Abstract
Cilia-based locomotion is the major form of locomotion for microscopic planktonic organisms in the ocean. Given their negative buoyancy, these organisms must control ciliary activity to maintain an appropriate depth. The neuronal bases of depth regulation in ciliary swimmers are unknown. To gain insights into depth regulation we studied ciliary locomotor control in the planktonic larva of the marine annelid, Platynereis. We found several neuropeptides expressed in distinct sensory neurons that innervate locomotor cilia. Neuropeptides altered ciliary beat frequency and the rate of calcium-evoked ciliary arrests. These changes influenced larval orientation, vertical swimming, and sinking, resulting in upward or downward shifts in the steady-state vertical distribution of larvae. Our findings indicate that Platynereis larvae have depth-regulating peptidergic neurons that directly translate sensory inputs into locomotor output on effector cilia. We propose that the simple circuitry found in these ciliated larvae represents an ancestral state in nervous system evolution.
Keywords: neural circuit, zooplankton, sensory-motor neuron, FMRFamide-related peptides
Two different types of locomotor systems are present in animals, one muscle based and the other cilia based. The neuronal control of muscle-based motor systems is well understood from studies on terrestrial model organisms. In contrast, our knowledge of the neuronal control of ciliary locomotion is limited, even though cilia-driven locomotion is prominent in the majority of animal phyla (1).
Ciliary swimming in open water is widespread among the larval stages of marine invertebrates, including sponges, cnidarians, and many protostomes and deuterostomes (2–5). Freely swimming ciliated larvae often spend days to months as part of the zooplankton (1, 6). The primary axis for ciliated plankton is vertical, and body orientation is maintained either by passive (buoyancy) or active (gravitaxis, phototaxis) mechanisms. When cilia beat, larvae swim upward, and when cilia cease beating, the negatively buoyant larvae sink. During swimming, the thrust exerted on the body is proportional to the beating frequency of cilia (7–9). The alternation of active upward swimming and passive sinking, together with swimming speed and sinking rate, is thought to determine vertical distribution in the water (8). Because several environmental parameters, including water temperature, light intensity, and phytoplankton abundance, change with depth, swimming depth will influence the speed of larval development, the magnitude of UV damage, and the success of larval feeding and settlement. To stay at an appropriate depth, planktonic swimmers must therefore sense environmental cues and regulate ciliary beating.
The ciliated larvae of the marine annelid Platynereis dumerilii provide an accessible model for the study of ciliary swimming in marine plankton (10). Platynereis can be cultured in the laboratory, and thousands of synchronously developing larvae can be obtained daily year-round (11). Platynereis has emerged as a model for the study of the evolution of development (evo-devo) and neurobiology. Its neuronal development is more representative of the ancestral bilaterian condition than that of conventional protostome models (flies, nematodes) and shares many features with vertebrate neurodevelopment (12). Platynereis has also retained ancestral neuron types, including ciliary photoreceptors and vasotocin–neurophysin-producing sensory–neurosecretory cells, shared with vertebrates but absent from flies and nematodes (13, 14). Such conservation makes Platynereis an interesting model for the reconstruction of the ancestral state of the bilaterian nervous system. The larval nervous system of Platynereis also shows surprising simplicity in its circuitry. The photoreceptor cell of the larval eyespot was shown to directly synapse on the ciliated cells and regulate phototactic turning (10). Such a sensory-motor system, directly regulating cilia, may be a relic from the earliest stages of the evolution of eyes and neural circuits (10, 15).
Planktonic ciliated larvae also adjust their ciliary activity in response to several environmental cues other than light (16–20). It is unclear, however, how other cues affect cilia and whether the innervation of ciliary bands by other neurons is as simple as that of the larval eyespots. Anatomical studies have revealed that larval ciliary bands receive extensive innervation from the nervous system, both in Platynereis and in other species (21). In protostome larvae, neurons expressing the neuropeptide FMRF-amide often contribute to this innervation (22–24). Neurons with related F-amide neuropeptides also innervate ciliary bands in sea urchin larvae (25), suggesting that neuropeptides may have a general role in the regulation of larval locomotion in both protostomes and deuterostomes. However, these limited studies have not revealed the general neural circuit architecture of ciliated larvae and the role of neuropeptides in regulating ciliary swimming.
Neuropeptides are considered the oldest neuronal signaling molecules in animals (26). They are produced from inactive precursor proteins by proteolytic cleavage and further processing (e.g., amidation) (27–29) and are released into the hemolymph to act as hormones or at synapses to regulate target cells. Neuropeptides have a wide range of functions in the control of neural circuits and physiology, including the modulation of locomotion and rhythmic pattern generators (30–33), presynaptic facilitation and remodeling of sensory networks (34, 35), and the regulation of reproduction (36, 37). We have only limited information about the role of neuropeptides in the regulation of ciliary beating (38, 39).
To gain further insights into ciliary locomotor control we characterized neuropeptide functions and the associated neural circuits in the larvae of Platynereis. We found diverse neuropeptides expressed in larval sensory neurons that directly innervate the ciliary band. Application of synthetic neuropeptides altered both ciliary beat frequency and the rate and duration of ciliary arrests, indicating that neuropeptides act as neurotransmitters on the ciliated cells. The neuropeptide-induced changes in ciliary activity altered the swimming trajectories and shifted the vertical distribution of the larvae. Our results suggest that in planktonic swimmers, neuropeptides released upon sensory stimulation at neurociliary synapses modulate ciliary activity, ultimately resulting in changes in swimming depth. The control of ciliary swimming, the ancestral form of animal locomotion, by a simple sensory-motor nervous system suggests that these locomotor circuits may represent an ancestral stage of neural circuit evolution.
Results
Identification of Platynereis Neuropeptides.
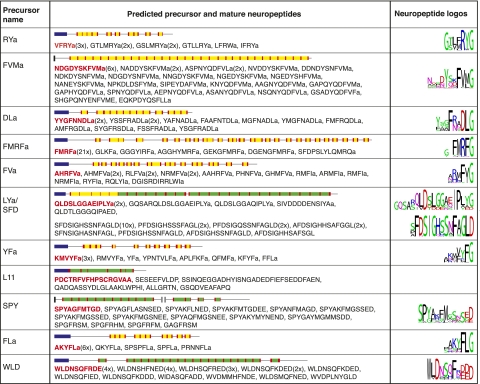
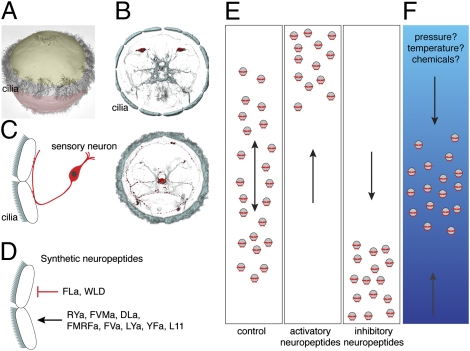
Given the widespread role of neuropeptides in regulating animal locomotion (30–32), we set out to characterize neuropeptides of the Platynereis larval nervous system. Here we describe 11 neuropeptide precursors identified in a Platynereis larval transcriptome resource using a combination of BLAST and pattern searches. On the basis of the 11 precursor sequences we predicted 120 Platynereis neuropeptides forming 11 distinct groups of similar peptides (Fig. 1). Full-length precursor sequences have an N-terminal signal peptide and contain repetitions of similar short neuropeptide sequences flanked by dibasic cleavage sites (KR, RK, or KK) for prohormone convertases (27, 28). We deduced the structure of mature Platynereis neuropeptides using NeuroPred (40) and manual curation (Fig. 1). In 8 precursors most peptides contain a Gly residue before the dibasic cleavage site. These peptides are expected to be further processed by α-amidating enzymes (29) and to terminate in an α-amide (RYa, FVMa, DLa, FMRFa, FVa, LYa, YFa, and FLa; “a”, “amide”). Other precursors give rise to peptides with a carboxyl terminus (L11, SPY, and WLD).
Fig. 1.
Neuropeptide precursors and their predicted neuropeptides in Platynereis. Schematic drawings of Platynereis neuropeptide precursors are shown with the location of the signal peptide (blue), the amidated (yellow) and nonamidated (green) neuropeptides, and cleavage sites (red). Sequences and the number of neuropeptides predicted from each precursor are shown. Sequence logos were generated on the basis of alignments of all neuropeptides from one precursor. The amidation signature C-terminal Gly is included in the logos. For the L11 precursor no logo is shown because the peptides are not similar. Neuropeptides used for the pharmacological experiments and immunizations (with an extra N-terminal Cys) are shown in red. For SPY and WLD we did not obtain a working antibody.
The Platynereis neuropeptide precursors are related to the widely distributed family of RFamide–like neuropeptide precursors. Members of this family are present in all eumetazoans, and their mature peptides can have diverse functions in the regulation of neuronal circuits (31, 37, 41–44). Many Platynereis neuropeptides have close relatives in other lophotrochozoan species (mollusks and annelids) and some also outside the lophotrochozoans (RYa, RFa, and L11). Conservation of amidated neuropeptides is restricted to a few residues N-terminal to the cleavage site and amidation signature (Fig. S1). Amidated neuropeptides within the same precursor protein also show higher conservation close to the amidated terminus (Fig. 1), indicating that the functionally important residues are located there. In contrast, nonamidated neuropeptides often show stronger N-terminal conservation both between species and within the same precursor (Fig. 1 and Fig. S1).
Neuropeptides Are Expressed in Distinct Sensory Neurons in the Platynereis Larval Nervous System.
To map neuropeptide expression in the Platynereis larval nervous system we performed in situ hybridizations on whole larvae with precursor-specific RNA probes. We combined in situ hybridization with fluorescent antibody staining using an anti-acetylated tubulin antibody that stains stabilized microtubules in cilia, axons, and dendrites. The samples were scanned in a confocal microscope combining fluorescence and reflection microscopy, a technique that visualizes the in situ hybridization signal [alkaline phosphatase/nitroblue tetrazolium (NBT)/5-bromo-4-chloro-3-indolyl phosphate (BCIP) staining] by reflection of the laser beam (45). Using this technique we obtained high-resolution three-dimensional information on the expression patterns and also on the morphology of the labeled cells.
We found that all precursors were expressed 48 h postfertilization (hpf) in the larval episphere (the anterior part of the larva) in a cell-type–specific manner (Fig. 2) in patterns that were largely invariant for the same gene from larva to larva. For all precursors, expression was detected in 2–12 neurons in the larval episphere.
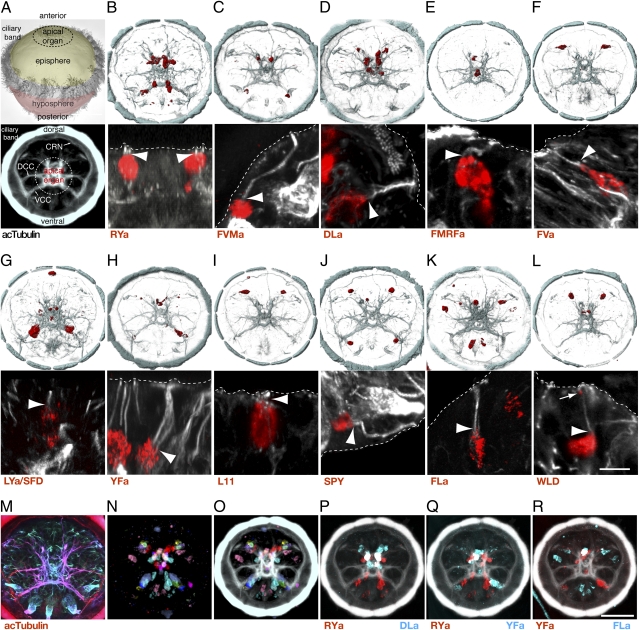
Fig. 2.
Expression of neuropeptide precursors in Platynereis larval sensory cells. (A) SEM image of a Platynereis larva with the main body regions indicated (Upper) and anterior view of the larval axonal scaffold with the main nerves indicated (Lower). (B–L) Whole-mount in situ hybridization for RYa (B), FVMa (C), DLa (D), FMRFa (E), FVa (F), LYa/SFD (G), YFa (H), L11 (I), SPY (J) FLa (K), and WLD (L) neuropeptide precursor mRNAs (red) counterstained with anti-acTubulin antibody (cyan, Upper; white, Lower). (Lower) close-up images of neuropeptide-expressing sensory cells. (M) Apical sensory dendrites (in cyan) in a depth-encoded acetylated tubulin confocal stack of a 48-hpf larva. (N and O) Expression of all neuropeptides (N) in the larval episphere, projected on a common reference scaffold (O) by image registration. (P–R) Overlap of RYa and DLa (P), RYa and YFa (Q) and YFa and FLa (R) expression, determined by image registration. All images are of 48-hpf larvae. A is a dorsal view, and B–R are anterior views. The in situ signal (red) labels the cell bodies. (Lower) Arrowheads point to the base of the apical sensory dendrites. Arrow in L points to WLD signal localized to the tip of the dendrite. Dashed lines indicate the apical margin of the neuroepithelium. [Scale bars: B–L (Lower), 5 μm; and B–L (Upper) and M–R, 50 μm.] CRN, ciliary ring nerve; DCC, dorsal branch of the circumesophageal connectives; VCC, ventral branch of the circumesophageal connectives.
To obtain a molecular map of all peptidergic neurons at cellular resolution we calculated an average expression pattern for each neuropeptide on the basis of three to five confocal scans and then registered these averages to a reference axonal scaffold. This procedure is possible because Platynereis larvae, like many spiralians, have a strict cell lineage (46) and there is very little cellular-level variation among larvae of the same developmental stage. For image registration we used a procedure similar to the one recently described for Platynereis (47), taking advantage of the highly stereotypic acetylated-tubulin reference channel.
Placing these average expression maps into the common acetylated-tubulin reference (Fig. 2A) revealed that neuropeptides are expressed broadly in the larval nervous system (Fig. 2 N and O and Movie S1). Many peptidergic neurons concentrated in the central region of the episphere, the apical organ (Fig. 2 B–E, N, and O). We further defined a bilaterally arranged dorso-lateral cluster of peptidergic neurons, in the region of the developing cerebral eyes, and a ventro-lateral cluster, above the ventral branch of the circumesophageal connectives (Fig. 2 F–L). The SD of cell center positions among different registered scans for the same gene ranged between 0.6 and 3.34 μm (median = 2.63; along the x axis), whereas the average cell diameter was 11.1 μm (SD = 4.11; along the x axis). This cellular resolution allowed us to show that most neuropeptides were not coexpressed and therefore define distinct peptidergic neurons in the Platynereis larva. We found only three pairs of neuropeptides with partially overlapping expression. DLa and RYa were coexpressed in three apical organ cells, whereas YFa overlapped with both FLa and RYa in two different cells of the apical organ (Fig. 2 P–R).
To characterize the morphology of the neuropeptidergic cells we performed high-resolution confocal scans of the NBT/BCIP signal together with the acetylated tubulin signal. The acetylated tubulin signal revealed that the larval episphere forms a polarized neural tissue with apical dendrites and a basal axonal scaffold (Fig. 2M). The neuropeptide precursors were all expressed in neurons with differentiated morphology, with cell bodies continuing in basally projecting axons that joined the axonal scaffold of the larval nervous system. Most of the peptidergic neurons were flask shaped and also had a dendrite extending apically (Fig. 2 B–L). Such flask-shaped sensory morphology has already been described with immunolabeling for the FMRFa-expressing cells in the Platynereis larva that we localize here with the FMRFa precursor (14). The ultrastructural reconstruction of these cells revealed that the apical dendrite has two sensory cilia. These cilia extend into the subcuticular space that has access to the external environment (14). Similar sensory cell morphologies have also been described ultrastructurally in various other annelids (48). The peptidergic sensory cells we found also had acetylated-tubulin positive projections that reach the apical-most side of the neuroepithelium and therefore are in direct contact with the external environment. The dendrites of different cells had terminals with various apical morphological modifications, including split (Fig. 2 B, D, and H) and bulbous endings (Fig. 2C), indicating that the neuropeptidergic neurons have various sensory modalities. We also observed that the mRNA of the WLD precursor localized to the tip of the sensory dendrite (Fig. 2L). We did not find apical dendrites on the large ventral LYa-expressing cells (Fig. 2G); these cells therefore likely do not have a sensory function.
Overall, our neuropeptide expression data revealed a cellular resolution molecular map of sensory neurons in the 48-hpf Platynereis larva.
Peptidergic Sensory Neurons Directly Innervate the Larval Ciliary Band.
Maturing neuropeptides can be transported to synaptic release sites via axonal transport. To visualize axonal projections of Platynereis sensory neurons, we developed antibodies against one mature peptide from each precursor (Fig. 1). We affinity purified the antibodies using the respective peptides immobilized to a resin and then performed immunostainings on 48-hpf larvae. The immunostainings revealed cell body and axonal signals for all neuropeptides. We always found a strong correlation in the number and position of the cell bodies between our in situ hybridization and immunostaining data, showing that the antibodies do not cross-react and label the same cells that express the respective precursors. (Fig. 3; compare with Fig. 2 and Movies S2 and S3). Although the cell body labeling was weak for FVMa and L11 (Fig. 3 B and H), upon inspection of 3D renderings of the data (Movies S2 and S3) we could nevertheless see them in positions corresponding to precursor expression. Using the FMRFa antibody, in addition to the two cells that also showed precursor expression (Fig. 2E) we observed three faintly labeled cells. Similarly, for LYa we observed two faint cells located more dorsally. These signals could be due either to the higher sensitivity of antibody stainings or to weak unspecific labeling.
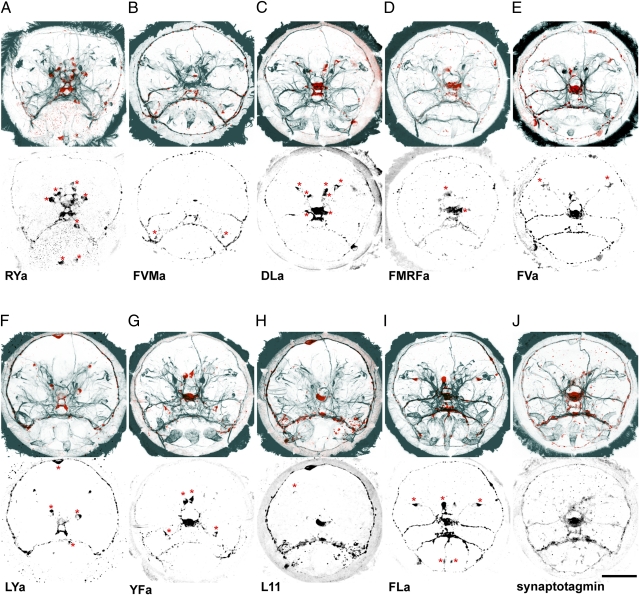
Fig. 3.
Neuropeptidergic sensory neurons directly innervate larval ciliary bands. Immunostainings with RYa (A), FVMa (B), DLa (C), FMRFa (D), FVa (E), LYa (F), YFa (G), L11 (H), FLa (I), and synaptotagmin (J) antibodies are shown in red (Upper) and in black (Lower), counterstained with anti-acTubulin (cyan). Labeled cell bodies that correspond to the cells labeled with in situ hybridization are marked with an asterisk. All images are anterior views of 48-hpf larvae. (Scale bar: 50 μm.)
We also observed staining of the axonal projections of the neuropeptidergic sensory neurons. Axons for all neuron types run along the ventral branch of the circumesophageal connectives (Fig. 3 A–I) and in some cases also along the dorsal branch (FVMa, DLa, FVa, YFa, and FLa; Fig. 3 B, C, E, G, and I). Axons joined the ciliary ring nerve (CRN) innervating the main ciliary band of the larva (Fig. 3 A–I; compare with Fig. 2A) and formed several varicose thickenings along its entire length (Fig. 3 A–I), indicating en passant synaptic contacts to the ciliated cells. Direct synaptic contact between ciliated cells and the ciliary ring nerve has also been described by electron microscopy for the larval eye photoreceptor axon (10).
To confirm that the ciliary ring nerve has synaptic zones all along its length and not only where the eyespot photoreceptor axon forms synapses (10) we generated an antibody against Platynereis synaptotagmin (12), a transmembrane protein involved in the fusion of synaptic vesicles and large dense-cored vesicles (49, 50). Anti-synaptotagmin antibody staining revealed strong signal in the median nervous system, along the circumesophageal connectives, and along the entire length of the ciliary ring nerve (Fig. 3J).
Many neuropeptides (DLa, FMRFa, FVa, YFa, L11, and FLa) also showed intense labeling in the apical neurosecretory plexus of the larva, indicating that they may also be released here (Fig. 3 C–E and G–I). A neurosecretory function has already been suggested for FMRFa (14). The innervation of the ciliary band by the diverse neuropeptidergic sensory neurons suggests that these cells, in addition to having a possible neurosecretory function, are involved in the direct motor control of the ciliary band and that there is extensive peptidergic regulation during ciliary swimming.
Neuropeptides Influence Ciliary Beating and Arrests.
The innervation of the ciliary band by neuropeptidergic cells suggested that neuropeptides could directly influence ciliary activity. To uncover neuropeptide effects we characterized ciliary activity using video microscopy on immobilized Platynereis larvae. We scored ciliary beat frequency and the rate and duration of ciliary arrests. Control larvae (50–60 hpf) beat with their cilia with a frequency of 16.2 beats/s (SEM = 0.26, n = 146 larvae). Complete arrests of all cilia occurred at regular intervals and lasted for several seconds (Movie S4 and Fig. 4E). The ratio of the total duration of arrested to beating periods was 0.22 (SEM = 0.02, n = 28 larvae).
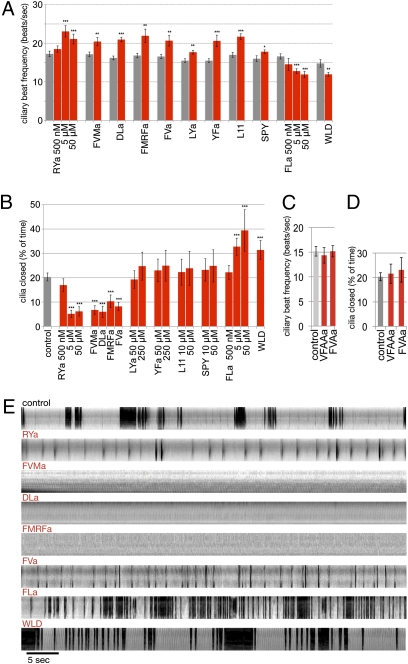
Fig. 4.
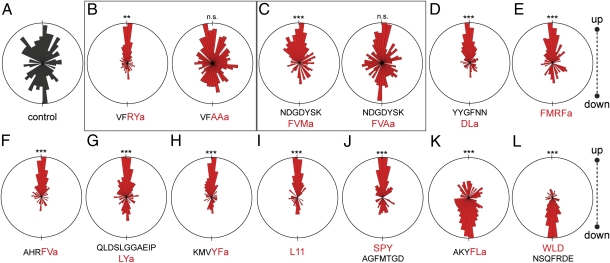
Neuropeptides regulate ciliary beating and arrests. (A) Ciliary beat frequency in the presence of neuropeptides. Gray bars represent untreated controls from the same batch of larvae. (B) Quantification of ciliary arrests in the presence of neuropeptides. (C) Ciliary beat frequency in control larvae (pooled from several batches) and in the presence of the Ala substituted neuropeptides. (D) Quantification of ciliary arrests in the presence of the Ala substituted neuropeptides. For B and D the same pooled control is shown. (E) Representative kymographs of ciliary beating (light gray sections) and ciliary arrests (dark gray sections) in the presence of neuropeptides, generated from 1-min videos on single immobilized larvae. The kymographs were generated from line selections perpendicular to the beating cilia (compare Movie S4). In A–D data are shown as mean ± SEM. P values of an unpaired t test are indicated: *P < 0.05; **P < 0.01; and ***P < 0.001. n > 10 larvae for A–D. In A the final concentrations are indicated for RYa and FLa; for the other peptides we used 5 μM (DLa), 10 μM (L11 and SPY), 20 μM (FVMa and WLD), and 50 μM (FMRFa, FVa, LYa, and YFa). In B we used concentrations as indicated or 5 μM (DLa), 20 μM (FVMa and WLD), and 50 μM (FMRFa and FVa), and in C and D we used 5 μM (VFAAa) and 20 μM (FVAa). For the full peptide sequences used see Fig. 1.
We next tested the effect of synthetic neuropeptides on ciliary beat frequency and ciliary arrests (500 nM to 250 μM concentration range, see Fig. 4 legend). Nine neuropeptides increased ciliary beat frequency (RYa, FVMa, DLa, FMRFa, FVa, LYa, YFa, L11, and SPY), and two decreased ciliary beat frequency (FLa and WLD, Fig. 4A).
The neuropeptides also affected ciliary arrests as quantified by the total duration of arrests during 1-min recordings (Fig. 4B) and visualized by kymographs (Fig. 4E and Movie S4). In the presence of RYa, FVMa, DLa, FMRFa, and FVa, ciliary arrests were strongly reduced (Fig. 4 B and E and Movie S4). In contrast, FLa and WLD led to very frequent and sustained arrests (Fig. 4 B and E and Movie S4). LYa, YFa, L11, and SPY had no effect on ciliary arrests even at concentrations five times higher than the concentrations significantly increasing ciliary beat frequency (Fig. 4B). These effects were dose dependent as determined for RYa and FLa (Fig. 4 A and B).
Next we tested the importance of conserved C-terminal residues in mature neuropeptides. Whereas the naturally occurring RYa (full sequence: VFRYa) increased ciliary beat frequency and abolished sustained ciliary arrests (Fig. 4 A, B, and E), changing the RY residues to Ala (VFAAa) resulted in a loss of these effects (Fig. 4 C and D). FVMa (full sequence: NDGDYSKFVMa) also increased ciliary beat frequency and inhibited arrests (Fig. 4 A, B, and E), but its effects were lost when the C-terminal Met residue was changed to Ala (Fig. 4 C and D).
These experiments show that Platynereis neuropeptides have strong and sequence-specific effects on two parameters of larval ciliary activity, ciliary beat frequency and ciliary arrests.
Neuropeptides Regulate Depth in the Water Column.
It has been proposed that changes in ciliary beat frequency and ciliary arrests regulate the depth of ciliary planktonic swimmers (8). To quantitatively test this hypothesis and to analyze the effects of neuropeptides on larval swimming behavior, we developed a vertical migration setup consisting of 25-cm–high tubes. This setup allowed us to record the swimming activity and the steady-state vertical distribution of large populations of larvae under different conditions.
First, we recorded swimming larvae at high resolution and found that they swam with a right-handed spiral, with their anterior end pointing upward. Occasionally, larvae sank with their anterior end pointing up, indicating that the center of gravity is closer to the posterior end (Movie S5). The analysis of swimming tracks in populations of untreated control larvae revealed that active upward and lateral swimming and passive sinking were balanced (no average displacement in the vertical direction). The angular plots of the displacement vectors of larval tracks showed a broad bimodal distribution, with the majority of tracks pointing upward or downward (Fig. 5A). Such nonbiased upward and downward displacement in the vertical tubes maintained a uniform vertical distribution as a steady state (Fig. 6A).
Fig. 5.
Neuropeptides regulate larval swimming directions. Angular plots are shown of the displacement vectors of larval swimming tracks for (A) control larvae and (B–L) larvae in the presence of the indicated neuropeptides. P values of a χ2-test comparing the number of upward and downward swimming larvae are indicated: **P < 0.01; ***P < 0.001. For each peptide the χ2-test was performed with a measurement on control larvae from the same batch. In A, a representative control is shown. n > 121 larvae for all measurements. The final concentrations were 5 μM (RYa, AAa, DLa, and FLa), 10 μM (L11 and SPY), 20 μM (FVMa, FVAa, and WLD), and 50 μM (FMRFa, FVa, LYa, and YFa).
Fig. 6.
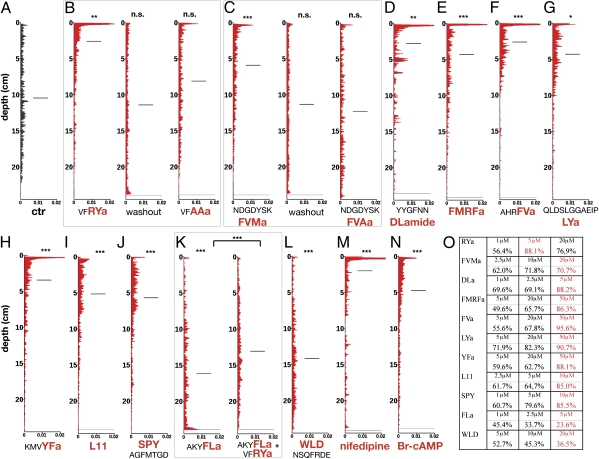
Neuropeptides regulate larval vertical distribution. Vertical distribution of control larvae (A) and larvae in the presence of VFRYa, washout, and VFAAa (B); NDGDYSKFVMa, washout, and NDGDYSKFVAa (C); YYGFNNDLa (D); FMRFa (E); AHRFVa (F); QLDSLGGAEIPLYa (G); KMVYFa (H); PDCTRFVFHPSCRGVAA or L11 (I); SPYAGFMTGD (J); AKYFLa and AKYFLa+VFRYa (K); WLDNSQFRDE (L); nifedipine (10 μM) (M); and Br-cAMP (5 μM) (N). (O) Percentage of larvae in the upper half of the tubes in the presence of different peptide concentrations. The concentrations shown in red were used in the other experiments. The horizontal lines in A–N indicate the mean. P values of a χ2-test are indicated: *P < 0.05; **P < 0.01; and ***P < 0.001. n > 69 larvae. The final concentrations of neuropeptides in B–L were 5 μM (RYa, AAa, DLa, and FLa), 10 μM (L11 and SPY), 20 μM (FVMa, FVAa, and WLD), and 50 μM (FMRFa, FVa, LYa, and YFa).
Next, we tested how changes in ciliary beat frequency and ciliary arrests induced by the neuropeptides alter the directionality of larval movement and the steady-state larval distribution. Neuropeptides that inhibited ciliary arrest and/or increased ciliary beat frequency led to biased upward swimming (RYa, FVMa, DLa, FMRFa, FVa, LYa, YFa, L11, and SPY; Fig. 5 B–J). This upward swimming was not observed when we used RYa and FVMa peptides with Ala substitutions (Fig. 5 B and C). In contrast, application of FLa and WLD, two peptides that caused lower ciliary beat frequency and frequent ciliary arrests, resulted in downward displacement of larvae (Fig. 5 K and L).
The changes triggered by the neuropeptides in the directionality of larval movement also shifted the mean vertical distribution at steady state in a dose-dependent manner. Activating peptides led to strong upward shifts, whereas inhibitory peptides caused downward shifts (Fig. 6). The peptide effects were lost following washout, as shown for RYa and FVMa (Fig. 6 B and C). When we combined RYa and FLa, two peptides with opposing effects, the effects canceled out, indicating that neuropeptide effects are additive (Fig. 6K). All neuropeptides affected vertical shifts in a concentration-dependent manner (Fig. 6O).
By influencing ciliary beat frequency and ciliary arrests, neuropeptides can change the net directionality of movement, leading to large shifts in the vertical distribution of populations of ciliated larvae.
Ciliary Arrests and Beat Frequency Have Distinct Contributions to Larval Swimming.
All neuropeptides that affected ciliary closures also affected ciliary beat frequency (Fig. 4 A and B). To uncouple the contribution of ciliary arrests and beat frequency on larval swimming, we tried to influence these parameters separately. To interfere with ciliary arrests without changing beat frequency we first characterized the mechanism of arrests.
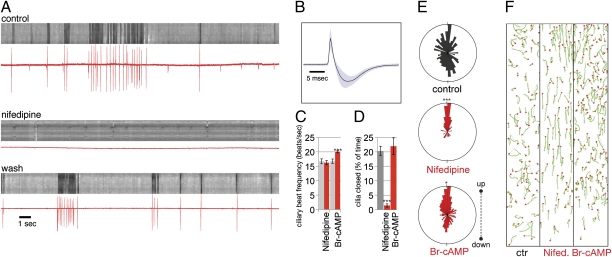
When performing extracellular current measurements with a recording pipette penetrated into the center of Platynereis larvae, we observed regular spiking activity. Combined extracellular recordings and video microscopy of cilia showed that the spikes correlated with ciliary arrests, with the first spike in a spike train preceding the arrest (Fig. 7A). Sustained arrests of cilia were accompanied by repeated spiking events of a frequency between 5 and 10 Hz, during the entire duration of the arrest episode. The long duration of the spiking events (full width at half max = 1.45 ms, SD = 0.17; Fig. 7B) indicated that they were likely evoked by calcium, rather than sodium. Calcium spikes were also shown to trigger ciliary arrests in mollusk larvae (19, 51). To test whether voltage-dependent calcium channels were responsible for the recorded spike events, we applied the L-type calcium-channel blocker nifedipine. In the presence of nifedipine, beat frequency was unaffected (Fig. 7C), but spikes and ciliary arrests were completely abolished (Fig. 7 A and D). These observations indicate that calcium spikes generated by L-type calcium channels induce and sustain ciliary arrests and also provided a tool to specifically block arrests without affecting ciliary beat frequency.
Fig. 7.
Calcium-regulated arrests and ciliary beat frequency both affect larval swimming. (A) Kymographs of ciliary beating (light gray sections) and ciliary arrest (dark gray sections) with parallel extracellular current recordings (red traces) for control, nifedipine addition, and washout. (B) Average of 179 spikes from recordings with a 0.1-ms temporal resolution. (C) Ciliary beat frequency in the presence of nifedipine and Br-cAMP. (D) Quantification of ciliary arrests in the presence of nifedipine and Br-cAMP. (E) Angular plots of the displacement vectors of larval swimming tracks for control larvae and larvae in the presence of nifedipine and Br-cAMP. (F) Swimming trajectories of control larvae and larvae in the presence of nifedipine and Br-cAMP. Red dots indicate the end of the tracks. The beginning and end of each track is connected by a gray line. Tracks were generated from a 10-s section of 30-fps video recordings. In C and D data are shown as mean ± SEM. P values of an unpaired t test are indicated: *P < 0.05; and ***P < 0.001. n > 10 larvae for C and D. Final concentration was 10 μM for nifedipine and 5 μM for Br-cAMP.
To test the contribution of ciliary arrests to the overall directionality of larval movement, we then tested the effect of nifedipine in vertical swimming assays. Application of nifedipine led to consistent upward swimming (Fig. 7 E and F; average upward displacement = 0.92 mm/s; control = −0.03 mm/s). Larval trajectories were straighter than in control larvae (Fig. 7F), also shown by the narrower distribution in the angular histograms (Fig. 7E). The consistent upward swimming led to an upward-shifted steady-state distribution (Fig. 6M).
The up-regulation of ciliary beat frequency also led to upward swimming, as shown by the neuropeptides that increased beat frequency without affecting arrests (LYa, YFa, L11, and SPY; Fig. 4 A and B and Fig. 5 G–J). To influence beat frequency independent of neuropeptides, we used the cell-permeable cAMP analog, Br-cAMP. cAMP is known to increase ciliary beat frequency in a wide range of eukaryotes (52) and it also increased beat frequency in Platynereis larvae, without affecting arrests (Fig. 7 C and D). As with neuropeptides, the Br-cAMP–induced increase in beat frequency led to upward swimming and higher steady-state vertical distribution (Fig. 7 E and F and Fig. 6N), but the swimming tracks remained irregular, similar to those in control larvae (Fig. 7F).
These results show that ciliary arrests and beat frequency have distinct contributions to larval swimming. Arrests allow larvae to sink and contribute to the maintenance of an unbiased net vertical displacement. Arrests also result in irregular swimming tracks (i.e., frequent turning events; compare Movie S5). Ciliary beating promotes upward swimming. Neuropeptides can influence both parameters, thereby modulating swimming directionality, the frequency of sinking events, and swimming pattern.
Discussion
Neuropeptides Regulate Ciliary Swimming.
Animals moving with cilia can either crawl on a surface (e.g., flatworms) or swim freely in water. Ciliary swimming is characteristic of the larval stage of many marine invertebrates with a benthic adult. Such ciliated larvae are widespread in the animal kingdom (1). Among the bilaterians, the lophotrochozoans (e.g., annelids and molluscs) and the deuterostomes (e.g., echinoderms and hemichordates) often have ciliated larvae (2, 3), whereas the ecdysozoans (e.g., insects and nematodes) lack them. Outside the bilaterians, cilia-based locomotion is present in ctenophores and in the larval stages of many cnidarians. Additionally, sponges, the basal-most animal group relative to the eumetazoans, often have ciliated larvae (4, 5). This trait, together with the lack of muscles in sponges, indicates that ciliary swimming is likely the ancestral form of locomotion in animals. Despite its importance for marine life and our understanding of the evolution of locomotion, we know very little about the control of ciliary swimming.
Here we described an unexpected diversity of neuropeptides influencing ciliary activity in larvae of the annelid Platynereis. The peptides can either activate ciliary swimming (increased ciliary beat frequency and reduced ciliary arrests) or inhibit swimming (reduced ciliary beat frequency and more arrests), leading to upward and downward shifts in larval vertical distribution. The regulation of ciliary beat frequency and ciliary arrests both contribute to changes in larval distribution, with arrests also contributing to the irregularity of swimming tracks.
Our vertical larval swimming experiments support the model that upward swimming and sinking determine the vertical distribution of marine plankton (8). It is remarkable that actively swimming Platynereis larvae maintain a uniform vertical distribution at steady state. Such a distribution can be maintained only if the upward and downward net movement of the larvae is balanced. To achieve this, swimming and sinking parameters (ciliary beat frequency, ciliary arrests, directionality, and buoyancy) must be fine-tuned. We showed how neuropeptide signaling can alter some of these parameters, shifting the overall larval vertical distribution.
Our results provide a general framework for how neuropeptides and the neurons that express them can regulate the depth of ciliated zooplankton.
Sensory Neurons Directly Control Cilia.
The morphology of the neuropeptidergic neurons suggests that these cells are dual-function sensory-motor cells directly translating sensory inputs into motor output on effector ciliated cells. The diversity of sensory neurons with cilia-regulating neuropeptides in the Platynereis larva is consistent with the multitude of environmental cues annelid and other ciliated larvae can respond to. These cues include light, pressure, salinity, and temperature, as well as settlement-inducing chemicals (6, 17, 18, 53). Some of these cues were shown to change the distribution of larvae, either by promoting upward swimming (e.g., increased pressure) (53) or by inhibiting cilia (e.g., settlement cues) (17). The sensory-motor neurons described here are potentially involved in similar responses in Platynereis larvae. Our results therefore suggest a model where certain aversive sensory inputs (e.g., high pressure or low temperature) can elicit neuropeptide release at the ciliary band, thereby promoting upward swimming. Such simple depth-regulating escape circuits could contribute to the accumulation of many planktonic organisms at water interfaces (54).
An analogous serotonergic sensory-motor neuron, mediating a response to an aversive cue, has also been described in the embryo of the pond snail, Helisoma trivolvis. Here the sensory neuron directly innervates embryonic cilia and increases ciliary beat frequency upon hypoxic stimulation in the egg capsule (55).
As with these previous studies, our results suggest that ciliated larvae harbor simple sensory-motor reflex circuits of various sensory modalities, able to directly influence ciliary activity and thus vertical position in the water.
Differences Between Muscle- and Ciliary-Control Circuits.
A direct sensory-motor regulation of locomotion, implicated by our findings, has not been described in muscle-based motor circuits in bilaterians. In such systems, distinct sensory, inter-, and motoneurons are always present, even in the simplest examples, such as the gill-withdrawal reflex of Aplysia (56) or stretch-receptor–mediated proprioception in Caenorhabditis (57). Muscle-regulating circuits always have interneurons because motor control can entail the contraction of several muscles in a strict temporal order (e.g., locomotor central pattern generators) (58), the integration of conflicting sensory inputs into different motor outputs (e.g., anterior and posterior stimulation during the nematode tap withdrawal reflex) (59), or the coordinated contraction and relaxation of antagonistic muscles (e.g., tendon jerk reflex in tetrapods) (60).
In contrast, the nervous system of the Platynereis larval episphere is organized predominantly as a direct sensory-motor system. This system of simple organization is not due to the simplicity or young age of these larvae, because at the corresponding stage the trunk nervous system already harbors distinct types of interneurons, likely involved in regulating the trunk musculature (12). The structural difference between the trunk and episphere nervous system therefore seems to reflect the different functionality of the two systems, in that the trunk system regulates muscles whereas the episphere system regulates cilia. Because ciliary locomotion does not require a complex coordination of various motor structures, simple sensory-motor neurons may be adequate to perform all activatory and inhibitory functions on a single ciliary band. If the innervation runs to all ciliated cells, as found for the neurons described here, regulation can occur uniformly along the entire ciliary band. Alternatively, regulation can be differential, due to local innervation of a segment of the ciliary band, allowing tactic turning behavior (10).
Are Ciliary Locomotor Circuits Ancestral?
Our results show that ciliated larvae use a very simple functional circuitry, regulating swimming by direct sensory-motor innervation. Such sensory-motor neurons are common in cnidarians (61–63); however, in bilaterians they have to date been described only in ciliated larvae (10, 55). This observation raises the interesting possibility that ciliary locomotor circuits in bilaterian larvae have retained an ancestral state of nervous system organization. If this scenario is true, ciliated larval circuitry may give insights into the evolutionary origin of the first nervous systems (15).
The cilia regulatory neuropeptides described here are widely conserved among marine invertebrates, including other annelids, mollusks, and cnidarians. Because all of these groups have ciliated larvae, our results have broad implications for the understanding of ciliary locomotor control in a wide range of invertebrate larvae.
Methods
Bioinformatics.
For the identification of neuropeptides we used BLAST searches and pattern searches using repetitions of the motive K[K/R]-x(3-10). The GenBank accession numbers are JF811323–JF811333.
Antibodies and Staining.
For antibody production, rabbits were immunized with neuropeptides coupled to a carrier via an N-terminal Cys. Sera were affinity purified on a SulfoLink resin (Pierce) coupled to the Cys-containing peptides. The bound antibodies were washed extensively with PBS and with 0.5 M NaCl to remove weakly bound antibodies. Fractions were collected upon elution with 100 mM glycine, pH 2.7 and 2.3. Inmunostainings and whole-mount in situ hybridization were performed on precisely staged larvae raised at 18 °C. Imaging of in situ hybridization samples was performed as previously described (45).
Microscopy and Image Processing.
Confocal images were taken on an Olympus Fluoview-1000 confocal microscope with a 60× water-immersion objective using 488- and 635-nm laser lines and a pinhole of airy unit 1. For volume scans 512 × 512-pixel image stacks were recorded at a Z-distance of 1 μm. Recordings of ciliary beating and arrest were performed with a Zeiss Axioimager microscope and a DMK 21BF04 camera (The Imaging Source) at 60 frames/s or 15 frames/s to quantify arrests. Larvae were immobilized between a slide and a coverslip spaced with adhesive tape. Freely swimming larvae in vertical tubes were recorded with a DMK 21BF03 camera (The Imaging Source) at 30 frames/s. The tubes were illuminated laterally with red light-emitting diode (LED) lights that larvae are unable to detect at the stages studied. Stacks of confocal images or videos were processed using ImageJ 1.42 and Imaris 5.5.1. For the whole-mount in situ and immunostaining samples we generated projections using Imaris. Some images were filtered with a median filter (3 × 3 × 1 filter size), using Imaris. Contrast was always adjusted equally across the entire image. Larval swimming videos were analyzed using custom ImageJ macros and Perl scripts.
Image Registration.
The expression patterns of the neuropeptides were registered to a reference axonal scaffold to obtain a combined molecular map of the peptidergic neurons. We established an image registration procedure similar to that in ref. 47, using the ITK toolkit (64). We first generated a reference scaffold by aligning 20 individual larvae to a representative scaffold, using affine and deformable transformations, and averaging them. Three to five gene expression patterns per gene were registered to the reference, using the acetylated tubulin channel. First affine (rotation, scaling, translation, shearing; itkAffineTransform class) and then deformable (nonuniform warp; itkBSplineDeformableTransform class) transformations were applied. For both steps the Mattes mutual information metric (itkMattesMutualInformationImageToImageMetric class) and the gradient step optimizer (itkRegularStepGradientDescentOptimizer class) were used. The multiresolution registration method was applied for both steps to speed up the process and make it more robust (itkMultiResolutionImageRegistrationMethod class).
Behavior.
Platynereis larvae were obtained from a breeding culture, following ref. 11. Behavioral experiments were performed in seawater, using 48- to 60-hpf larvae (raised at 18 °C). Neuropeptides were added to the seawater and larvae were incubated for 2–5 min before recordings. For each peptide first we tested a range of different concentrations in the vertical migration assay and scored the effects by quantifying the larvae in the upper versus the lower half of the tubes (Fig. 6O). The lowest concentration with maximum effect was used in the single-larva assays.
Electrophysiology.
Extracellular recordings were performed at room temperature with 29- to 33-hpf larvae immobilized with a holding pipette (opening ∼30 μm). For recordings we used borosilicate capillaries (Science Products GB150F-8P) pulled with a Sutter P-1000 Flaming/Brown micropipette puller (Heka Elektronik) to 2–4 MΩ resistance. To facilitate penetration of the cuticle 0.002% trifluoromethanesulfonic acid was added to the larvae 10 min before the experiments. The recording solution contained 70 mM CsCl, 10 mM Hepes, 11 mM glucose, 10 mM glutathione, 5 mM EGTA, 500 mM aspartic acid, 5 mM ATP, and 0.1 mM GTP, and the pH was set to 7.3 with CsOH. The sample was illuminated with a Sharp DLP projector (PG-F212X-L) through a 740/10 band-pass filter, and ciliary beating was recorded at 6.5 frames/s. Recordings were done with the pipette in close proximity to the ciliated cells.
Supplementary Material
Acknowledgments
We thank Elizabeth Williams, Andrew Renault, and Fulvia Bono for comments on the manuscript and Heinz Schwarz for advice on immunostainings. This work was supported by a Sequencing Grant to G.J. (M.IF.A.ENTW8050) from the Max Planck Society. Work of T.A.M. and S.-L.O. is supported by the Centre for Integrative Neuroscience (Deutsche Forschungsgemeinschaft, EXC 307). The research leading to these results received funding from the European Research Council under the European Union's Seventh Framework Programme (FP7/2007-2013)/European Research Council Grant Agreement 260821.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JF811323–JF811333).
See Author Summary on page 18593.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109085108/-/DCSupplemental.
References
- 1.Young CM, editor. Atlas of Marine Invertebrate Larvae. 1st Ed. San Francisco: Academic; 2002. [Google Scholar]
- 2.Nielsen C. Trochophora larvae: Cell-lineages, ciliary bands, and body regions. 1. Annelida and Mollusca. J Exp Zoolog B Mol Dev Evol. 2004;302:35–68. doi: 10.1002/jez.b.20001. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen C. Trochophora larvae: Cell-lineages, ciliary bands and body regions. 2. Other groups and general discussion. J Exp Zoolog B Mol Dev Evol. 2005;304:401–447. doi: 10.1002/jez.b.21050. [DOI] [PubMed] [Google Scholar]
- 4.Maldonado M. The ecology of the sponge larva. Can J Zool. 2006;84:175–194. [Google Scholar]
- 5.Nielsen C. Six major steps in animal evolution: Are we derived sponge larvae? Evol Dev. 2008;10:241–257. doi: 10.1111/j.1525-142X.2008.00231.x. [DOI] [PubMed] [Google Scholar]
- 6.Thorson G. Light as an ecological factor in the dispersal and settlement of larvae of marine bottom invertebrates. Ophelia. 1964;1:167–208. [Google Scholar]
- 7.Hamasaki T, Barkalow K, Richmond J, Satir P. cAMP-stimulated phosphorylation of an axonemal polypeptide that copurifies with the 22S dynein arm regulates microtubule translocation velocity and swimming speed in Paramecium. Proc Natl Acad Sci USA. 1991;88:7918–7922. doi: 10.1073/pnas.88.18.7918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chia F, Buckland-Nicks J, Young C. Locomotion of marine invertebrate larvae: A review. Can J Zool. 1984;62:1205–1222. [Google Scholar]
- 9.Satir P. The cilium as a biological nanomachine. FASEB J. 1999;13(Suppl 2):S235–S237. doi: 10.1096/fasebj.13.9002.s235. [DOI] [PubMed] [Google Scholar]
- 10.Jékely G, et al. Mechanism of phototaxis in marine zooplankton. Nature. 2008;456:395–399. doi: 10.1038/nature07590. [DOI] [PubMed] [Google Scholar]
- 11.Hauenschild C, Fischer A. Platynereis dumerilii. Mikroskopische Anatomie, Fortpflanzung, Entwicklung. Stuttgart: Gustav Fischer; 1969. [Google Scholar]
- 12.Denes AS, et al. Molecular architecture of annelid nerve cord supports common origin of nervous system centralization in bilateria. Cell. 2007;129:277–288. doi: 10.1016/j.cell.2007.02.040. [DOI] [PubMed] [Google Scholar]
- 13.Arendt D, Tessmar-Raible K, Snyman H, Dorresteijn AW, Wittbrodt J. Ciliary photoreceptors with a vertebrate-type opsin in an invertebrate brain. Science. 2004;306:869–871. doi: 10.1126/science.1099955. [DOI] [PubMed] [Google Scholar]
- 14.Tessmar-Raible K, et al. Conserved sensory-neurosecretory cell types in annelid and fish forebrain: Insights into hypothalamus evolution. Cell. 2007;129:1389–1400. doi: 10.1016/j.cell.2007.04.041. [DOI] [PubMed] [Google Scholar]
- 15.Jékely G. Origin and early evolution of neural circuits for the control of ciliary locomotion. Proc Biol Sci. 2011;278:914–922. doi: 10.1098/rspb.2010.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bayne B. Responses of Mytilus edulis larvae to increases in hydrostatic pressure. Nature. 1963;198:406–407. [Google Scholar]
- 17.Hadfield MG, Koehl MA. Rapid behavioral responses of an invertebrate larva to dissolved settlement cue. Biol Bull. 2004;207:28–43. doi: 10.2307/1543626. [DOI] [PubMed] [Google Scholar]
- 18.Hidu H, Haskin HH. Swimming speeds of oyster larvae Crassostrea virginica in different salinities and temperatures. Estuaries. 1978;1:252. [Google Scholar]
- 19.Mackie GO, Singla CL, Thiriot-Quievreux C. Nervous control of ciliary activity in gastropod larvae. Biol Bull. 1976;151:182–199. doi: 10.2307/1540713. [DOI] [PubMed] [Google Scholar]
- 20.Lacalli TC, Gilmour T. Ciliary reversal and locomotory control in the Pluteus larva of Lytechinus pictus. Philos Trans R Soc B. 1990;330:391–396. [Google Scholar]
- 21.Lacalli TC. Structure and organization of the nervous system in the Trochophore larva of Spirobranchus. Philos Trans R Soc B. 1984;306:79–135. [Google Scholar]
- 22.Dickinson A, Nason J, Croll R. Histochemical localization of FMRFamide, serotonin and catecholamines in embryonic Crepidula fornicata (Gastropoda, Prosobranchia) Zoomorphology. 1999;119:49–62. [Google Scholar]
- 23.Voronezhskaya EE, Tsitrin EB, Nezlin LP. Neuronal development in larval polychaete Phyllodoce maculata (Phyllodocidae) J Comp Neurol. 2003;455:299–309. doi: 10.1002/cne.10488. [DOI] [PubMed] [Google Scholar]
- 24.Gruhl A. Serotonergic and FMRFamidergic nervous systems in gymnolaemate bryozoan larvae. Zoomorphology. 2009;128:135–156. [Google Scholar]
- 25.Beer A-J, Moss C, Thorndyke M. Development of serotonin-like and SALMFamide-like immunoreactivity in the nervous system of the sea urchin Psammechinus miliaris. Biol Bull. 2001;200:268–280. doi: 10.2307/1543509. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe H, Fujisawa T, Holstein TW. Cnidarians and the evolutionary origin of the nervous system. Dev Growth Differ. 2009;51:167–183. doi: 10.1111/j.1440-169X.2009.01103.x. [DOI] [PubMed] [Google Scholar]
- 27.Chun JY, Korner J, Kreiner T, Scheller RH, Axel R. The function and differential sorting of a family of aplysia prohormone processing enzymes. Neuron. 1994;12:831–844. doi: 10.1016/0896-6273(94)90336-0. [DOI] [PubMed] [Google Scholar]
- 28.Hook V, et al. Proteases for processing proneuropeptides into peptide neurotransmitters and hormones. Annu Rev Pharmacol Toxicol. 2008;48:393–423. doi: 10.1146/annurev.pharmtox.48.113006.094812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eipper BA, Stoffers DA, Mains RE. The biosynthesis of neuropeptides: Peptide alpha-amidation. Annu Rev Neurosci. 1992;15:57–85. doi: 10.1146/annurev.ne.15.030192.000421. [DOI] [PubMed] [Google Scholar]
- 30.McVeigh P, Kimber MJ, Novozhilova E, Day TA. Neuropeptide signalling systems in flatworms. Parasitology. 2005;131(Suppl):S41–S55. doi: 10.1017/S0031182005008851. [DOI] [PubMed] [Google Scholar]
- 31.Nelson LS, Rosoff ML, Li C. Disruption of a neuropeptide gene, flp-1, causes multiple behavioral defects in Caenorhabditis elegans. Science. 1998;281:1686–1690. doi: 10.1126/science.281.5383.1686. [DOI] [PubMed] [Google Scholar]
- 32.Marder E, Bucher D. Central pattern generators and the control of rhythmic movements. Curr Biol. 2001;11:R986–R996. doi: 10.1016/s0960-9822(01)00581-4. [DOI] [PubMed] [Google Scholar]
- 33.Marder E, Richards KS. Development of the peptidergic modulation of a rhythmic pattern generating network. Brain Res. 1999;848:35–44. doi: 10.1016/s0006-8993(99)02118-6. [DOI] [PubMed] [Google Scholar]
- 34.Root CM, Ko KI, Jafari A, Wang JW. Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. Cell. 2011;145:133–144. doi: 10.1016/j.cell.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chalasani SH, et al. Neuropeptide feedback modifies odor-evoked dynamics in Caenorhabditis elegans olfactory neurons. Nat Neurosci. 2010;13:615–621. doi: 10.1038/nn.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ringstad N, Horvitz H. FMRFamide neuropeptides and acetylcholine synergistically inhibit egg-laying by C. elegans. Nat Neurosci. 2008;11:1168–1176. doi: 10.1038/nn.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collins, JJ, 3rd, et al. Genome-wide analyses reveal a role for peptide hormones in planarian germline development. PLoS Biol. 2010;8:e1000509. doi: 10.1371/journal.pbio.1000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willows AO, Pavlova GA, Phillips NE. Modulation of ciliary beat frequency by neuropeptides from identified molluscan neurons. J Exp Biol. 1997;200:1433–1439. doi: 10.1242/jeb.200.10.1433. [DOI] [PubMed] [Google Scholar]
- 39.Katsukura Y, Ando H, David CN, Grimmelikhuijzen CJP, Sugiyama T. Control of planula migration by LWamide and RFamide neuropeptides in Hydractinia echinata. J Exp Biol. 2004;207:1803–1810. doi: 10.1242/jeb.00974. [DOI] [PubMed] [Google Scholar]
- 40.Southey BR, Amare A, Zimmerman TA, Rodriguez-Zas SL, Sweedler JV. NeuroPred: A tool to predict cleavage sites in neuropeptide precursors and provide the masses of the resulting peptides. Nucleic Acids Res. 2006;34(Web Server issue):W267–W272. doi: 10.1093/nar/gkl161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gustafsson MKS, et al. Neuropeptides in flatworms. Peptides. 2002;23:2053–2061. doi: 10.1016/s0196-9781(02)00193-6. [DOI] [PubMed] [Google Scholar]
- 42.Hewes RS, Taghert PH. Neuropeptides and neuropeptide receptors in the Drosophila melanogaster genome. Genome Res. 2001;11:1126–1142. doi: 10.1101/gr.169901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plickert G, et al. The role of alpha-amidated neuropeptides in hydroid development—LWamides and metamorphosis in Hydractinia echinata. Int J Dev Biol. 2003;47:439–450. [PubMed] [Google Scholar]
- 44.Nathoo AN, Moeller RA, Westlund BA, Hart AC. Identification of neuropeptide-like protein gene families in Caenorhabditis elegans and other species. Proc Natl Acad Sci USA. 2001;98:14000–14005. doi: 10.1073/pnas.241231298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jékely G, Arendt D. Cellular resolution expression profiling using confocal detection of NBT/BCIP precipitate by reflection microscopy. Biotechniques. 2007;42:751–755. doi: 10.2144/000112462. [DOI] [PubMed] [Google Scholar]
- 46.Dorresteijn AWC. Quantitative analysis of cellular differentiation during early embryogenesis of Platynereis dumerilii. Rouxs Arch Dev Biol. 1990;199:14–30. doi: 10.1007/BF01681530. [DOI] [PubMed] [Google Scholar]
- 47.Tomer R, Denes AS, Tessmar-Raible K, Arendt D. Profiling by image registration reveals common origin of annelid mushroom bodies and vertebrate pallium. Cell. 2010;142:800–809. doi: 10.1016/j.cell.2010.07.043. [DOI] [PubMed] [Google Scholar]
- 48.Purschke G. Sense organs in polychaetes (Annelida) Hydrobiologia. 2005;535:53–78. [Google Scholar]
- 49.Walch-Solimena C, et al. Synaptotagmin: A membrane constituent of neuropeptide-containing large dense-core vesicles. J Neurosci. 1993;13:3895–3903. doi: 10.1523/JNEUROSCI.13-09-03895.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voets T, et al. Intracellular calcium dependence of large dense-core vesicle exocytosis in the absence of synaptotagmin I. Proc Natl Acad Sci USA. 2001;98:11680–11685. doi: 10.1073/pnas.201398798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arkett S, Mackie G, Singla C. Neuronal control of ciliary locomotion in a gastropod veliger (Calliostoma) Biol Bull. 1987;173:513–526. doi: 10.2307/1541697. [DOI] [PubMed] [Google Scholar]
- 52.Salathe M. Regulation of mammalian ciliary beating. Annu Rev Physiol. 2007;69:401–422. doi: 10.1146/annurev.physiol.69.040705.141253. [DOI] [PubMed] [Google Scholar]
- 53.Knight-Jones EW, Qasim SZ. Responses of some marine plankton animals to changes in hydrostatic pressure. Nature. 1955;175:941–942. doi: 10.1038/175941a0. [DOI] [PubMed] [Google Scholar]
- 54.Harder W. Reactions of plankton organisms to water stratification. Limnol Oceanogr. 1968;13:156–168. [Google Scholar]
- 55.Kuang S, Doran SA, Wilson RJA, Goss GG, Goldberg JI. Serotonergic sensory-motor neurons mediate a behavioral response to hypoxia in pond snail embryos. J Neurobiol. 2002;52:73–83. doi: 10.1002/neu.10071. [DOI] [PubMed] [Google Scholar]
- 56.Castellucci V, Pinsker H, Kupfermann I, Kandel ER. Neuronal mechanisms of habituation and dishabituation of the gill-withdrawal reflex in Aplysia. Science. 1970;167:1745–1748. doi: 10.1126/science.167.3926.1745. [DOI] [PubMed] [Google Scholar]
- 57.Li W, Feng Z, Sternberg PW, Xu XZS. A C. elegans stretch receptor neuron revealed by a mechanosensitive TRP channel homologue. Nature. 2006;440:684–687. doi: 10.1038/nature04538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grillner S. The motor infrastructure: From ion channels to neuronal networks. Nat Rev Neurosci. 2003;4:573–586. doi: 10.1038/nrn1137. [DOI] [PubMed] [Google Scholar]
- 59.Wicks SR, Rankin CH. Integration of mechanosensory stimuli in Caenorhabditis elegans. J Neurosci. 1995;15:2434–2444. doi: 10.1523/JNEUROSCI.15-03-02434.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liddell E, Sherrington C. Reflexes in response to stretch (myotatic reflexes) Proc R Soc Lond B. 1924;96:212–242. [Google Scholar]
- 61.Westfall JA. Ultrastructural evidence for a granule-containing sensory-motor-interneuron in Hydra littoralis. J Ultrastruct Res. 1973;42:268–282. doi: 10.1016/s0022-5320(73)90055-5. [DOI] [PubMed] [Google Scholar]
- 62.Westfall JA, Kinnamon JC. A second sensory–motor–interneuron with neurosecretory granules in Hydra. J Neurocytol. 1978;7:365–379. doi: 10.1007/BF01176999. [DOI] [PubMed] [Google Scholar]
- 63.Westfall JA, Wilson JD, Rogers RA, Kinnamon JC. Multifunctional features of a gastrodermal sensory cell in Hydra: Three-dimensional study. J Neurocytol. 1991;20:251–261. doi: 10.1007/BF01235543. [DOI] [PubMed] [Google Scholar]
- 64.Yoo TS, et al. Engineering and algorithm design for an image processing Api: A technical report on ITK—the Insight Toolkit. Stud Health Technol Inform. 2002;85:586–592. [PubMed] [Google Scholar]