Abstract
DNA copy number variations (CNVs) play an important role in the pathogenesis and progression of cancer and confer susceptibility to a variety of human disorders. Array comparative genomic hybridization has been used widely to identify CNVs genome wide, but the next-generation sequencing technology provides an opportunity to characterize CNVs genome wide with unprecedented resolution. In this study, we developed an algorithm to detect CNVs from whole-genome sequencing data and applied it to a newly sequenced glioblastoma genome with a matched control. This read-depth algorithm, called BIC-seq, can accurately and efficiently identify CNVs via minimizing the Bayesian information criterion. Using BIC-seq, we identified hundreds of CNVs as small as 40 bp in the cancer genome sequenced at 10× coverage, whereas we could only detect large CNVs (> 15 kb) in the array comparative genomic hybridization profiles for the same genome. Eighty percent (14/16) of the small variants tested (110 bp to 14 kb) were experimentally validated by quantitative PCR, demonstrating high sensitivity and true positive rate of the algorithm. We also extended the algorithm to detect recurrent CNVs in multiple samples as well as deriving error bars for breakpoints using a Gibbs sampling approach. We propose this statistical approach as a principled yet practical and efficient method to estimate CNVs in whole-genome sequencing data.
Keywords: structural variation, genomic alterations, model selection, semiparametric model
Copy number variations (CNVs), which are gains or deletions of genomic segments, account for a substantial proportion of human genetic variations. CNVs have been shown to be associated with a wide spectrum of human disorders such as autoimmune diseases (1), autism (2), schizophrenia (3, 4), and obesity (5, 6). CNVs can also occur in the form of somatic alterations. For example, cancer genomes often acquire dosage or structural alteration of cancer-related genes, some of which may confer tumor cells a selective growth advantage over normal cells.
Microarray-based platforms have been widely used in genome-wide studies to identify CNVs in cancer genomes (somatic CNVs) (7–10) as well as in the genomes of normal population (germ-line CNVs) (11–16). These efforts have led to the discovery of previously unrecognized oncogenes and tumor suppressors. However, due to the limited resolution of microarrays, this approach has suffered from poor sensitivity in detecting small CNVs. More recently, the advent of next-generation sequencing and the rapid increase in its throughput have led to the possibility that CNVs can be characterized in much finer detail via whole-genome sequencing. With mate-pair reads, sequencing-based approaches can also be used for the discovery of structure variations (SV) such as insertions, deletions, inversions, and translocations (17–19). Current mate-pair-based algorithms for SV detection cannot estimate the magnitude of copy number change accurately, tend to perform poorly in genomic regions rich in tandem or inverted duplications (20), and have limited power in detecting insertions larger than the insert size (the distance between the two ends of a paired-end read) (21).
Although the density of aligned reads along the genome generally corresponds to the DNA copy number, it is also affected by many sources of bias. These include the higher likelihood of sequencing GC-rich fragments, the uneven representation of genomic regions in library preparation due to variability in DNA fragmentation, and the regional variation in the fraction of short reads that can be aligned to a given position. Thus, somatic CNVs in the tumor genome can be best identified by comparing the read distribution in the tumor genome with that of the matched normal genome. The genomic regions with disproportionate read counts indicate potential CNVs—e.g., the prevalence of tumor reads over normal ones suggests a somatic copy gain in the tumor genome. This approach was adopted by SegSeq (22) and CNV-seq (23). However, both of these methods perform statistical tests based on a Poisson model, in which reads are assumed to be distributed uniformly across the genome. This assumption hardly holds even for normal genomes (SI Appendix, Fig. S1) due to the biases mentioned above. In addition, the results are sensitive to the window size that must be specified a priori in these methods, and approaches with initial local detection of breakpoints followed by copy number estimation tend to perform poorly compared to more global approaches (24).
In this paper, we describe a statistically rigorous and computationally efficient algorithm called BIC-seq for detecting CNVs from whole-genome sequencing data. This algorithm does not assume a Poisson or other parametric models on the read distribution as is done in currently available methods, and it is thus more robust to outliers and datasets that cannot be well approximated with a parametric model. It is also fast and able to handle high-coverage genomes effectively. Furthermore, the statistical framework behind BIC-seq can be extended to the problem of identifying recurrent CNVs in multiple cancer genomes. To test the performance of the algorithm, we applied BIC-seq on the newly sequenced cancer genome of a patient with glioblastoma multiforme (GBM) and its matched control. Large CNVs (e.g., > 15 kb) identified by BIC-seq were largely concordant with those identified from the two microarray platforms on which the same genomes were profiled. However, many small CNVs identified by BIC-seq were not detected on the arrays. Subsequent validation on a subset of small CNVs by quantitative PCR (qPCR) showed > 80% true positive rates and confirmed the accuracy of the magnitude of copy number ratios.
Results
CNV Detection Algorithm Based on the Bayesian Information Criterion.
Current high-throughput sequencing platforms generate short (36–100 bp) sequenced tags from one or both ends of hundreds of millions of DNA fragments. These short reads can then be aligned to the reference genome using alignment tools such as MAQ (Mapping and Assembly with Quality) (25), Bowtie (26), and BWA (Burrows-Wheeler Aligner) (27). The basic idea behind CNV detection is that larger or smaller than the expected number of tags in a genomic region corresponds to gain or loss of DNA, respectively. Although it is tempting to design an algorithm to detect such a variation in a single sample, this would result in a large number of false positives because the tag distribution along the genome fluctuates considerably due to biases in sequencing and genome content as described above. Thus, the “read-depth” methods including ours use the disease-vs.-matched control approach.
In the BIC-seq algorithm, the short reads are aligned onto the reference genome and the uniquely aligned reads are sorted according to their genomic coordinates. Single genomic positions having many orders of magnitude more mapped reads than their neighboring positions (SI Appendix, Fig. S2) are filtered out because they are likely to be a result of amplification bias (Materials and Methods). Then, tumor and normal reads are mixed and binned (1-bp bins are allowed). The idea of BIC-seq is to iteratively identify and merge the most similar pair of bins (Fig. 1). Here, we choose to use the Bayesian Information Criterion (28) (BIC) as the merging and stopping criterion. The BIC, which is widely used as a model selection criterion in statistics, consists of two terms: One term is the negative log likelihood, which measures how well the model fits the data; and the second term is the penalty for the model complexity, which prevents model overfitting. The model complexity is measured by the product of the number of variables in the model and the logarithm of the number of observations (Materials and Methods and SI Appendix). For CNV detection, the number of variables is equal to the number of breakpoints plus one. Generally, the model with a smaller BIC is preferred. To accommodate more flexibility to BIC-seq, we introduce a parameter λ in the penalty term of the BIC. This parameter allows the user to control the smoothness (the number of breakpoints) of the final profile generated by BIC-seq and reflects the user’s prior belief about the CNV profile. The larger this parameter is, the smoother the final profile will be.
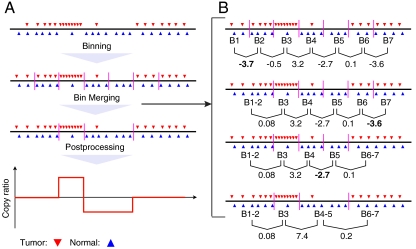
Fig. 1.
A schema for BIC-seq. (A) The dataflow of BIC-seq. First, the short reads are aligned to the reference genome and the outliers are removed. Then, the short reads are binned into small bins (e.g., 10 bp bins), and the initial bins are iteratively merged using the BIC. The vertical purple bars in the plot are the boundaries between neighboring bins. Lastly, copy ratios are calculated based on the segmentation given by BIC-seq. (B) A bin-merging procedure based on the BIC. We demonstrate the procedure for λ = 1. Given a list of initial bins, BIC-seq first calculates the BIC differences between the current configuration and all possible configurations in which two adjacent bins are merged. In the plot, the numbers under the bin pairs are their corresponding BIC differences. Then, BIC-seq identifies the pair with the smallest BIC difference. If this BIC difference is less than zero, the corresponding bin pair will be merged; otherwise, BIC-seq will stop merging bin pairs. In this example, the bin pair B1 and B2 have the smallest BIC difference (-3.7) and they are merged, giving a new bin B1-2. BIC-seq then updates the BIC differences for the bin pairs. As shown in the plot, we only need to update the BIC difference for the bin pair B1-2 and B3, because all other BIC differences remain the same as before the merging of B1 and B2. This fact holds in general and we used it to expedite BIC-seq (SI Appendix). The above process is then repeated until the BIC cannot be improved further—i.e., until no BIC difference is less than zero. After the merging of bin pairs, BIC-seq also tries to merge three or more neighboring bins if their merging can improve the BIC (SI Appendix). For this example, the merging of three or more neighboring bins cannot improve the BIC.
Given an initial configuration of bins, BIC-seq attempts to reduce the overall BIC by merging the appropriate neighboring bins. Briefly, for each pair of neighboring bins, BIC-seq calculates the BIC difference as if the pair were merged. Then, BIC-seq identifies the pair of bins with the smallest BIC difference and merges the bin pair if this BIC difference is less than zero. The above process is repeated until the overall BIC cannot be further reduced by merging pairs of neighboring bins. After merging bin pairs, BIC-seq also attempts to merge three or more neighboring bins if some of them can be merged (SI Appendix). By using a red-black tree (29) to efficiently store BIC differences, it is computationally efficient (SI Appendix). For instance, if the initial bins are chosen as equally spaced bins of 100 bp, it only takes BIC-seq a few seconds to finish the merging process for a chromosome on a single-processor computer. Based on the segments obtained from the bin merging process, the copy ratios between tumor and normal genome of each segment can then be easily calculated (SI Appendix).
BIC-seq can be easily extended to the multisample case by modifying the log likelihood term and the penalty term in the definition of the BIC (Materials and Methods and SI Appendix). In addition to giving point estimates of the breakpoints, it is also desirable to assign confidence to the point estimate. Here, we developed a Bayesian model and a corresponding Gibbs sampler (30, 31) to assign credible intervals to the breakpoints given by BIC-seq (Materials and Methods and SI Appendix). This error estimation for CNV boundaries is helpful in selecting candidate genes and other genomic elements for further examination.
After the segmentation of the genome, one can apply a copy ratio and/or a p-value threshold to get a set of CNV candidates. To give a false discovery rate (FDR) estimate of this CNV call set, we pool the tumor and normal reads together, randomly sample with replacement from the pooled data as tumor/normal reads, and run BIC-seq on the resulting pseudotumor/normal data (SI Appendix). CNVs are called with the same criterion as used in the original dataset. Because the randomly sampled data are generated under the null hypothesis of no CNV in the genome, any CNV call from the pseudotumor/normal data would be a false positive. The ratio between the numbers of CNV calls from the resampled data and the original data is then an FDR estimate. A more stable FDR estimate can be obtained by repeating this resampling procedure many times and taking the mean as the final FDR estimate.
BIC-seq has a single parameter λ that the user can specify, based on the desired level of confidence in the CNV calls and the genome coverage (also see section Sequencing Coverage and Statistical Power). For all levels of coverage, a copy ratio threshold should first be used (e.g., log2 ratio > 0.2 or < -0.2) to remove low-amplitude changes that are likely to a result of random fluctuations. Then, for low-coverage data (< 1×), λ should be small (e.g., 1 or 1.2). To further reduce FDR, one can apply a p-value threshold to remove the less significant, small CNVs. For medium coverage (e.g., 2–5×), a larger λ (e.g., 2) should work well without the additional p-value filtering. For high-coverage (e.g., 10–30×), λ = 4 should give very confident calls while still detecting many small CNVs (e.g., 100–1,000 bp). SI Appendix, Fig. S4 confirms that λ = 4 has good sensitivity at detecting CNVs as small as 500 bp. We evaluated the effect of λ on the performance of BIC-seq in a simulation study, which showed that both true positive rate and FDR increase as λ gets smaller (SI Appendix, Fig. S3). Simulation studies also confirm that BIC-seq is more sensitive than SegSeq for a given FDR level (SI Appendix, Fig. S5).
We also note that the merging process of BIC-seq is equivalent to performing a series of likelihood ratio tests. Thus, the choice of the tuning parameter λ is equivalent to setting a type I error rate (a type I error in a merging step is the event that two neighboring bins should be merged but fail to merge). The likelihood ratio test statistic asymptotically follows a χ2 distribution, which allows us to provide a more intuitive way of specifying the tuning parameter λ—i.e., the user can specify the expected number of type I errors in the merging process (SI Appendix).
Copy Number Profiling of Human Solid Tumor (GBM) by BIC-seq.
We applied BIC-seq to profiles from a cancer patient, sequenced as part of the Cancer Genome Atlas (TCGA) project (Materials and Methods). Tumor and matched control (blood) DNA were obtained from the same individual, a 53-y-old woman who was diagnosed with primary GBM, and sequenced on the Applied Biosystems SOLiD (Sequencing by Oligonucleotide Ligation and Detection) platform. We obtained 833 million (10×) and 603 million (7×) uniquely aligned 35-bp single-end reads for the tumor and its matched normal genome, respectively. To be conservative, we set the tuning parameter λ = 4; a smaller value results in identification of smaller feature sizes at the expense of lower confidence level. The initial bins were chosen as 10-bp bins, which was a reasonable compromise between resolution and memory usage for this dataset. After filtering for segments with copy ratios greater (tumor/matched normal) than 1.5 or less than 0.5, we found 306 putative somatic CNVs, ranging from 10 bp to 5.7 Mb with a median size of 59 kb. Because deletions or insertions that are about the length of the sequenced read cannot be distinguished from each other by a read-depth method, we focused on CNVs larger than the read length. After removing 15 such CNVs, we obtained 291 putative CNVs (Fig. 2), which covered 88.51 Mb (2.95%) of the human genome. Among the 291 somatic CNVs, 192 (73.56%) showed at least a partial overlap with 1,926 reference sequence (Refseq) genes (including intronic sequences) and 170 (58.42%) with coding sequences (Fig. 2). We further compared these CNVs with cancer genes listed in Futreal et al. (32) and found that 19 out of 291 somatic CNVs showed overlap with 22 out of 288 cancer genes. These 22 genes include the well-known cancer-related genes EGFR and CDKN2A, whose amplification and deletions have been frequently observed in GBM (33, 34).
Fig. 2.
CNVs detected by BIC-seq in the GBM genome. (A and B) The distribution of putative CNVs detected in GBM with tuning parameter λ = 2 and 4, respectively. Here, the x axis notes the CNV sizes. (C and D) Overlaps of GBM CNVs with Refseq genes for λ = 2 and λ = 4. Intergenic, no overlap with any gene; whole gene, covering an entire gene; exon, overlapping with at least one exon but not covering an entire gene; Intron, overlapping with introns but not exons.
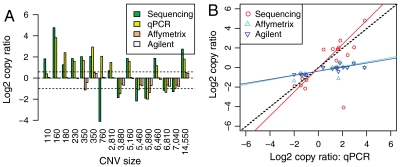
We compared these CNVs with those detected by two microarray platforms, Agilent 244K comparative genomic hybridization (CGH) microarrays and Affymetrix SNP 6.0 arrays (see Materials and Methods). When we plotted the log2 ratios of the 291 putative CNVs as estimated by array platforms and sequencing (SI Appendix, Fig. S6), we found that a large fraction of the BIC-seq ratios were larger than the array ratios (which cluster around zero), indicating that these CNVs detected by BIC-seq were missed by arrays. The slopes of the linear median regression lines are 0.557 and 0.554 with SDs of 0.110 and 0.101. To determine whether CNV size is related to the difference in the platform performance, we classified the 291 CNVs into two sets according to their sizes, one set consisting of 184 CNVs larger than 15 kb and the other consisting of 107 CNVs less than 15 kb. We found that, for the large CNVs, the log2 copy ratios given by BIC-seq and two microarray-based platforms are close, especially between the log2 copy ratios given by BIC-seq and the Agilent array (SI Appendix, Fig. S7 A and B and Fig. S8). But for the small CNVs, the log2 ratios were dramatically different between the platforms. (SI Appendix, Fig. S7 C and D).
To test the accuracy of the copy ratio estimates given by BIC-seq for small CNVs (less than 15 kb), we selected 16 CNVs for which good primers could be designed, ranging from 110 bp to 14 kb, from the 107 small CNVs for qPCR validation. The experimental validation confirmed 14 out of 16 CNV calls (true positive 87.5%; Fig. 3), assuming that a copy gain or loss is a true positive if its copy ratio estimate given by qPCR is greater than 1.5 or less than 0.5, respectively. Fig. 4 shows two examples of validated CNVs discovered by BIC-seq but missed by array platforms. To further investigate the accuracy of copy ratio estimates, we fitted three linear median regression models, using the log2 copy ratios given by qPCR as the predictor and the log2 copy ratios given by BIC-seq and the two array platforms as the responses. For BIC-seq ratios, the estimated slope of the linear model is not significantly different from one (1.21 with SD 0.20) and the estimated intercept is not significantly different from zero (-0.062 with SD 0.33). However, for Affymetrix ratios, the slope and the intercept of the model are 0.23 (SD 0.06) and -0.36 (SD 0.09); for Agilent ratios, the slope and the intercept are 0.21 (SD 0.05) and -0.38 (SD 0.08). This regression analysis suggests that the copy ratios given by BIC-seq are more concordant with qPCR-based estimates than those given by array platforms.
Fig. 3.
Experimental validation of 16 BIC-seq CNVs. (A) Bar plot of the log2 copy ratios estimated from the four platforms. The two dashed lines correspond to copy ratios 1.5 and 0.5, respectively. (B) Scatter plot of the log2 copy ratios given by the sequencing data and two array-based platforms versus the log2 copy ratios given by qPCR. The red, cyan, and blue solid lines are the fitted linear median regression models using the log2 copy ratios given by qPCR as a predictor and the log2 copy ratios given by sequencing data and two array-based platforms as responses, respectively. The slopes of the linear model for sequencing, Affymetrix, and Agilent are 1.21 (SD 0.20), 0.23 (SD 0.06), and 0.21 (SD 0.05), showing that copy ratios given by sequencing data are more accurate than that given by array platforms.
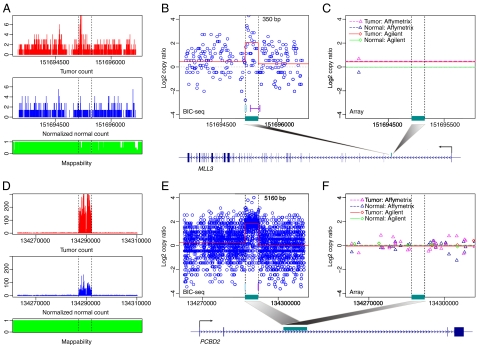
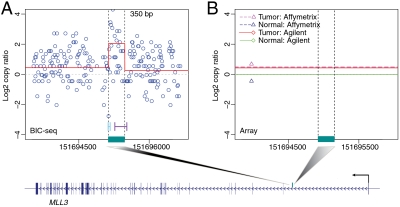
Fig. 4.
Two qPCR validated CNVs that were missed by the array-based platforms. (A–C) A 350-bp focal CNV. (A) The distribution of tumor (Top) and normal (Bottom) reads near the identified CNV at nucleotide resolution. The normal reads are rescaled such that the summation of the transformed values is the same as the total tumor read count. (B) The local profile given by BIC-seq (red line). The circles are the copy ratios calculated based on 10-bp bins. The regions marked by cyan and purple lines are the 95% credible intervals for the left and right breakpoints of the CNV. The CNV overlaps with the intronic region of the gene MLL3 and the position of the CNV in the gene is marked by the dark cyan bar. (C) The profiles given by Affimetrix and Agilent platforms. (D–F) Another validated CNV missed by the array platforms. The CNV overlapped with the gene PCBD2. Both tumor and normal genomes are enriched in this CNV region, but the enrichment magnitude of the tumor genome is even greater than its matched normal genome.
We observed that several of the validated small CNVs overlapped with cancer-related genes, including INPP1, ADAM12, MGMT, MLL3, PCBD2, and AMY2A, whose copy number alteration may influence cancer development and progression. For example, INPP1 encodes one of the enzymes involved in the phosphatidylinositol (PI) signaling pathway, which is frequently altered in glioblastoma (33). Thus, in addition to well-known events such as PIK3CA mutations, genomic alterations involving the INPP1 locus may contribute to glioblastoma pathogenesis. Overexpression of ADAM12 has been reported in a variety of human cancers and the expression level of ADAM12 mRNA showed a correlation with glioblastoma cell proliferation (35). MGMT promoter methylation occurs frequently in glioblastoma and is associated with a better treatment response and a more favorable prognosis, but it is generally believed that MGMT is not commonly mutated or deleted (36). However, we observed a focal deletion involving the MGMT locus, which suggests that a focal deletion involving this locus may act as an additional inactivating mechanism in glioblastoma. This phenomenon may contribute to the occasional discordance between MGMT promoter status and treatment response. Indeed, a recent report suggests that treatment response correlates more closely with MGMT mRNA expression level than with MGMT promoter methylation (37). The gene MLL3, which encodes components of a histone H3 lysine 4 methyltransferase complex (38), is frequently mutated in various cancer types suggesting that MLL3 may play a role in gliomagenesis (39–41). Homozygous deletion involving AMY2A was observed in approximately 20% of primary gastric cancers (42) and the amplification of PCBD2 was previously observed in mesothelioma (43).
We observed that, in some CNV regions, there is read enrichment in the tumor genome compared to the normal genome, yet both tumor and normal genomes have several magnitude more reads than in their neighboring regions (Fig. 4 D–F). This read distribution might be due to a complex genomic configuration (e.g., tumor-specific somatic copy gain occurring within patient-specific germ-line CNVs). This phenomenon is also observed in the genomes of acute myeloid leukemia (AML) patients described below and deserves further investigation.
Copy Number Profiling of Human Hematologic Tumor (AML) by BIC-seq.
We further applied BIC-seq on two AML genomes (44, 45). For the first AML genome (44) (AML1), we obtained 1.9 (20×) and 0.8 (9×) billion uniquely aligned 32 bp single-end reads from the tumor and its matched normal genomes, respectively (SI Appendix, Fig. S9). Most of the second AML genome (45) (AML2) were paired-end reads (SI Appendix). We aligned the two ends of paired-end reads independently and obtained 588 (10×) and 655 (11×) million uniquely aligned 49-bp reads from the tumor and its matched normal genome (SI Appendix, Fig. S10). Similar to the analysis of the GBM genome, the initial bins were chosen as 10-bp bins and the tuning parameter λ was set as 4. BIC-seq identified 985 and 143 putative CNVs for AML1 and AML2, respectively. After removing the putative CNVs smaller than the read length, AML1 had 943 putative CNVs with median size 260 bp (ranging from 40 bp to 127 kb), and covered 0.6 Mb (0.02%) of the human genome; AML2 had 107 putative CNVs with median size 1,160 bp (ranging from 50 bp to 19 kb) and covered 0.2 Mb (0.006%) of the human genome. Four hundred seventy-one out of 943 AML1 CNVs overlapped with 479 Refseq genes (SI Appendix, Fig. S11) including nine genes in the cancer gene list (32). For example, a 1.1-kb CNV (copy ratio 0.5) overlapped with the coding region of RUNX1, which was reported to be involved in AML (46, 47). For AML2 CNVs, 20 out of 107 CNVs overlapped with 19 Refseq genes (SI Appendix, Fig. S11) and 2 of 19 genes are on a previously published list of cancer-relevant genes (32).
Between AML1 and AML2, there was one common CNV, which overlapped ANKRD30BL. If CNVs smaller than the read length are included, there were three more, involving PCBD2 and MAN1A1. Interestingly, PCBD2, which was previously reported to be amplified in a mesothelioma tumor genome (43), is also detected in the GBM genome (Fig. 4 D–F). BIC-seq predicts that all three tumor genomes are amplified at this locus, and the CNV sizes are 5,160, 4,520, and 40 bp for GBM, AML1, and AML2, respectively.
Sequencing Coverage and Statistical Power.
The statistical power for detecting CNVs is dependent on the sequencing depth. Generally, the greater the number of reads sequenced, the higher the statistical power. However, it is important to characterize the performance of the algorithm as a function of sequencing depth because this is a key consideration in designing a study. The cost of sequencing is roughly proportional to the coverage and one must determine, e.g., whether it is more beneficial to sequence each individual at 20× or to sequence twice the number of individuals at 10×. In addition to the robustness of the algorithm, intrinsic properties of the CNV, such as its size, magnitude of change in copy number, and alignability of the genomic location, can also have substantial influence on the statistical power of CNV detection.
Here, we use simulation to depict the relationship between these factors and the statistical power. We first simulated 100 “tumor” chromosomes based on the human reference chromosome 22 (see Materials and Methods). Each of the simulated tumor chromosomes contained 42 CNVs (7 CNV sizes × 6 copy ratios) ranging from 100 bp to 100 kb with varying magnitudes of change. Then, short reads were randomly placed on the 100 template tumor chromosomes and the human reference chromosome 22 using MetaSim (48), and these short reads were aligned back to the reference genome using Bowtie (25). Finally, we applied BIC-seq to evaluate the statistical power for CNV detection under many different scenarios (Fig. 5 A–C). The CNV regions were chosen as regions with log2 copy ratios less than -0.2 or greater than 0.2 and further filtered using a p-value cutoff such that the estimated FDR is less than 0.01.
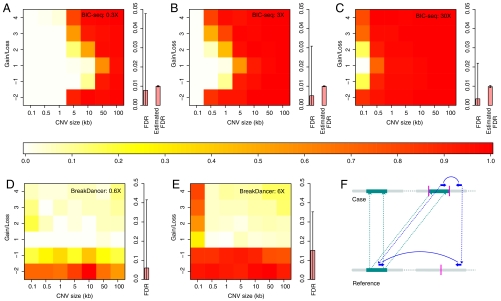
Fig. 5.
Statistical power for CNV detection. The colors represent the power of CNV detection under different scenarios, with red regions indicating the greatest power. The y axis denotes the number of copies gained or lost. (A–C) The CNV detection power of BIC-seq at 0.3×, 3×, and 30× coverage, respectively. The bar plot on the right shows the mean FDR of the 100 simulations and the corresponding mean estimated FDR. Here, 0.3× coverage means that both tumor and normal sequencing are 0.3× and similarly for the others. The bars in the box plot are the error bars of the true and the estimated FDRs (2 SD around the mean with lower bound of zero). (D and E) The SV detection power of BreakDancer at 0.6× and 6× coverage. Because BreakDancer only uses sequencing data from the tumor genome, we doubled the sequencing reads to make the comparison fair (Materials and Methods). (F) The signature of a PEM resulting from a duplication that would be misleading. A DNA segment (the dark cyan segment in the reference genome and the left dark cyan segment in the case genome) is duplicated and inserted to another position on the same chromosome (the dark cyan segment on the right part of the case genome). The purple bar in the plot represents the breakpoint of the insertion. A paired-end read (the blue arrows) that spans the breakpoint in the case genome is sequenced. When the pair is mapped back to the reference genome, the mapped distance between the two ends is significantly larger than the insert size, which is similar to a paired-end read resulting from a large deletion in the case genome.
Overall, copy loss is much easier to detect than copy gain. For example, the power to detect a two-copy loss CNV is roughly the same as that for detecting four-copy gain CNVs. One-copy gain CNVs have the lowest statistical power, sometimes dramatically; e.g., at 3× coverage (Fig. 5B), the power for a 500-bp CNV of two-copy loss is around 90%, but the power for a one-copy gain CNV of the same size is less than 1%.
For comparison, we also tested the performance of a well-known algorithm BreakDancer (19), which is based on paired-ends mapping (PEM). As above, we first simulated 100 tumor chromosomes, each of which contained 42 CNVs with predefined sizes and copy numbers (Materials and Methods). Then, we applied MetaSim to generate paired-end reads. The insert size and the SD of the insert size were set as 220 and 20 bp, respectively, as estimated from the AML2 data. Then, we applied BreakDancer on the simulated data to call CNVs. Because BIC-seq needs sequencing data from both tumor and normal genomes to call the somatic CNVs, but BreakDancer only needs sequencing data from tumor genomes to call the SVs, we required the tumor reads for BreakDancer to be at least twice as many as that for BIC-seq to make the comparison fair (Materials and Methods). For BreakDancer, we used a lenient criterion to determine whether an SV call is a positive discovery (Materials and Methods). As shown in Fig. 5 D and E, BreakDancer performs well when detecting deletions and small insertions, but it has limited power for detecting insertions larger than 500 bp. The reason is that, for an insertion larger than the insert size, the signature of the PEMs is complex. Some of PEMs may appear to contain a deletion between the pairs instead (Fig. 5F), making it difficult for the algorithm to make a prediction. In contrast, BIC-seq performs very well at detecting larger CNVs. For low coverage data (Fig. 5 A and D), BIC-seq performs well for detecting large deletions (e.g., one-copy loss of size > 50 kb) in addition to large insertions, but BreakDancer does poorly because there are few or no supporting PEMs for the SVs. This simulation suggests that BIC-seq and the PEM-based algorithms are complementary methods and that a combination of the two methods would give higher sensitivity, especially for low-coverage data.
In practice, it is important to determine which sequencing depth will allow for the detection of nearly all CNVs. The simulation studies here suggest that read-depth methods should be able to detect almost all CNVs larger than 500 bp at 30× coverage. Combined with a PEM-based method, it should be possible to detect all but very small deletions and insertions. In general, one should first use a read-depth method followed by a PEM-based method to discover large CNVs and a PEM-based method followed by a read-depth method to capture smaller CNVs.
Discussion
We described a statistically rigorous and computationally efficient algorithm for detecting CNVs from high-throughput sequencing data. BIC-seq was able to detect CNVs as small as 40 bp in three human cancer genomes (one GBM and two AMLs) with medium-to-high coverage. This algorithm is fast: Once the reads are aligned, it can resolve CNVs in a 30× genome with 10-bp resolution in approximately 2 h. Subsequent qPCR validation demonstrated that the sequencing-based CNV calls, including the estimated copy number magnitude, are more accurate than those from array platforms. Taking advantage of the discrete sequencing data, we also developed a method to assign error bars for the breakpoints, which would be important for subsequent localization of the breakpoints. To facilitate detection of recurrent CNVs in multiple tumor genomes, we extended BIC-seq to the multisample case. There are several algorithms, such as GISTIC (Genomic Identification of Significant Targets in Cancer) (49), for finding recurrent CNVs in a group of samples. These algorithms combine information from multiple samples after each profile is segmented individually. Multisample BIC-seq, on the other hand, performs multiprofile segmentation directly on the raw data and therefore is more powerful for detecting recurrent small or low-copy-change CNVs that may not be captured on a sample-by-sample segmentation.
Paired-end sequencing with an insert size ranging from 200–300 bp to approximately 3–5 kb is becoming the standard in whole-genome sequencing. The only exceptions are when the DNA is sufficiently degraded that a paired-end library cannot be constructed. PEM is advantageous in several aspects, including allowing one to infer the location of duplicated segments and to detect balanced rearrangements such as translocations and inversions. However, its performance in genomic regions enriched with repeats or segmental duplications is limited. Further, as shown in the simulation study above, one should combine the PEM algorithms with a read-density approach to improve the sensitivity and specificity of CNV detection. For example, if the read-density-based methods detect a copy loss region, there should be paired-end reads surrounding the detected region whose mapped distances are significantly larger than expected. Assuming sufficient sequencing depth, the copy loss calls that are not supported by such paired-end reads are then likely to be false positives. Likewise, the methods based on PEM have limited power in detecting large insertions, but the read-density-based methods are better at detecting larger CNV events. Thus, one may first apply the read-density-based methods to detect the large copy gain regions, and then use PEM to confirm the existence of the copy gain region or to refine the breakpoint positions.
In the 1,000 Genomes Project (50), the investigators identified thousands of SVs in a normal population based on the consensus of many SV/CNV detection algorithms, but they only focused on deletions. They found that, for deletions, Genome STRiP (Structure in Populations) (51), which combines the read-depth and the paired-end information, gave the lowest false discovery rate. However, this method relies on population-wide information and has limited power for detecting rare CNVs. Furthermore, because this algorithm first uses a PEM-based method to call the candidate CNVs and then uses read depth to remove false positives, it also suffers from the similar problem as BreakDancer—i.e., low sensitivity for detecting large insertions. For the same reason, this algorithm also has limited power for detecting large deletions using low-coverage data, e.g., at 0.3× coverage as shown in Fig. 5 A and D.
Array CGH methods have brought tremendous progress in characterizing copy number profiles in many diseases. But, as sequencing technology continues to improve, the cost of sequencing is rapidly declining and a multitude of samples will be subject to whole-genome sequencing. Given the power to detect CNVs and other genomic changes at unprecedented resolution, the next algorithmic challenge will be to determine the biological significance of the aberrations and identify the “driving” variants from the “passenger” variants. For more subtle variants, this problem will require delineating the phenotype more precisely and collecting appropriate samples as well as relevant controls. For some phenotypes, it will also require a population-scale sequencing effort to reach an acceptable sensitivity and specificity. In all cases, it will be important to handle the challenges of managing large amounts of data and to apply efficient and statistically powerful algorithms.
Materials and Methods
Data Availability.
TCGA accession numbers for the GBM data are TCGA-06-0208-01 (tumor) and TCGA-06-0208-10 (normal). The sequencing data are available from the database of Genotypes and Phenotypes (dbGaP). Because these are individual-level data, authorization is required for data access. The segmented profiles (level III) for the Agilent and Affymetrix data were obtained from the Cancer Genome Atlas data portal (http://tcga-data.nci.nih.gov/tcga/). These profiles had been generated by applying the Circular Binary Segmentation (52) method, one of the leading CNV detection algorithms for microarray platforms.
Outlier Removal.
Some genomic positions can have mapped short reads several orders of magnitude more than their neighboring regions. We remove these outliers before further analysis. To determine whether a genomic position s0 is an outlier, we choose a local window of s0 and calculate the pth quantile q (e.g., 0.95th quantile) of the read counts at the genomic positions in the local window. If read count ns0 at s0 is more than m (e.g., m = 5) times of the quantile q, this genomic position will be viewed as an outlier position and the read count at this position will be set as mq. Here, the local window is chosen as the window (a, b) such that both the left window (a, s0) and the right window (s0, b) contain w/2 genomic positions that have at least one read, where w is a parameter determined by the user. In the CNV analysis on the GBM and AML genomes, we set w = 200, m = 5, and p = 0.95.
The Statistical Model and the Segmentation Algorithm.
Given a short read R that was mapped to the reference genome, let Y be one if the read is from the tumor genome and zero if from the normal genome, and S be the mapped position of the read. We view the short read R as equivalent to the two-dimensional random variable (Y, S)—i.e., R = (Y,S). Suppose that the joint distribution of R = (Y,S) is f(y,s), and the marginal distribution function of S is f(s). Then, given a set of short reads R1 = (y1,s1),…,Rn = (yn,sn) on a reference chromosome c, the joint likelihood of these short reads are 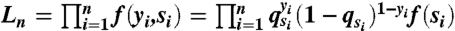 , where qsi = Pr(Y = 1|S = si) is the probability of a read being a tumor read conditional on the read being mapped to the genomic position si.
, where qsi = Pr(Y = 1|S = si) is the probability of a read being a tumor read conditional on the read being mapped to the genomic position si.
If the tumor genome is identical to the normal genome, the conditional probabilities qsi would be the same genome wide. However, if the tumor genome has any somatic CNV regions, the conditional probabilities qsi will be different for different CNV regions, but for any two points between two consecutive breakpoints, the conditional probabilities qsi would still be the same. To determine which set of breakpoints is the best, we use the BIC as the criterion, and the set of the breakpoints with the smaller BIC is preferred. The BIC of a model is in general defined as
where L is the likelihood function evaluated at the maximum likelihood estimate (MLE), k is the number of parameters of the model, and n is the total number of observations. For the profile with m breakpoints, there are m + 1 parameters p0,p1,…,pm, where pi are the common probabilities qs for s between two consecutive breakpoint τi and τi+1. We thus define the BIC as
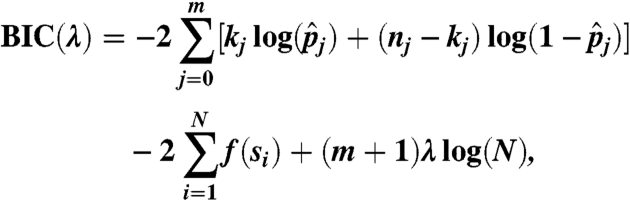 |
where kj and nj are the number of tumor reads and the total number of sequencing reads between the breakpoints τj and τj+1, and 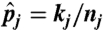 is the MLE of the parameter pj given the breakpoints. Note that though the distribution function f is unknown, the term involving f is the same for all models. Thus, we can compare two models’ BIC without specifying any parametric form about f. (See SI Appendix for the BIC-seq algorithm.)
is the MLE of the parameter pj given the breakpoints. Note that though the distribution function f is unknown, the term involving f is the same for all models. Thus, we can compare two models’ BIC without specifying any parametric form about f. (See SI Appendix for the BIC-seq algorithm.)
The above statistical model can be easily extended to the multisample case. Specifically, suppose that we have sequencing data from G pairs of tumor/normal genomes and that there are nk short reads from the kth pair of the genomes on a reference chromosome c. Let Lnk be the joint likelihood of the kth pair as above. Then, the joint likelihood of the short reads on c from G pairs of tumor/normal genomes is just the product of the individual likelihood Lnk—i.e., 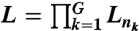 . The BIC becomes
. The BIC becomes
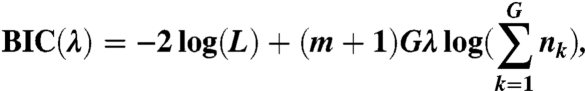 |
where m is the number of breakpoints. Then, we can easily extend the corresponding algorithm for CNV detection to the multisample case.
Credible Intervals to Breakpoints.
We develop a Gibbs sampler to assign credible intervals to the breakpoints given by BIC-seq. Given a genomic window a, b, assume that there is only one breakpoint τ in the window. Suppose that the reads in the window a, b are D = (Ri1,…,Ri2). Let p1, p2 be the probabilities of a read being a tumor read before and after the breakpoint τ. Then, conditional on the breakpoint τ and the probabilities p1, p2, the joint distribution of D = (Ri1,…,Ri2) is
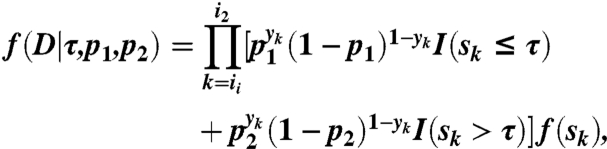 |
where I(·) is the indicator function. Put uniform priors on p1, p2, and τ. We have that the full conditional distributions of p1, p2 are β-distributions and the full conditional distribution of τ can be easily sampled using the inverse of its cumulative distribution function (SI Appendix). Given a breakpoint τj predicted by BIC-seq, we can take a, b to be the breakpoints before and after the breakpoint τj. However, to expedite the Gibbs sampler, we choose a, b such that there are at most 1,000 normal reads in the intervals (a, τj) and (τj, b) (SI Appendix).
Experimental Validation Using qPCR.
All CNVs detected by BIC-seq were validated with qPCR. Primers were constructed internal to the CNV with Primer3 (53). See SI Appendix, Table S1 for primer sequences and sizes of qPCR amplification. For all CNVs, reactions were performed in duplicate. The reaction mixture (25 µL) contained 1× Power SYBR green (catalog no. 4367659, Applied Biosystems), 2 µM of each primer, and 10 ng of DNA. Reactions mixtures were run on a 7900HT fast real-time PCR machine (Applied Biosystems) with the following thermal conditions: one cycle of 95 °C for 10 min, 40 cycles of 94 °C for 15 s, 60 °C for 30 s, followed by data collection. Data were initially analyzed with 7900HT Fast System Software at a threshold determination of 0.02 for TCGA-06-0208 tumor and blood. Threshold cycle (CT) values were exported to Excel for further statistical analysis. The average ΔCTs were calculated and compared against the BIC-seq copy ratios, to assess the accuracy of the program.
Simulation.
We used chromosome 22 (hg18) as the template to generate tumor chromosomes containing CNVs. The CNV sizes were set as 100 or 500 bp, 1, 5, 10, 50, and 100 kb. The copy number of the CNV segments were chosen as 0, 1, 3, 4, 5, 6, which corresponds to homozygous deletion, heterozygous deletion, one-, two-, three-, and four-copy gain, respectively. The positions of the CNVs were randomly selected and the only constraint was that the CNV region should not contain more than 20% Ns. In each simulation, we generated two tumor chromosomes which contained 42 (equal to 7 CNV sizes × 6 copy ratios) CNVs with different sizes and copies of CNV segments. The two chromosomes are identical except in the CNV regions. For homozygous deletion both copies of the chromosomes had the randomly selected segments deleted, but for heterozygous deletion, only one of the two simulated chromosomes had the deletion. For copy gain regions, if the copy number was even, the two simulated chromosomes would be homologous in the CNV region; but if the copy number was odd, one of the two simulated chromosomes had one more copy than the other chromosome. Duplicated segments were placed next to its original location. After the tumor chromosomes were generated, we used them as templates and applied the sequencing simulator MetaSim (48) to generate 36 bp short reads. The sequencing error model was the empirical error model of Illumina sequencing platform. We generated 600 million short reads from each pair of simulated chromosomes and approximately 57% of them can be uniquely mapped back to chromosome 22, resulting in about 30× coverage of chromosome 22. The short reads from normal genome were generated using chromosome 22 as template. For 0.3× and 3× coverage simulation, we randomly sampled 1% and 10% of the 30× simulated data, respectively. A CNV gain (loss) region is viewed as detected if at least 50% of the region is covered by CNV gain (loss) regions predicted by BIC-seq.
For the comparison of BIC-seq and BreakDancer, we generated another set of tumor chromosomes using chromosome 22 as the template. The synthetic tumor chromosomes contained 42 CNVs whose sizes and copy numbers were set as above. The simulation setup for deletions was the same as before. Duplications, on the other hand, were randomly inserted to the synthetic chromosome rather than being placed next to the original segments. After the synthetic tumor chromosomes were generated, we applied MetaSim to generate 36-bp paired-end reads assuming that the insert sizes are normally distributed with the mean of 220 bp and the SD of 20 bp. The insert size and the SD of the insert size were estimated from AML2 paired-end sequencing data. For each synthetic tumor chromosome, we simulated two sets of paired-end reads, one with 6 million paired-end reads and the other with 0.6 million. We used BWA (27) with the default parameters to map the reads back to the reference genome; the resulting sequence coverage is about 6× for the 6 million dataset and 0.6× for the 0.6 million dataset. BreakDancer was then applied to call the insertions and deletions. For the copy loss, if there is a deletion called by BreakDancer reciprocally overlapping at least 50% with the “true” CNV region, we say that the copy loss is correctly predicted. For copy gain regions, we used a lenient criterion to determine if the region is correctly predicted by BreakDancer. We say a copy gain region is correctly predicted by BreakDancer as long as there is some insertion predicted by BreakDancer that overlaps with any copy of the duplicated segment. Furthermore, sometimes BreakDancer predicts an insertion as a translocation (SI Appendix, Fig. S12), which is much larger than the original size of the insertion. These predictions were also counted as correct predictions. If we count these predictions as false positives, the performance of BreakDancer for detecting insertions larger than 500 bp would drop (SI Appendix, Fig. S13). For comparison, we also generated normal paired-end reads using chromosome 22 as a template and applied BIC-seq to call the CNVs. To make the comparison fair, BIC-seq is applied only based on half of the tumor paired-end reads. The performance of BIC-seq based on the paired-end reads drops slightly compared to its performance based on the single-end reads, but overall they are very similar (SI Appendix, Fig. S14). The common CNVs detected by both algorithms are shown in SI Appendix, Fig. S15.
Software.
The R-package BICseq can be obtained from http://compbio.med.harvard.edu/Supplements/PNAS11.html.
Supplementary Material
Acknowledgments.
This work was supported by the US National Institutes of Health Grants RC1 HG005482 and R01 GM082798 (to P.J.P.) and U24 CA144025 (to R.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.T. is a guest editor invited by the Editorial Board.
See Author Summary on page 18583.
Data deposition: The sequences reported in this paper have been deposited in the Genotypes and Phenotypes (dbGaP) database, http://www.ncbi.nlm.nih.gov/gap (accession nos. phs000178.v5.p5 and phs000159.v2.p1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110574108/-/DCSupplemental.
References
- 1.Fanciulli M, et al. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet. 2007;39:721–723. doi: 10.1038/ng2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stone J, et al. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stefansson H, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walters R, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11. Nature. 2010;463:671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bochukova E, et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463:666–670. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walter M, et al. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc Natl Acad Sci USA. 2009;106:12950–12955. doi: 10.1073/pnas.0903091106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bredel M, et al. High-resolution genome-wide mapping of genetic alterations in human glial brain tumors. Cancer Res. 2005;65:4088–4096. doi: 10.1158/0008-5472.CAN-04-4229. [DOI] [PubMed] [Google Scholar]
- 9.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bignell G, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCarroll S, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 13.Díaz de Ståhl T, et al. Profiling of copy number variations (CNVs) in healthy individuals from three ethnic groups using a human genome 32 K BAC-clone-based array. Hum Mutat. 2008;29:398–408. doi: 10.1002/humu.20659. [DOI] [PubMed] [Google Scholar]
- 14.Zogopoulos G, et al. Germ-line DNA copy number variation frequencies in a large North American population. Hum Genet. 2007;122:345–353. doi: 10.1007/s00439-007-0404-5. [DOI] [PubMed] [Google Scholar]
- 15.Shaikh T, et al. High-resolution mapping and analysis of copy number variations in the human genome: A data resource for clinical and research applications. Genome Res. 2009;19:1682–1690. doi: 10.1101/gr.083501.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itsara A, et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009;84:148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sindi S, Helman E, Bashir A, Raphael B. A geometric approach for classification and comparison of structural variants. Bioinformatics. 2009;25:i222–230. doi: 10.1093/bioinformatics/btp208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hormozdiari F, Alkan C, Eichler E, Sahinalp S. Combinatorial algorithms for structural variation detection in high-throughput sequenced genomes. Genome Res. 2009;19:1270–1278. doi: 10.1101/gr.088633.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen K, et al. BreakDancer: An algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medvedev P, Stanciu M, Brudno M. Computational methods for discovering structural variation with next-generation sequencing. Nat Methods. 2009;6:S13–S20. doi: 10.1038/nmeth.1374. [DOI] [PubMed] [Google Scholar]
- 21.Tuzun E, et al. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 22.Chiang D, et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2008;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie C, Tammi M. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics. 2009;10:80. doi: 10.1186/1471-2105-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai W, Johnson M, Kucherlapati R, Park P. Comparative analysis of algorithms for identifying amplifications and deletions in array CGH data. Bioinformatics. 2005;21:3763–3770. doi: 10.1093/bioinformatics/bti611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langmead B, Trapnell C, Pop M, Salzberg S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwarz G. Estimating the dimension of a model. Ann Stat. 1978;6:461–464. [Google Scholar]
- 29.Bayer R. Symmetric binary B-trees: Data structure and maintenance algorithms. Acta Inf. 1972;1:290–306. [Google Scholar]
- 30.Tanner M, Wong W. The calculation of posterior distributions by data augmentation. J Am Stat Assoc. 1987;82:528–540. [Google Scholar]
- 31.Gelfand A, Smith A. Sampling-based approaches to calculating marginal densities. J Am Stat Assoc. 1990;85:398–409. [Google Scholar]
- 32.Futreal P, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLendon R, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu H, et al. Oncogenic EGFR signaling cooperates with loss of tumor suppressor gene functions in gliomagenesis. Proc Natl Acad Sci USA. 2009;106:2712–2716. doi: 10.1073/pnas.0813314106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kodama T, et al. ADAM12 is selectively overexpressed in human glioblastomas and is associated with glioblastoma cell proliferation and shedding of heparin-binding epidermal growth factor. Am J Pathol. 2004;165:1743–1753. doi: 10.1016/S0002-9440(10)63429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Esteller M, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 37.Kreth S, et al. O-Methylguanine-DNA methyltransferase (MGMT) mRNA expression predicts outcome in malignant glioma independent of MGMT promoter methylation. PLoS One. 2011;6:e17156. doi: 10.1371/journal.pone.0017156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ansari K, Mandal S. Mixed lineage leukemia: Roles in gene expression, hormone signaling and mRNA processing. FEBS J. 2010;277:1790–1804. doi: 10.1111/j.1742-4658.2010.07606.x. [DOI] [PubMed] [Google Scholar]
- 39.Parsons D, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2010;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, et al. Somatic mutations of the mixed-lineage leukemia 3 (MLL3) gene in primary breast cancers. Pathol Oncol Res. 2011;17:429–433. doi: 10.1007/s12253-010-9316-0. [DOI] [PubMed] [Google Scholar]
- 41.Balakrishnan A, et al. Novel somatic and germline mutations in cancer candidate genes in glioblastoma, melanoma, and pancreatic carcinoma. Cancer Res. 2007;67:3545–3550. doi: 10.1158/0008-5472.CAN-07-0065. [DOI] [PubMed] [Google Scholar]
- 42.Kang J, Koo S, Kwon K, Park J. AMY2A: A possible tumor-suppressor gene of 1p21. 1 loss in gastric carcinoma. Int J Oncol. 2010;36:1429–1435. [PubMed] [Google Scholar]
- 43.Bueno R, et al. Second generation sequencing of the mesothelioma tumor genome. PLoS One. 2010;5:e10612. doi: 10.1371/journal.pone.0010612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ley T, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mardis E, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dicker F, et al. Mutation analysis for RUNX1, MLL-PTD, FLT3-ITD, NPM1 and NRAS in 269 patients with MDS or secondary AML. Leukemia. 2010;24:1528–1532. doi: 10.1038/leu.2010.124. [DOI] [PubMed] [Google Scholar]
- 47.Shukla A, Green J. Bowling plexopathy. N Engl J Med. 1991;324:928–928. doi: 10.1056/NEJM199103283241320. [DOI] [PubMed] [Google Scholar]
- 48.Richter D, Ott F, Auch A, Schmid R, Huson D. MetaSim: A sequencing simulator for genomics and metagenomics. PLoS One. 2008;3:e3373. doi: 10.1371/journal.pone.0003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beroukhim R, et al. Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proc Natl Acad Sci USA. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mills R, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Handsaker R, Korn J, Nemesh J, McCarroll S. Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nat Genet. 2011;43:269–276. doi: 10.1038/ng.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 53.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]