Abstract
Rab GTPases are key regulators of membrane traffic pathways within eukaryotic cells. They are specifically activated by guanine nucleotide exchange factors (GEFs), which convert them from their “inactive” GDP-bound form to the “active” GTP-bound form. In higher eukaryotes, proteins containing DENN-domains comprise a major GEF family. Here we describe at 2.1-Å resolution the first structure of a DENN-domain protein, DENND1B-S, complexed with its substrate Rab35, providing novel insights as to how DENN-domain GEFs interact with and activate Rabs. DENND1B-S is bi-lobed, and interactions with Rab35 are through conserved surfaces in both lobes. Rab35 binds via switch regions I and II, around the nucleotide-binding pocket. Positional shifts in Rab residues required for nucleotide binding may lower its affinity for bound GDP, and a conformational change in switch I, which makes the nucleotide-binding pocket more solvent accessible, likely also facilitates exchange.
In eukaryotic cells, material is transported between membrane-bound organelles or the plasma membrane by vesicles. These bud from the membrane of the donor organelle, travel to and are recognized at the target compartment, and finally fuse with it to deliver their cargo. Vesicle traffic is orchestrated by a large number of proteins, including coat proteins that facilitate budding, tethering complexes involved in vesicle recognition, SNARE proteins that drive membrane fusion, and small GTPases in the Rab/Ypt family that have regulatory roles (1). Different subsets of these proteins are involved in different transport pathways.
Rab GTPases are key determinants of organelle identity and hence in ensuring that vesicular cargo is delivered to the correct destination (2–4). The Rabs themselves are activated at the appropriate membranes by Rab-specific guanine nucleotide exchange factors (GEFs), which facilitate the conversion of the Rab from its inactive, GDP-bound, to the active, GTP-bound state. The GEF interacts with its Rab substrate to lower its nucleotide-binding affinity, accelerating the departure of bound GDP and allowing GTP, which is 10-fold more abundant in the cell than GDP, to bind (5, 6). In their GTP-bound membrane associated form, Rabs then recruit additional proteins to mediate vesicle recognition, tethering, and fusion events.
Mammals have more than 60 different Rabs (7), with GEFs that activate most of these yet to be identified. Recent data suggest that a large subset of mammalian Rabs, or at least 10 different Rab GTPases, are activated by proteins containing a DENN domain (8). In humans, eighteen different DENN-domain proteins have been identified, including members of the DENND1 through DENND5 families, the myotubularin-related proteins MTMR5 and 13, and the MAP-kinase activating death domain MADD (8, 9). Studies of the DENND1 family have shown that the DENN domain itself is sufficient for GEF activity (10, 11), where both DENND1A and 1B activate Rab35 for its role in the endocytic pathway (8, 10, 11). The substrate specificity of DENND1C remains unclear (8), although it has also been proposed as a Rab35 GEF (8, 11).
Despite their importance in regulating membrane traffic in higher eukaryotes, little is known regarding the structure of DENN domains or the mechanisms by which they function. Here, to obtain first insights regarding how these proteins recognize and activate their Rab partners, we have determined at 2.1-Å resolution the structure of the DENN domain from DENND1B in complex with the nucleotide-free form of its substrate Rab35. The complex represents an intermediate on the reaction pathway, after GDP has left the Rab35 nucleotide-binding pocket and before GTP binding.
Results and Discussion
Overview.
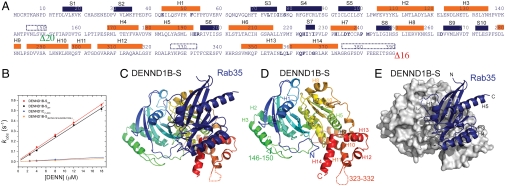
Crystals were obtained for a complex consisting of Rab35, lacking the unstructured C terminus (residues 181–201), and the DENN domain of human DENND1B. The DENN-domain construct used for crystallization corresponds to the shortest isoform of DENND1B, DENND1B-S, except that 16 residues were truncated at the C terminus and that one loop was shortened (Fig. 1A). The loop between residues 140–170 in the full-length DENND1B sequence is longer than in other DENND family proteins, and it likely interferes with crystallization because we were able to obtain crystals of DENN1B-S/Rab35 complex only when 20 residues were deleted from this region. Importantly, truncating the C terminus and shortening the loop did not affect GEF activity for Rab35, as assessed by monitoring GDP release. The catalytic efficiencies (analogous to kcat/Km of enzymatic reactions) (12) for the intact DENND1B-S and the crystallization construct are 0.0291 ± 0.0011 μM-1 s-1 and 0.0306 ± 0.0008μ μM-1 s-1, respectively, in the range observed for other Rab GEFs (Fig. 1B) (12–15).
Fig. 1.
Structure of the DENND1B-S/Rab35 complex. (A) Sequence of DENND1B-S in crystallization construct. Secondary structure as in the crystal structure is indicated. Helices are orange bars and strands blue. Disordered regions are indicated by dotted lines. Sixteen residues at the C terminus and a 20-residue loop in wild-type protein were deleted; their locations are indicated by green and red arrowheads. (B) The DENND1B-S construct used for crystallization is equally efficient in catalyzing guanine nucleotide exchange for Rab35 as wild-type protein. MantGDP dissociation from Rab35 was continuously monitored by FRET (λex = 280 nm, λem = 435 nm) with increasing concentrations of either the crystallization construct or wild-type DENN domain. The DENND1C DENN domain is approximately 10-fold less efficient, as is a DENND1B mutant, where residues at the Rab35 interface were changed to corresponding residues in DENND1C. See Fig. S5A for representative time course data. (C) Ribbons diagram of DENND1B-S, colored with blue at the N terminus and red at the C terminus, complexed with Rab35 (blue). (D) DENND1B-S alone with secondary structure elements labeled. (E) Rab35 is rendered as a ribbon, DENND1B-S as a surface representation.
Single wavelength anomalous dispersion (SAD) methods with crystals containing selenomethionine- substituted DENND1B-S were used for phasing. There are two DENN-domain/Rab35 complexes (A, B) in the asymmetric unit. The DENN domains are similar with an rms distance of 0.226 Å over 269 of 374 Cα atoms (the largest differences are in loops comprising residues 57–67, 87–93, 140–160), and the DENND1B/Rab35 interfaces are practically identical. Interactions and distances referred to below are for complex A, for which the model is slightly more complete. Complex A includes residues 4–33 and 35–177 of Rab35 and residues 1–373 of DENND1B-S, except for residues 146–151 and 326–332. In complex B, residues 33–35 in Rab35 and 22–26, 144–159, and 324–332 in DENND1B-S were poorly ordered and were not modeled. Data collection and refinement statistics are in Table 1.
Table 1.
Data collection and refinement statistics
| Data collection | ||
| Crystal | Native | Se |
| Space group | C2 | C2 |
| Unit cell dimensions | a = 122.59 Å, b = 56.56 Å, c = 175.31 Å, β = 95.63° | a = 122.18 Å, b = 56.49 Å, c = 174.85 Å, β = 95.95° |
| Wavelength (Å) | 0.9795 | 0.9795 |
| Resolution (Å) | 25.0−2.1(2.18−2.10) | 25.0−2.5(2.59−2.50) |
| Rsym(%) | 5.3 (32.4) | 8.6 (45.1) |
| I/σ | 24.1 (3.6) | 38.3 (9.2) |
| Completeness (%) | 94.0 (99.3) | 100.0 (100.0) |
| Redundancy | 3.3 (3.1) | 14.9 (14.6) |
| Refinement | ||
| Resolution (Å) | 25.0−2.1 | |
| No. unique reflections | 59538 | |
| Rwork/Rfree (%) | 22.63/26.91 | |
| No. of protein atoms | 8241 | |
| No. of waters | 186 | |
| Average B (Å2) | 52.88 | |
| Average B (protein) | 52.99 | |
| Average B (water) | 48.06 | |
| rmsd | ||
| Bond lengths (Å) | 0.019 | |
| Bond angles (°) | 1.62 | |
| Ramachandran plot | ||
| Allowed (% residues) | 97.3 | |
| Generously allowed (% residues) | 2.7 | |
| Disallowed (% residues) | 0 | |
Numbers in parentheses denote values in highest resolution bin.
Structure of DENND1B-S.
The DENN domain forms a heart-shaped structure, with the N-terminal residues 1–169 forming one lobe and C-terminal residues 170–377 forming the second one (Fig. 1 A and C–E). The N-terminal half folds into a longin module, consisting of a central antiparallel β-sheet (strands S1-S5) layered between helix H1 and helices H2 and H3. A long random-coil region (residues 141–160) links the two lobes. A β-sheet (strands S6–S10) also forms the core of the second lobe. Helices H7 and H8 are on one side, and a helical bundle consisting of helices H5–H6 and H9–H10 on the other. Helices H11–H14 at the C terminus cap one end of the helical bundle. The linker between helix H11 and H12 (residues 324–332) is disordered in the structure. Based on bioinformatics analyses, DENN domains have been divided into three segments (16), termed upstream DENN (u-DENN), core DENN, and downstream DENN (d-DENN), which are separated by linker regions that vary in length in different DENN-domain proteins. The boundaries between these segments do not coincide with domain boundaries observed in the crystal structure. The u-DENN segment corresponds to the N-terminal half of the longin domain (S1–S2–H1–S3–S4), and the core DENN to the remainder of the longin domain (S5–H2–H3) and most of the C-terminal lobe, except for the C-terminal helices H12–H14. Helices H12–H14 constitute the d-DENN segment.
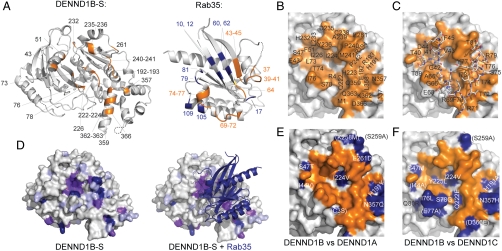
The Rab35 binding surface of the DENN domain is formed primarily by residues in the C-terminal lobe (Fig. 2 A–C). The DENN-domain/Rab35 interface occludes approximately 3,040 Å2 of solvent-accessible surfaces (17). Specifically, it includes residues in helices H7, H13, and H14, in strand S7, and in the loops H5–S6, H6–S7, H7–S8, H8–H9, and H13–H14. In the N-terminal longin module, Rab35 interacts with residues in helix H1 and in the loop S3–S4.
Fig. 2.
Interactions between DENND1B-S and Rab35. (A) Residues in DENND1B-S within 5 Å of Rab35 are labeled (left), as are residues in Rab35 within 5 Å of the DENN-domain (right). In Rab35, interacting residues in switches I or II are orange and residues outside the switch region blue. A 180° rotation about a vertical axis would position the Rab to interact with DENND1B-S. (B) DENND1B-S oriented as in A with residues within 4.5 Å of Rab35 in orange and labeled. (C) DENND1B-S as in B, with Rab35 switch regions I and II bound. Rab35 residues are labeled. (D) Sequence conservation in 18 human DENN domains mapped onto the surface of DENND1B-S. Strictly, highly, and weakly conserved residues (identical in 16–18, 11–15, and 8–10 sequences, respectively) are purple, magenta, and lilac, respectively. (E) The Rab35 interaction surface is similar in DENND1B and −1A. The Rab35 interaction surface on DENND1B is as in B, with residues in DENND1B within 10 Å of Rab35 that are different in DENND1A indicated (blue). Residues further than 5 Å from Rab35 are in parentheses. (F) As in E but comparing DENND1B and −1C.
Longin domains occur in a number of proteins involved in membrane traffic, including several that interact with small GTPases. The Mon1/Ccz1 heterodimeric GEF for Ypt7 is predicted to have two longin modules (18), and in the TRAPPI complex, a multimeric GEF for Rab1 (19), three subunits have a longin-fold (14, 20). One of these, Trs20, does not interact with the Rab, whereas two others, Bet5 and Trs23, form most of the Rab binding surface (14, 20). We note that different surfaces in the DENND1B longin module, Bet5, and Trs23 mediate interactions with Rab substrate (Fig. S1). In the signal recognition particle receptor, a longin domain in the α subunit mediates its interaction with the β subunit, a Ras-like GTPase, via still another surface (Fig. S1) (14, 20). Therefore, although longin modules form versatile GTPase binding domains, the actual binding surface for small GTPases is not conserved.
In contrast, the Rab interaction surface is highly conserved across the DENN-domain family (Fig. 2D), and in DENND1B and −1A, which both activate Rab35, the surfaces are almost identical, with conservative changes at the periphery (Fig. 2E). The Rab binding surface is less well conserved in DENND1C (Fig. 2F), with four nonconservative mutations: S47M, H191A, Q222E, Y225L. Although DENND1C is still able to bind Rab35 (Fig. S2), binding is weaker (11), and DENND1C is an order of magnitude less efficient in catalyzing nucleotide exchange for Rab35 (Fig. 1B). Replacing the four variant DENND1B residues with their DENND1C counterparts lowers the catalytic efficiency of DENND1B by approximately 10-fold (Fig. 1B), so that these residues likely account for the lower exchange activity of DENND1C as compared to DENND1A or −B. The lower exchange activity for Rab35 observed for DENND1C as compared with DENND1A or −B raises the possibility that another Rab, not Rab35, is its preferred substrate.
Comparison of DENND1B-Bound Rab35 and Nucleotide-Bound Rab.
Like other Rab GTPases (21), Rab35 has a central six-stranded β-sheet surrounded by five alpha helices (Fig. 1 C and E). Interactions with the DENN domain are mediated by residues in the central beta sheet and helix H4 as well as in switch regions I and II, the elements near the GTPase nucleotide-binding pocket that differ in its GDP- and GTP-bound forms (Fig. 2 A and C). A surface that encompasses the switch regions is commonly used by Rabs in interacting with other proteins, including GDP dissociation inhibitor (GDI) as well as Rab effectors (21).
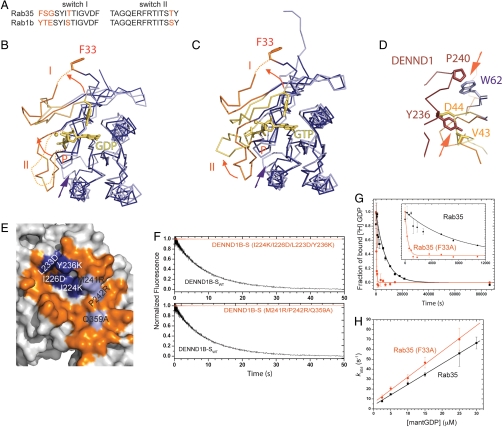
Structures for the nucleotide-bound forms of Rab35 are not known, but Rab35 shares high sequence identity with Rab1, for which both GDP- and GTP-bound structures have been determined (22). By analogy with Rab1, residues 33–45 and 64–77 in Rab35 correspond to switch regions I and II, respectively, and these are 81% identical and 96% similar (Fig. 3A). The structures of nucleotide-bound Rab1 (PDB ID codes 2FOL and 1YZN) are thus the best approximations available for nucleotide-bound forms of Rab35, and indeed, the rms distance between nucleotide-free Rab35 and GDP-bound Rab1 is 1.20 Å over 137 nonloop Cα positions.
Fig. 3.
Rearrangement of Rab35 and switches I and II in the Rab/GEF complex. (A) Sequences in the switch regions of Rab35 and Rab1 are highly conserved. See also Fig. S2F for a comparison of Rab1 and Rab35 surfaces. (B) Comparison of Rab35 (purple) in the complex with GDP-bound Rab1 (lilac; PDB ID code 2FOL ). Switches I and II are orange or yellow, respectively, and GDP and Mg2+ are yellow. F33 (Y33 in Rab1) moves out of the nucleotide-binding pocket as switch I alters its conformation. F33 is not well ordered and that the position of its side chain is uncertain. Arrows indicate conformational changes. (C) As in B but comparing Rab35 in the complex with GTP-bound Rab1 (PDB ID code 1YZN). (D) Switch I in Rab35 must rearrange as Rab35 binds DENND1B because it would otherwise clash sterically with H7 in the DENN domain (brown). Rab35 in the Rab/GEF complex is purple/orange; GDP-bound Rab is lilac/yellow. Arrows indicate clashes. (E) DENND1B residues predicted to affect either switch I or II binding were mutated, as indicated (blue and lilac for I and II, respectively). Compare this panel to Fig. 2 B–C. (F) The catalytic efficiency of the mutants is reduced compared to wild type as assessed by monitoring mantGDP release. A representative time course with DENN domains at 4 μM. (G) [3H]GDP dissociates approximately 20-fold faster from the Rab35 F33A mutant than from Rab35. A time course of [3H]GDP dissociating from Rab35 or Rab35-F33A (both at 200 nM concentration) is shown. The solid line is the best fit of a single exponential function, yielding a GDP dissociation rate constant (k-GDP) of 1.35 ± 0.07 × 10-4 s-1 for Rab35 and 25.6 ± 2.8 × 10-4 s-1 for Rab35-F33A. (H) mantGDP association rates are similar for Rab35 and the F33A mutant. MantGDP binding to nucleotide-free Rab was monitored by FRET (Fig. S5B). The concentration dependence of the observed GDP association rate constant (k+GDP) on [mantGDP] is shown. The rate constants from the slope of the best linear fit are 2.09 ± 0.03 μM-1 s-1 for Rab35 and 2.59 ± 0.09 μM-1 s-1 for Rab35 (F33A). Data were generated considering only the fast kobs from the double-exponential fit. The true concentration dependence may be hyperbolic, but we were not able to work at sufficiently high mantGDP concentration to see saturation.
A comparison of the DENND1B-bound Rab35 and nucleotide-bound forms of Rab1 shows that there are subtle changes in regions of the Rab that directly interact with Mg2+-associated nucleotide in the nucleotide-bound form. The P loop (residues 17–22Rab35), which interacts with the β- and γ- phosphate groups and Mg2+ in nucleotide-bound structures of Rab1, shifts slightly into the nucleotide-binding pocket (Cα displacement < 1.9 Å), reorienting residues K21Rab35 and Ser22Rab35. Instead of stabilizing nucleotide binding, the strictly conserved K21Rab35 interacts with D63Rab35 and Q67Rab35. Ser22Rab35 moves away from the Mg2+ binding site (Cβ moves by 2 Å). In addition, D123Rab35 and S150Rab35, which interact with the guanosine base and are involved in its recognition, are shifted slightly out of the nucleotide-binding site (< 1.5 Å) in the Rab/GEF complex. The positional shifts in the nucleotide-binding pocket may occur only after nucleotide has departed and so may not be a cause for its departure. Alternatively, they may be induced by DENN-domain binding to lower the affinity for nucleotide, thereby facilitating its release.
The most dramatic differences between Rab35 and nucleotide-bound Rab1 are in the switch regions (Fig. 3 B–D). Switch I must rearrange in order for Rab35 to bind the DENN domain because in its nucleotide-bound conformations it would clash sterically with helix H7 in the DENN domain (Fig. 3D), and in Rab35/DENN-domain structure, the nucleotide-binding pocket is opened by a conformational change in switch I (Fig. 3B). Thus, in the Rab/GEF complex, residues 37–45Rab35 are folded against the surface of the DENN domain, and residues 33–35Rab35 in switch I are not well ordered. F33Rab35 in switch I is displaced from the pocket, and its Cα position moves by approximately 10 Å. In most Rabs, an aromatic residue corresponding to F33Rab35 is conserved and stabilizes bound nucleotide via an edge-to-face interaction between the aromatic side chain and the guanosine base. Its removal from the nucleotide-binding pocket would be expected to diminish the affinity of Rab35 for nucleotide (see below).
Switch II is not well ordered in the Rab1–GDP structure but the Rab35 switch II region becomes ordered against the DENND1B-S surface in the Rab35/DENN GEF complex (Fig. 3B). Switch II is ordered in the GTP-bound form of Rab1, but its conformation differs from that in the Rab/GEF complex (Fig. 3B). In fact, switch II cannot assume its GTP-bound conformation in the complex because it would sterically clash with DENND1B (loops S3–S4, H6–S7, and H7–S8). Nevertheless, it is likely that the folding of switch II, even if it is into a different conformation, lowers the entropic cost associated with GTP binding.
Finally, removed from the nucleotide-binding pocket, there is a significant conformational change at the C terminus of helix H3 and in the H3–S5 loop. The Cα positions are displaced by 3.6–6.3 Å in this region in Rab35 as compared with nucleotide-bound forms of Rab1. We cannot exclude the possibility that Rab35 simply differs from Rab1 in this region and that the observed shift in H3 and the H3–S5 loop is unrelated to the nucleotide-bound state of Rab35 or DENND1B binding. However, shifting these residues may also accommodate the switch II conformation in the Rab/GEF complex.
DENND1B-S interactions with switch regions are important for Rab binding.
We mutated DENND1B-S surface residues predicted to affect either switch I (Fig. 3E, I224K/I226D/L233D/Y236K; numbering as in Fig. 1F) or switch II (M241R/P242R/Q359A) binding. Both sets of mutations lower the apparent affinity of DENND1B for Rab35, so that neither mutant forms an observable complex with the Rab, as monitored by size exclusion chromatography under conditions where the wild-type DENN domain associates stably with Rab35 (Fig. S2). We also did not detect any measurable GEF activity of the mutants for Rab35, which under our conditions implies that the catalytic efficiency of both is reduced by more than two orders of magnitude as compared to wild-type DENND1B-S (Fig. 3F). Our results demonstrate that interactions with both switch regions are important for binding.
Additionally, the interaction with switch I may be important for GEF activity in inducing a conformational change in switch I. Complex formation displaces F33Rab35 in switch I from the nucleotide-binding pocket. A Rab35 F33A mutant binds GDP with approximately 15-fold lower affinity than wild-type Rab35, as calculated from GDP association and dissociation rate constants (Fig. 3 G and H), and F33Rab35 rearrangement due to switch I reorganization during DENND1B-S binding would be expected to have a similar effect. F33Rab35 displacement alone does not account for the more than 1,000-fold enhancement in the nucleotide exchange rate due to DENND1B (Fig. 1B), as the intrinsic nucleotide dissociation rate of the F33A mutant is only approximately 20 times faster than that of wild-type Rab35 (Fig. 3G). Further, DENND1B also accelerates exchange for the F33A mutant (Fig. S3), again suggesting that there are additional factors important for catalytic efficiency. Indeed, the conformational change in switch I may play another role as it opens the nucleotide-binding pocket, allowing for a direct path to solvent and facilitating both GDP exit and GTP reentry.
Because the switch regions in Rab35 and Rab1b are highly related in sequence (Fig. 3A and Fig. S2F), our structure suggests that Rab1b should bind DENND1B. And indeed, we find that DENND1B associates stably with the nucleotide-free form of Rab1b, as monitored by gel-filtration chromatography (Fig. S2). Nevertheless, neither we nor others (8) were able to detect any measurable exchange activity for DENND1B-S with respect to Rab1b. One possibility is that the differences in the Rab1 and Rab35 switch residues, although limited, lead to nonproductive binding of Rab1b. Or differences in core residues and subtle changes in core packing are transmitted to the DENND-binding surface, thereby affecting activity. Yet a third possible explanation is that, as discussed above, the conformation of helix H3 and the H3–S5 loop differs in Rab35 and Rab1, and that the Rab1 conformation interferes with productive binding by DENND1B.
Model for the Guanine Nucleotide Exchange Mechanism.
Our structure suggests several complementary mechanisms by which DENN-domain binding may enhance GDP dissociation from Rab35. The affinity of Rab35 for nucleotide may be lowered as residues that are critical for nucleotide binding are shifted out of the nucleotide-binding pocket. Residues Ser22Rab35, D123Rab35, and S150Rab35 move slightly, by < 2 Å, whereas F33, the conserved aromatic residue that interacts with the nucleotide base, moves by approximately 10 Å as a result of the change in switch I conformation. Further, switch I refolding results in an opening of the nucleotide-binding pocket, facilitating both GDP departure and also GTP association. That switch II folds, thus more closely resembling its GTP- than its GDP-bound conformation, may enhance GTP binding.
The DENN domain differs in fold from the other Rab GEFs for which structures are known, including the coiled-coil Sec2 (23, 24), the Vps9 domain (25), and DrrA (26), a Legionella protein that acts as a GEF for Rab1 (27). The DENN domain also differs from the TRAPP complex, the cellular GEF for Rab1 (19), in its overall architecture, although as noted previously, both have one or more longin modules.
Nevertheless, despite their differing structures, accumulating data suggest that Rab GEFs use varying combinations of the same general strategies in facilitating nucleotide exchange. They all remodel the nucleotide-binding site of their Rab substrate, and all bind switches I and II, inducing large conformational changes in these regions. Switch I opens, and an aromatic residue that normally stabilizes bound nucleotide is displaced. TRAPP and Vps9 additionally insert structural elements into the nucleotide-binding site: In TRAPP, the C terminus of the Bet3 subunit is inserted and has been proposed to initiate switch I refolding (14), and the Vps9 domain inserts an acidic residue that is thought to sterically clash with and electrostatically repel the nucleotide phosphate group (25). In contrast, Sec2 (23, 24), DrrA (26) and the DENN domain do not protrude into the Rab nucleotide-binding pocket, although steric clashes between DENND1B (helix H7) and portions of switch I may contribute to switch I refolding as the Rab/GEF complex forms.
Materials and Methods
Cloning of Recombinant Proteins.
Coding sequences for full-length DENND1B-S (residues 1–426), the modified DENND1B-S used in crystallization (DENND1B-Sxtal, containing residues 1–149 and 170–410 of DENND1B-S), and DENND1C (residues 1–424) were cloned into a modified pET-28a vector, which has an N-terminal His6-SUMO tag. Mutants of full-length DENND1B-S were prepared using site-directed mutagenesis (Stratagene).
C-terminally truncated Rab35 (residues 1–180) used in crystallization was cloned into a modified pCOLADuet-1 vector, after an N-terminal His6 tag and a TEV protease cleavage site. The full-length Rab35 and Rab1b proteins were cloned into pETDuet-1 vector with a noncleavable His6 tag, and the Rab35 F33A mutant was generated by site-directed mutagenesis (Stratagene).
Protein Expression, Purification, and Complex Formation.
Plasmids containing the coding sequences were transformed into Escherichia coli BL21(DE3) cells. The cells were grown to an OD600 0.7, then shifted to 25 °C, and protein expression was induced by adding 0.5 mM IPTG. Cells were harvested 20 h after induction. Selenomethionine-substituted DENND1B-Sxtal was produced similarily, according to ref. 28. Cells were collected by centrifugation, resuspended in lysis buffer (20 mM Tris [pH 8.0], 300 mM NaCl, 2 mm DTT, 10 mM imidazole) supplemented with protease inhibitors (Complete EDTA-free, from Roche), DnaseI (Sigma), and lysozyme (American Bioanalytical), and lysed using a cell disruptor (Avestin).
DENN-domain proteins were isolated by Ni-NTA chromatography. The His6-SUMO tag was cleaved using SUMO protease and removed by passing the protein solution over Ni-NTA resin again. The protein was further purified by anion exchange chromatography (MonoQ, GE Healthcare) and gel filtration (Superdex 200, GE Healthcare). It was eluted in buffer containing 20 mM Tris (pH 8.0), 300 mM NaCl, and 2–5 mM DTT, and concentrated to approximately 10 mg/mL.
Rab35 used in crystallization was isolated by Ni-NTA chromatography (Qiagen), and its hexahistidine tag was removed with TEV protease. Rab35 was further purified by gel filtration on a Superdex 200 column and concentrated to approximately 25 mg/mL. For full-length Rab35 and the Rab35 F33A mutant, the TEV cleavage step was omitted.
To form the Rab35/DENN-domain complex for crystallization, a 3x molar excess of Rab35 was mixed with DENND1B-Sxtal and incubated overnight at room temperature in the presence of 20 mM EDTA. EDTA strips nucleotide-Mg2+ from the Rab to facilitate the formation of the Rab/GEF complex. The protein mixture was applied to a Superdex 75 gel-filtration column (GE Healthcare) to separate complex from excess Rab35.
Crystallization.
Crystals of native or selenomethionine-substituted DENND1B-S/Rab35 complex were grown at 16 °C by the hanging drop vapor diffusion method. Drops contained 1.35 μL each of protein (4 mg mL-1) and mother liquor (0.1 M Magnesium formate, 19% (w/v) polyethylene glycol 3350, 0.1 M HEPES at pH 7.4), and 0.3 μL of a 40 mM solution of n-Nonyl-β- D-glucoside. Crystals were quick-soaked in mother liquor supplemented with 15% ethylene glycol, loop mounted, and flash-frozen in liquid nitrogen.
Data Collection and Structure Determination.
Data were collected at beamline X25 at NSLS, Brookhaven National Laboratory, and processed using HKL2000 (29) (Table 1). Selenomethionine-substituted crystals diffracted to 2.5 Å and native crystals to 2.1 Å.
The selenomethionine-substituted crystals were used in a SAD experiment to obtain phases, with data collected at the selenium edge. Selenium positions were identified using SHELXD (30), and the positions were refined and density modified phases calculated in SHARP (31). Twofold noncrystallographic averaging as implemented in the RAVE suite of programs (32) yielded an easily interpretable electron density map (Fig. S4), and the Rab/GEF complex was modeled using O (33) and COOT (34). The model was refined against data from the native crystals. An initial round of torsion angle dynamics in CNS (35) was followed by cycles of manual rebuilding in COOT and positional and TLS refinement with REFMAC5 (36, 37). Figures were made using Pymol (DeLano Scientific, LLC).
GDP Release.
The rate-limiting step in guanine nucleotide exchange is GDP dissociation from the Rab, which GEFs accelerate (5). Nucleotide dissociation from Rabs was measured as in refs. 12 and 15 by monitoring the fluorescence associated with Förster resonance transfer (FRET) from Rab35 tryptophan residues to bound (N-methylanthraniloly)-GDP (mantGDP) after rapidly mixing under pseudo-first order conditions ([DENN]≫[Rab35]). To prepare mantGDP-bound Rab35, Rab35 was incubated with 1 mM labeled GDP in the presence of 5 mM EDTA for 10 min at 37 °C. Mg2+ was then added to a final concentration of 10 mM to stabilize the GDP and allow for its binding to the Rab. Excess mantGDP was removed using a Biospin column (Bio-Rad), and the protein was transferred to reaction buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 5 mM MgCl2, 2 mM DTT). The Rab was at a concentration of 0.25 μM, and multiple GEF concentrations were used (2.5, 4.0, 8.0, 12, 18 μM). Measurements were carried out at 25 ± 0.1 °C as described in ref. 15, using an Applied Photophysics SX.18MV-R stopped-flow apparatus equipped with a 400-nm long-pass filter. All concentrations stated are after mixing. Time courses were fitted with a sum of exponential functions by nonlinear least squares regression to determine kobs. The kobs values from multiple time courses were averaged. The rate of intrinsic nucleotide release by Rab35 and Rab35 F33A was measured similarly but quantitating release of tritiated GDP (45 Ci/mmol, PerkinElmer) using a filter-based binding assay (14).
GDP Association.
GDP association with nucleotide-free Rab35 or the F33A mutant was measured by monitoring the fluorescence enhancement associated with FRET after rapid mixing under pseudo-first order condition ([mantGDP]≫[Rab35]). Nucleotide-free Rab35 or mutant was prepared by addition of EDTA and incubation at 25 °C for less than 10 min and used immediately for measurement. Nucleotide-free Rab35 or mutant (0.25 μM) was rapidly mixed with mantGDP (2.5, 5, 10, 15, 25, or 30 μM) using the same stopped-flow apparatus as described above for GDP release assays. All concentrations stated are after mixing. The final buffer condition was the same as for GDP dissociation experiments. Multiple time courses were averaged and fitted into a double exponential function.
Supplementary Material
Acknowledgments.
The authors thank Louise Lucast at Yale for help with Fig. 3G and Fig. S3. We are also grateful to the staff at beamline X25 at NSLS for help with data collection. K.M.R. was supported by the National Institutes of Health (NIH) (GM080616); E.M.D.L.C. by NIH (GM097348), the National Science Foundation (MCB-0546353), the American Heart Association (0940075N), and Hellman Family Foundation; and F.A.B. by the Wellcome Trust (082467/Z/07/Z). X.W. was supported by an NIH predoctoral training grant.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3TW8).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110415108/-/DCSupplemental.
References
- 1.Cai H, Reinisch K, Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell. 2007;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Behnia R, Munro S. Organelle identity and the signposts for membrane traffic. Nature. 2005;438:597–604. doi: 10.1038/nature04397. [DOI] [PubMed] [Google Scholar]
- 3.Pfeffer SR, Aivazian D. Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol. 2004;5:886–896. doi: 10.1038/nrm1500. [DOI] [PubMed] [Google Scholar]
- 4.Zerial M, McBride H. Rab proteins as membrane organizers. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 5.Goody RS, Hofmann-Goody W. Exchange factors, effectors, GAPs and motor proteins: Common thermodynamic and kinetic principles for different functions. Eur Biophys J. 2002;31:268–274. doi: 10.1007/s00249-002-0225-3. [DOI] [PubMed] [Google Scholar]
- 6.Itzen A, Goody RS. GTPases involved in vesicular trafficking: structures and mechanisms. Semin Cell Dev Biol. 2011;22:48–56. doi: 10.1016/j.semcdb.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Colicelli J. Human RAS superfamily proteins and related GTPases. Science’s Stke. 2004;2004:R13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshimura S, Gerondopoulos A, Linford A, Rigden D, Barr F. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J Cell Biol. 2010;191:367–381. doi: 10.1083/jcb.201008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marat AL, Dokainish H, McPherson PS. DENN domain proteins: Regulators of Rab GTPases. J Biol Chem. 2011;286:13791–13800. doi: 10.1074/jbc.R110.217067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allaire PD, et al. The Connecdenn DENN domain: A GEF for Rab35 mediating cargo-specific exit from early endosomes. Mol Cell. 2010;37:370–382. doi: 10.1016/j.molcel.2009.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marat AL, McPherson PS. The connecdenn family, Rab35 guanine nucleotide exchange factors interfacing with the clathrin machinery. J Biol Chem. 2010;285:10627–10637. doi: 10.1074/jbc.M109.050930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esters H, et al. Vps9, Rabex-5 and DSS4: Proteins with weak but distinct nucleotide-exchange activities for Rab proteins. J Mol Biol. 2001;310:141–56. doi: 10.1006/jmbi.2001.4735. [DOI] [PubMed] [Google Scholar]
- 13.Itzen A, Rak A, Goody RS. Sec2 is a highly efficient exchange factor for the Rab protein Sec4. J Mol Biol. 2007;365:1359–1367. doi: 10.1016/j.jmb.2006.10.096. [DOI] [PubMed] [Google Scholar]
- 14.Cai Y, et al. The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes. Cell. 2008;133:1202–1213. doi: 10.1016/j.cell.2008.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chin HF, et al. Kinetic analysis of the guanine nucleotide exchange activity of TRAPP, a multimeric Ypt1p exchange factor. J Mol Biol. 2009;389:275–88. doi: 10.1016/j.jmb.2009.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levivier E, et al. uDENN, DENN, and dDENN: Indissociable domains in Rab and MAP kinases signaling pathways. Biochem Biophys Res Commun. 2001;287:688–695. doi: 10.1006/bbrc.2001.5652. [DOI] [PubMed] [Google Scholar]
- 17.Nicholls A. New York: Columbia University; 1993. GRASP: Graphical representation and analysis of surface properties. [Google Scholar]
- 18.Kinch LN, Grishin NV. Longin-like folds identified in CHiPs and DUF254 proteins: Vesicle trafficking complexes conserved in eukaryotic evolution. Protein Sci. 2006;15:2669–2674. doi: 10.1110/ps.062419006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W, Sacher M, Ferro-Novick S. TRAPP stimulates guanine nucleotide exchange on Ypt1p. J Cell Biol. 2000;151:289–296. doi: 10.1083/jcb.151.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim YG, et al. The architecture of the multisubunit TRAPP I complex suggests a model for vesicle tethering. Cell. 2006;127:817–30. doi: 10.1016/j.cell.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 21.Lee MT, Mishra A, Lambright DG. Structural mechanisms for regulation of membrane traffic by rab GTPases. Traffic. 2009;10:1377–1389. doi: 10.1111/j.1600-0854.2009.00942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eathiraj S, Pan X, Ritacco C, Lambright DG. Structural basis of family-wide Rab GTPase recognition by rabenosyn-5. Nature. 2005;436:415–419. doi: 10.1038/nature03798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong G, Medkova M, Novick P, Reinisch KM. A catalytic coiled coil: structural insights into the activation of the Rab GTPase Sec4p by Sec2p. Mol Cell. 2007;25:455–462. doi: 10.1016/j.molcel.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satoh A, Fukai S, Ishitani R, Nureki O. Crystal structure of the Sec4p.Sec2p complex in the nucleotide exchanging intermediate state. Proc Natl Acad Sci USA. 2007;104:8305–8810. doi: 10.1073/pnas.0701550104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delprato A, Lambright DG. Structural basis for Rab GTPase activation by VPS9 domain exchange factors. Nat Struct Mol Biol. 2007;14:406–412. doi: 10.1038/nsmb1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schoebel S, Oesterlin LK, Blankenfeldt W, Goody RS, Itzen A. RabGDI displacement by DrrA from Legionella is a consequence of its guanine nucleotide exchange activity. Mol Cell. 2009;36:1060–1072. doi: 10.1016/j.molcel.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 27.Murata T, et al. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol. 2006;8:971–977. doi: 10.1038/ncb1463. [DOI] [PubMed] [Google Scholar]
- 28.Doublie S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. Carter CW, Sweet R, editors. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 30.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 31.De La Fortelle E, Bricogne G. Carter CW, Sweet R, editors. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwave anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 32.Kleywegt GJ, Zou JY, Kjeldgaard M, Jones TA. International Tables for Crystallography F. London: Kluwer; 2001. Model building and computer graphics; pp. 353–356. [Google Scholar]
- 33.Kleywegt GJ, Jones AT. Model building and refinement practice. Methods Enzymol. 1997;277:208–230. doi: 10.1016/s0076-6879(97)77013-7. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Brunger AT, et al. Crystallography & NMR System: A new software for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 36.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 37.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. 2001;D57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.