL, which nonnative North American bullfrogs have introduced to Japan. Therefore, it appears that we can now provisionally recognize at least four lineages of Bd, two of which are possibly endemic (BdCH and Japan), one of which may have been previously endemic to South Africa but was then vectored to Mallorca (BdCAPE), and one of which has a pan-global distribution (BdGPL). This diversity was uncovered from sampling only 20 genomes from a cohort biased toward sampling amphibian populations experiencing chytridiomycosis (and hence infected with the BdGPL); therefore, our data suggest that a more extensive diversity of amphibian-associated chytrid lineages is pending discovery. Whether these are B. dendrobatidis sensu stricto, or represent cryptic species, remains to be determined. However, observations that populations infected with non-BdGPL lineages do not undergo epizootics suggest that the diversity of Bd represents a patchwork of genetically and phenotypically diverse lineages. In this case, determining the geographical origin(s) of the parental genotypes of BdGPL will likely remain an elusive challenge until a much wider sample of amphibian-associated chytrid lineages has been sampled to identify the full phylogeographic range of diversity held within the order Rhizophydiales.
The origin of novel virulence in fungal species via recombination/hybridization is a well-recognized pathway underpinning disease emergence for increasing numbers of plant and animal pathogens (1, 23). Genomic rearrangement between allopatric fungal lineages that have not evolved reproductive barriers is promoted when anthropogenically mediated dispersal increases the rate of lineage mixing. The resulting novel interlineage recombinants exhibit a diversity of virulence profiles, some of which can initiate epidemics; contemporary examples include the evolution of hypervirulence in the Vancouver Island outbreak of Cryptococcus gattii (24), and many novel aggressive plant pathogens that increasingly threaten global food security (25) as well as natural populations (26).
We postulate that the anthropogenic mixing of allopatric lineages of Bd has led to the generation of the hypervirulent BdGPL via an ancestral meiosis, and that, as previously suggested (9), this lineage is undergoing further diversification by either mitotic or sexual recombination. We show that the global trade in amphibians is resulting in contact and cross-transmission of Bd among previously naive host species, resulting in intercontinental pathogen spread. As the rate of interlineage recombination between fungi will be proportional to their contact rates, we predict that such globalization will increase the frequency that recombinant genotypes are generated. Theory and experimentation has shown that in genetically diverse infections, virulent lineages can have a competitive advantage, resulting in increased transmission (27, 28). As a consequence, we predict the evolution of further hypervirulent fungal lineages across a diverse range of host species and biomes in the absence of tighter biosecurity (1).
Materials and Methods
Full details are given in SI Appendix. Libraries were constructed according to the protocols provided by Life Technologies (Fragment Library kit). Fragment library sequencing was performed on two flowcells on an Applied Biosystems SOLiD 3 machine. Two pools of libraries were required: pool 1 contained libraries 1–8 with barcodes 1–8, and pool 2 contained libraries 9–20 with barcodes 1–12 (Table 1). Pooled barcoded libraries were united, and ePCR was performed according to Life Technologies’ (“full scale”) specification (templated bead preparation kits). After a run, a total amount of 260–290 million beads was loaded onto the flowcells. The output read length was 50 bp. The genome sequence and feature file for the chytrid fungus Batrachochytrium dendrobatidis (Bd) strain JEL423 was downloaded from http://www.broadinstitute.org/ (GenBank project accession no. AATT00000000). The feature file for JEL423 had all but the longest splice variants removed for each gene leaving 8,794/8,819 genes. We trimmed the ABI SOLiD reads to 30 nt to remove low-quality bases from the 3′-end and aligned to the nuclear genome and mitochondrial sequence using Burrows-Wheeler Aligner (BWA) v0.5.8 (29) with default parameters.
The method we used for SNP calling was chosen after assessing the false discovery rate of 97 different settings and three separate methods, and was based on three depth-dependent binomial distributions (SI Appendix, Fig. S2B). For each base in the genome, we asked, given the number of bases agreeing with the reference base (k), and the depth of coverage (n), and a set prior probability of finding the right base, 90% (p), what is its probability f(k; n, p)? We considered that over any given nucleotide, there are three potential circumstances, each with its own probabilities: (i) agree with reference/homozygous agree P(0.9), (ii) heterozygous allele P(0.45), and (iii) disagree with ref/homozygous SNP P(0.03). Therefore, the nucleotide identified by which possible allele had the highest probability. In addition, we required a minimum depth of at least four reads to be considered different or the same as the reference (SI Appendix, Fig. S2C).
Trees were made using Bayesian Monte Carlo Markov chain (MCMC) analysis implemented by BEAST v1.6.1 (15), with the Hasegawa–Kishino–Yano substitution model with a γ-distribution of rates, under a strict clock. After discarding the first 10% as burn-in, we ran three separate chains of over 50 million generations, sampling every 1,000th generation. We combined the estimated topologies from the three different runs to construct the maximum clade consensus tree shown in Fig. 1. GARD analysis was run on the alignment of all variable and covered positions in the genome, with ambiguous character codes used to represent heterozygous SNPs. The analysis treated such positions as partially missing data during phylogenetic analyses. General time reversible (30) model of nucleotide substitution with a four-bin general discrete distribution to account for site-to-site rate variation was used to evaluate the goodness-of-fit for competing models. The best model was selected by small-sample corrected Akaike information criterion (31, 32). Because recombination was detected in the nuclear genomes, we used the unweighted pair group method with arithmetic mean (UPGMA) algorithm in phylogenetic analysis using parsimony (PAUP) to construct a dendrogram for the nuclear genome.
Experimental assessment of host response to challenge by Bd isolates were done according and subject to ethical review at the Institute of Zoology and Imperial College and licensed by the UK Home Office. Common toad (B. bufo) larvae (Gosner stage 24, Gosner 1960) hatched from 12 egg strings were randomly allocated to 1 of 10 experimental treatment groups (9 Bd isolates and 1 negative control). The experiment was undertaken in a climate-controlled (approximately 16 °C constant temperature) room with a 12:12 h day/night light schedule. Tadpoles in Bd+ treatments were exposed to high doses (3,000–17,000 active zoospores per exposure in liquid media) of Bd zoospores every 4 d for a total of eight times. Zoospore counts and volume of media were standardized among isolates for each exposure. Tadpoles in the negative control treatment were exposed on the same schedule to an equivalent volume of sterile media as was used for Bd+ treatments, but lacking any Bd. All animals surviving to the end of the experiment were humanely killed.
Photography of Bd used in assessing phenotypic differences between lineages was conducted using a Canon EOS 350D (3,456 × 2,304 pixel field). Initial photographs were taken of the floor of the tissue culture flask where sporangia had settled 3 d postculture. We subcultured the isolates into 12-well tissue culture plates (Nunc), with three replicas per isolate at a concentration of ∼8,000 zoospores per well (calculated using an improved Neubauer hemocytometer). The bottoms of the wells were photographed at 10, 15, and 20 d postinitial culture. Two to three images were obtained from each well for each isolate. Using ImageJ software (33) we measured the diameter of the largest sporangia contained in the field of view. We determined the MC50 to itraconazole for 12 isolates (estimated 5,750 Bd zoospores in 100 μL of culture) using concentrations of itraconazole ranging from 0.07 μg/mL to 2.1875 ng/mL (7). Each isolate was incubated at 18 °C and replicated three times. Optical densities were read on days 3, 5, 8, 10, and 12 using a BioTek Absorbance microplate reader (ELx808), using an absorbance of 450 nm.
Supplementary Material
Acknowledgments
We thank Joyce Longcore for supplying isolates of Bd that we sequenced in this project, Beni Schmidt and Ursina Tobler for facilitating the collection of animals from which BdCH was isolated, and Andrew Rambaut and Philippe Lemey for advice on the phylogenetic analysis. The government of Montserrat issued permits for the isolation and export of Bd from L. fallax. Computational analysis was supported by the University of California San Diego's Center for AIDS Research BEAST Core (National Institutes of Health AI 036214). SOLiD sequencing was performed by International Society for Systems Biology and Centre for Integrative Systems Biology, Imperial College. This project was funded by the UK Natural Environmental Research Council Grant NE/E006701/1, the UK Department for Environment, Food and Rural Affairs Grant FC1195, the Biotechnology and Biological Sciences Research Council Grant BB/H008802/1, the European Research Council Grant 260801-BIG_IDEA, and the Biodiversa project RACE: Risk Assessment of Chytridiomycosis to European Amphibian Biodiversity (http://www.bd-maps.eu).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: Sequences are deposited in the NCBI Short Read Archive (accession no. SRA030504).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111915108/-/DCSupplemental.
References
- 1.Fisher MC, Garner TWJ, Walker SF. Global emergence of Batrachochytrium dendrobatidis and amphibian chytridiomycosis in space, time, and host. Annu Rev Microbiol. 2009;63:291–310. doi: 10.1146/annurev.micro.091208.073435. [DOI] [PubMed] [Google Scholar]
- 2.Frick WF, et al. An emerging disease causes regional population collapse of a common North American bat species. Science. 2010;329:679–682. doi: 10.1126/science.1188594. [DOI] [PubMed] [Google Scholar]
- 3.Crawford AJ, Lips KR, Bermingham E. Epidemic disease decimates amphibian abundance, species diversity, and evolutionary history in the highlands of central Panama. Proc Natl Acad Sci USA. 2010;107:13777–13782. doi: 10.1073/pnas.0914115107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.James TY, et al. Rapid global expansion of the fungal disease chytridiomycosis into declining and healthy amphibian populations. PLoS Pathog. 2009;5:e1000458. doi: 10.1371/journal.ppat.1000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker SF, et al. Invasive pathogens threaten species recovery programs. Curr Biol. 2008;18:R853–R854. doi: 10.1016/j.cub.2008.07.033. [DOI] [PubMed] [Google Scholar]
- 6.Weldon C, du Preez LH, Hyatt AD, Muller R, Spears R. Origin of the amphibian chytrid fungus. Emerg Infect Dis. 2004;10:2100–2105. doi: 10.3201/eid1012.030804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher MC, et al. Proteomic and phenotypic profiling of the amphibian pathogen Batrachochytrium dendrobatidis shows that genotype is linked to virulence. Mol Ecol. 2009;18:415–429. doi: 10.1111/j.1365-294X.2008.04041.x. [DOI] [PubMed] [Google Scholar]
- 8.Morehouse EA, et al. Multilocus sequence typing suggests the chytrid pathogen of amphibians is a recently emerged clone. Mol Ecol. 2003;12:395–403. doi: 10.1046/j.1365-294x.2003.01732.x. [DOI] [PubMed] [Google Scholar]
- 9.Morgan JA, et al. Population genetics of the frog-killing fungus Batrachochytrium dendrobatidis. Proc Natl Acad Sci USA. 2007;104:13845–13850. doi: 10.1073/pnas.0701838104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skerratt LF, et al. Spread of chytridiomycosis has caused the rapid global decline and extinction of frogs. EcoHealth. 2007;4:125–134. [Google Scholar]
- 11.Fisher MC, Stajich J, Farrer RA. Emergence of the chytrid fungus Batrachochytrium dendrobatidis and global amphibian declines. In: Heitman J, Sibley D, Howlett B, editors. Evolution of Virulence in Eukaryotic Microbes. Washington, DC: American Society for Microbiology; 2012. in press. [Google Scholar]
- 12.Stoletzki N, Eyre-Walker A. Estimation of the neutrality index. Mol Biol Evol. 2011;28:63–70. doi: 10.1093/molbev/msq249. [DOI] [PubMed] [Google Scholar]
- 13.Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- 14.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW. GARD: A genetic algorithm for recombination detection. Bioinformatics. 2006;22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- 15.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vredenburg VT, Knapp RA, Tunstall TS, Briggs CJ. Dynamics of an emerging disease drive large-scale amphibian population extinctions. Proc Natl Acad Sci USA. 2010;107:9689–9694. doi: 10.1073/pnas.0914111107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lips KR, et al. Emerging infectious disease and the loss of biodiversity in a Neotropical amphibian community. Proc Natl Acad Sci USA. 2006;103:3165–3170. doi: 10.1073/pnas.0506889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berger L, et al. Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of Australia and Central America. Proc Natl Acad Sci USA. 1998;95:9031–9036. doi: 10.1073/pnas.95.15.9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houlahan JE, Findlay CS, Schmidt BR, Meyer AH, Kuzmin SL. Quantiative evidence for global amphibian population declines. Nature. 2000;412:499–500. doi: 10.1038/35008052. [DOI] [PubMed] [Google Scholar]
- 20.Walker SF, et al. Factors driving pathogenicity vs. prevalence of amphibian panzootic chytridiomycosis in Iberia. Ecol Lett. 2010;13:372–382. doi: 10.1111/j.1461-0248.2009.01434.x. [DOI] [PubMed] [Google Scholar]
- 21.Fisher MC, Garner TWJ. The relationship between the introduction of Batrachochytrium dendrobatidis, the international trade in amphibians and introduced amphibian species. Fungal Biol Rev. 2007;21:2–9. [Google Scholar]
- 22.Goka K, et al. Amphibian chytridiomycosis in Japan: Distribution, haplotypes and possible route of entry into Japan. Mol Ecol. 2009;18:4757–4774. doi: 10.1111/j.1365-294X.2009.04384.x. [DOI] [PubMed] [Google Scholar]
- 23.Stukenbrock EH, McDonald BA. The origins of plant pathogens in agro-ecosystems. Annu Rev Phytopathol. 2008;46:75–100. doi: 10.1146/annurev.phyto.010708.154114. [DOI] [PubMed] [Google Scholar]
- 24.Fraser JA, et al. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437:1360–1364. doi: 10.1038/nature04220. [DOI] [PubMed] [Google Scholar]
- 25.Strange RN, Scott PR. Plant disease: A threat to global food security. Annu Rev Phytopathol. 2005;43:83–116. doi: 10.1146/annurev.phyto.43.113004.133839. [DOI] [PubMed] [Google Scholar]
- 26.Goss EM, Carbone I, Grünwald NJ. Ancient isolation and independent evolution of the three clonal lineages of the exotic sudden oak death pathogen Phytophthora ramorum. Mol Ecol. 2009;18:1161–1174. doi: 10.1111/j.1365-294X.2009.04089.x. [DOI] [PubMed] [Google Scholar]
- 27.Nowak MA, May RM. Superinfection and the evolution of parasite virulence. Proc Biol Sci. 1994;255:81–89. doi: 10.1098/rspb.1994.0012. [DOI] [PubMed] [Google Scholar]
- 28.de Roode JC, et al. Virulence and competitive ability in genetically diverse malaria infections. Proc Natl Acad Sci USA. 2005;102:7624–7628. doi: 10.1073/pnas.0500078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tavaré S. Some probabilistic and statistical problems on the analysis of DNA sequences. Lect Math Life Sci. 1986;17:57–86. [Google Scholar]
- 31.Sugiura N. Further analysis of the data by Akaike's information criterion and the finite corrections. Comm Statist Theory Methods. 1978;7:13–26. [Google Scholar]
- 32.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol. 2006;23:1891–1901. doi: 10.1093/molbev/msl051. [DOI] [PubMed] [Google Scholar]
- 33.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.