Abstract
Molecules differentially expressed in blood vessels among organs or between damaged and normal tissues, are attractive therapy targets; however, their identification within the human vasculature is challenging. Here we screened a peptide library in cancer patients to uncover ligand-receptors common or specific to certain vascular beds. Surveying ∼2.35 × 106 motifs recovered from biopsies yielded a nonrandom distribution, indicating that systemic tissue targeting is feasible. High-throughput analysis by similarity search, protein arrays, and affinity chromatography revealed four native ligand-receptors, three of which were previously unrecognized. Two are shared among multiple tissues (integrin α4/annexin A4 and cathepsin B/apolipoprotein E3) and the other two have a restricted and specific distribution in normal tissue (prohibitin/annexin A2 in white adipose tissue) or cancer (RAGE/leukocyte proteinase-3 in bone metastases). These findings provide vascular molecular markers for biotechnology and medical applications.
Keywords: human disease, phage display, obesity, angiogenesis, tumor
Despite methodological efforts to systematically map protein interactions in model organisms (1–4) including human-derived biological systems (2, 3), transfer of datasets from simple life forms to patient applications remains challenging. This issue becomes relevant in the setting of blood vessel formation and cancer metastasis, because inherent limitations of experimental models fail to recapitulate the multiple functional human ligand-receptors involved in processes such as angiogenesis, vasculogenesis, or site-specific metastasis (5–7).
To discover or analyze functional ligand-receptor interactions in blood vessels under disease conditions, we have used combinatorial screenings based on phage display, which has enabled the targeted delivery of agents to specific vascular beds (8, 9). This approach allows the selection of homing peptides to specific organs in vivo after systemic administration of random peptide libraries (10, 11). We have isolated ligand peptides and identified their tissue-specific receptors in rodents and in a patient, and have developed a ligand-receptor in prostate cancer (12, 13) that serves as the basis for an ongoing first-in-man trial.
Over the past few years we have developed a tripartite approach to enable serial combinatorial selections to humans. First, we established an ethical framework to ensure respectful research in patients who were brain-dead or whose families decided to terminate life support (14, 15). Second, we adapted techniques that were validated in rodents (16) to enable synchronous selection of ligands to multiple organs. Third, we integrated genomic tools, in which recovery of 106 peptides is ∼1,000-fold faster and ∼250-fold cheaper (17). Here, these quantitative and qualitative methods enabled refined combinatorial selections in patients and identified unique ligand-receptors.
Results
An Integrated Strategy for Combinatorial Selections in Patients.
Our approach (Fig. S1) involved serial rounds of selection in three cancer patients, which enabled the enrichment of targeting peptides for identifying ligand-receptors. After systemic delivery of a peptide library to the first human subject (12), phage were recovered from organs, pooled, and serially screened in two subsequent patients (Fig. S1, Step #1). Large-scale sequencing was performed after the third selection round.
To identify targeting peptides with tissue-specificity from this sequence dataset, we performed Monte Carlo simulation on the datasets (Fig. S1, Step #2) for each tissue (16). This approach allowed the predicted selection of targeting peptides over the random library. The simulation was followed by high-throughput analysis of peptides divided into tripeptides, which provided sufficient structure for motifs to mediate protein interactions (18). This step uses pattern-recognition software (12, 16) that analyzes the frequencies of n distinct motifs representing all possible combinations of three residue (n3) overlapping sequences in both directions (n ≪ n3). Statistical significance was assessed by one-tailed Fisher’s exact test. Once enriched 3-mers were selected, full 7-mers containing those tripeptides were identified from the original dataset, which served to isolate tissue-specific and shared ligand-receptors (Fig. S1, Step #3).
Next, three approaches were used for isolation of functional ligand-receptors (Fig. S1, Step #4): (i) protein purification and identification of candidate receptors were performed by affinity chromatography and mass spectrometry, (ii) identification of native ligands was achieved from protein arrays with antipeptide antibodies or BLAST analysis, and (iii) supervised online database searches to evaluate known ligand-receptor interactions. Biochemical validation of ligand-receptors was ultimately carried out (Fig. S1, Step #5).
Large-Scale Analysis and Combinatorial Selection.
A total of three rounds of synchronous combinatorial screening in patients produced 2,348,940 tripeptides, allowing assessment of virtually every peptide displayed in the selected tissues. For each round, we analyzed frequencies and tissue distribution of all tripeptides in each direction within the recovered full peptide sequences. The same procedure was applied to the unselected phage display library. Peptide sequences were evaluated by one-sided Fisher’s exact tests, which identified a pool of tripeptide motifs (n = 23) enriched (P < 0.05) in targeted tissues in round three relative to the unselected library (Table S1).
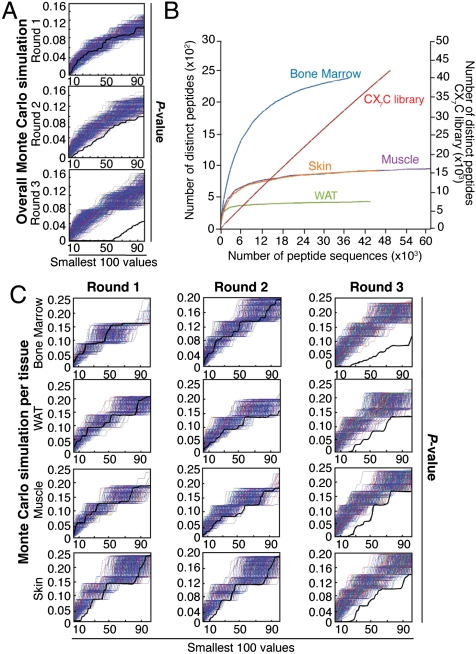
Analysis of large-scale derived sequences established a nonrandom distribution of tripeptides among tissues (Fig. 1A) with specific saturation curves (Fig. 1B), and revealed ∼78% agreement between the DNA sequencing methods (Table S1) used (17). Monte Carlo simulations confirmed a progressive accumulation of enriched motifs from the first to the third round (Fig. 1A), in a tissue-specific manner (Fig. 1C); such simulations also revealed that the analytical design used had a > 95% probability of detecting enriched motifs (P < 0.05). Saturation plots of high-throughput DNA pyrosequencing performed in the third round confirmed the identification of most peptides present within the selected tissues (Fig. 1B). Moreover, we observed that each tissue had a reproducible ligand saturation curve, a result suggestive of intrinsic tissue diversity among vascular receptor pools. In contrast, we could not reach ligand saturation with the unselected library, an indication of true randomness (Fig. 1B).
Fig. 1.
Combinatorial selection in patients. (A) Monte Carlo simulations with peptides analyzed in serial rounds of selection show nonrandom distribution of tripeptides. Thick black lines represent the Fisher’s exact test; thin blue lines represent the corresponding random permutated dataset. (B) A saturation plot (modified from ref. 17) shows the number of distinct peptides as a function of the total number of peptide sequences. All tissues reached saturation, as indicated by flattening of the slope; in contrast, unselected library showed no evidence of saturation. (C) Isolated peptides were grouped according to tissue-of-origin and subjected to Monte Carlo simulation. For every simulation, the pool of peptides was randomly distributed into groups corresponding to the number of sequences analyzed for each targeted tissue. Frequencies were calculated for each simulated organ, and Fisher’s exact test applied on the permutated dataset. A nonrandom selection of tripeptides was observed in all organs tested.
By comparing tripeptide frequencies in targeted vs. nontargeted tissues, we showed that these motifs were not evenly distributed and some were enriched with tissue-specificity (Table S1, underlined motifs). Of note, 9 of 23 motifs (39%) recovered from white adipose tissue (WAT) were found only in WAT, 14 of 17 bone marrow-homing motifs (82%) were found only in bone marrow, and all tripeptides selected in skin and muscle were unique to the tissue-of-origin. Full peptide sequences with apparent tissue-specificity (for tumor-containing bone marrow, skin, WAT, and muscle) were obtained from selected tripeptides. We also observed progressive enrichment of ligand peptides common to all tissues examined, a result consistent with selection of an ubiquitous ligand or one shared in the several targeted vasculatures; peptide-based affinity chromatography and mass spectrometry served to uncover candidate receptors for each selected peptide in the human vasculature (Table S2). Additionally, protein array screenings with polyclonal antibodies against the targeting peptides and similarity data mining guided the identification of native ligands. We reasoned that some ligand peptides likely mimic circulating proteins that bind to receptors exposed on vascular endothelial cells. Thus, we refined our similarity searches according to protein structure, glycosylation state, subcellular location, and membrane orientation (www.hprd.org). As a proof-of-concept (i.e., specific binding), we present examples of ligand-receptors in shared (n = 2) and tissue-specific (n = 2; WAT and bone marrow) settings selected from human blood vessels of normal and tumor-containing tissues. The validation of these four candidate ligand-receptors suggests a reasonable success rate for this approach; the remaining eleven candidate ligand-receptors were not pursued here.
Native Ligands and Shared Receptors in Human Endothelial Cells.
To identify shared molecular targets in human blood vessels, we analyzed peptides recovered from biopsies of multiple tissues. Statistical analysis revealed that the selected peptides CMRGFRAAC and CMGGHGWGC (Table S2) contained multiple tripeptides showing the highest frequencies of recovery in all sampled tissues by round three and were therefore chosen as representative ligands for further analysis.
We used peptide-based affinity chromatography to purify interacting molecules for CMRGFRAAC. Total protein extract from human tissue was loaded onto a peptide column, and interacting proteins were eluted with a solution of the synthetic peptide. Phage binding assays in vitro were used to identify eluted fractions containing the highest concentration of receptors. Proteins in fractions of interest were separated by SDS-polyacrylamide gel electrophoresis (PAGE), and protein bands were identified by mass spectrometry and database search. As a result, human annexin A4 (ANXA4) was isolated as a candidate receptor for CMRGFRAAC (Fig. S2A). Annexins are a highly conserved family of membrane-binding proteins with intracellular and extracellular functions (19, 20); ANXA4 function in angiogenesis is currently unknown.
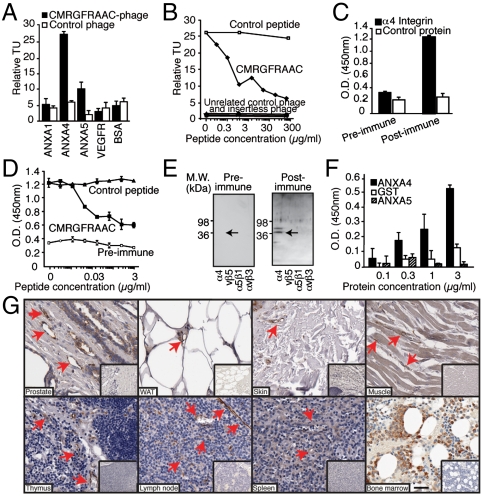
A series of in vitro phage- and protein-based assays was designed to determine whether ANXA4 is a receptor for CMRGFRAAC (Fig. 2). We assessed specificity by evaluating the binding of CMRGFRAAC-displaying phage or insertless phage to immobilized ANXA4 or controls. We observed binding of CMRGFRAAC-displaying phage to ANXA4, compared to the family related control proteins ANXA1 and ANXA5. The unrelated proteins vascular endothelial growth factor receptor (VEGFR) and bovine serum albumin (BSA) were additional controls, binding at background levels (Fig. 2A). Competition assays in the presence of the targeted synthetic peptide or controls confirmed binding specificity (Fig. 2B).
Fig. 2.
Discovery of integrin α4 subunit/ANXA4 as a shared ligand-receptor in the vasculature of multiple human tissues. (A) Binding of phage clones to the receptor ANXA4. Phage displaying the peptide CMRGFRAAC bound preferentially to its receptor, relative to negative controls. Experiments were performed three times in triplicate with similar results. Bars represent mean ± standard error of the mean (SEM). (B) Competition with the synthetic peptide shows that binding of selected phage to the purified receptor is specific. Binding of unrelated control phage, insertless phage, binding to BSA and inhibition with an unrelated peptide served as controls. (C) ELISA with preimmune and postimmune rabbit polyclonal antibodies against CMRGFRAAC and performed on recombinant integrin α4 or a control (shown is α5β1 integrin). (D) Binding of postimmune antibodies to recombinant integrin α4 is inhibited by CMRGFRAAC but not by an unrelated control peptide. (E) Binding specificity was confirmed by immunoblotting. Integrins αvβ5, α5β1, and αvβ3 were used as negative controls. Arrow points to integrin α4. (F) Direct interaction between ANXA4 and integrin α4. The binding is concentration-dependent, indicating specificity. (G) Immunostaining of sections of normal human tissues with an anti-ANXA4 polyclonal antibody. Arrows point to ANXA4-positive blood vessels. Insets show negative control staining. (Scale bar, 100 μm).
Having isolated ANXA4 as a potential shared human vascular marker, we next screened a representative panel of normal human tissues (n = 38) in tissue microarray (TMA) format as a broader survey of the expression and location of ANXA4 (Table S3). We found ANXA4-positive staining in the vasculature of 50% of tissues examined (Fig. 2G, Table S3), including the more intensively analyzed WAT, skin, muscle, and bone marrow. Other ANXA4-positive sites included the vascular endothelium of human thymus, lymph nodes, spleen, heart, stomach, colon, and various areas of the brain. Overall, expression of ANXA4 did not correlate with the embryologic origin of tissues/organs and in most cases was not restricted to endothelial cells.
Next, we sought native ligands for this vascular receptor. Similarity searches and data mining suggested that CMRGFRAAC is a mimic of the integrin α4 subunit (Table S2, residues 147–152), a cell adhesion molecule with a well established role in lymphocyte and monocyte transendothelial migration (21). Enzyme-linked immunosorbent assays (ELISA) in the presence or absence of the synthetic peptide (Fig. 2 C and D), and immunoblotting (Fig. 2E) with an antibody against a keyhole limpet hemocyanin (KLH)-conjugated CMRGFRAAC, confirmed integrin α4 subunit as a native ligand. Negative controls included integrins αvβ5, α5β1, and αvβ3. Protein interaction experiments showed that binding of soluble ANXA4 to immobilized α4 integrin is concentration-dependent (Fig. 2F). Evaluation of integrin α4 subunit expression in a representative panel of normal tissue samples (Fig. S2B) showed expression mostly restricted to the surface of leukocytes, suggesting a role for the ANXA4/integrin α4 in adhesion of leukocytes to the vascular endothelium.
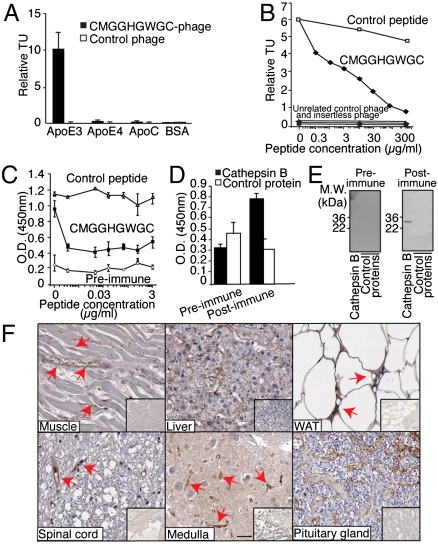
By a similar approach, we identified apolipoprotein E3 (ApoE3) as a potential native receptor for the ligand CMGGHGWGC (Fig. S2C). ApoE is a lipoprotein constituent and functions in lipid metabolism; it has a major role in cholesterol efflux and has been linked to atherosclerosis (22). However, no direct link has been described between ApoE3 and blood vessels formation in normal or pathological conditions. The relationship between CMGGHGWGC and ApoE3 was validated by in vitro assays, and binding specificity confirmed via phage binding to ApoE3 or controls (ApoE4 and ApoC; Fig. 3A), plus inhibition with the cognate synthetic peptide (Fig. 3B). Moreover, screening of normal human TMA revealed ApoE3-positive staining in ∼40% of examined organs, including muscle, liver, WAT, and distinct areas of the brain (Fig. 3F, Table S3). Similar to ANXA4, expression of ApoE3 in normal organs did not correlate with the embryologic origin of each tissue or organ, and it was not restricted to endothelial cells. Protein similarity search identified CMGGHGWGC as a mimic of cathepsin B. This finding is of interest as cathepsin B participates in the release of atherosclerotic plaque in animal models (23), supporting the hypothesis that ApoE3 and cathepsin B interact in vivo. Notably, structural analysis located the CMGGHGWGC peptide within the active catalytic site of cathepsin B (Fig. S3A). ELISA (Fig. 3D), inhibition assay (Fig. 3C), and immunoblotting with a polyclonal antibody against KLH-conjugated CMGGHGWGC (Fig. 3E), support the candidacy of cathepsin B as a ligand mimicked by CMGGHGWGC. Finally, evaluation of cathepsin B expression in a panel of normal tissue samples (Fig. S3B) showed widespread expression of the protein in different cellular compartments, including the blood vessels of WAT and muscle.
Fig. 3.
Discovery of cathepsin B/ApoE3 as a shared ligand-receptor in the vasculature of multiple human tissues. (A) Binding of CMGGHGWGC-phage to ApoE3. CMGGHGWGC- phage bound preferentially to its receptor relative to negative controls. Experiments were performed three times in triplicate with similar results. Bars represent mean ± SEM (B) Competition assay with the cognate synthetic peptide shows that binding of CMGGHGWGC-phage to the purified ApoE3 is specific. Binding of unrelated control phage, insertless phage, binding to BSA and inhibition with an unrelated peptide served as controls. (C and D) ELISA with preimmune and postimmune polyclonal antibodies against CMGGHGWGC (C) and performed on purified cathepsin B or control protein (D). (E) Binding specificity was confirmed by immunoblotting. (F) Immunostaining of human sections with an anti-ApoE3 antibody confirms that the candidate receptor ApoE3 is expressed in the normal vasculature of several human tissues (arrows). (Scale bar, 100 μm).
Selection and Validation of a Ligand-Receptor System in Human WAT.
In previous work, we have found that the peptide CKGGRAKDC was localized to and internalized by cells of mouse WAT vasculature, and that its native vascular target was prohibitin, a multifunctional protein expressed selectively on WAT endothelium (8).
Given that mouse and human prohibitin differ by only a single residue, we asked whether this protein might also serve as a target for WAT-homing peptides in patients, and evaluated this possibility here by analysis of the binding of a large pool of peptide-targeted phage clones (n = 851) isolated after three rounds of selection in human WAT to bind to immobilized prohibitin. A subset of nonredundant peptides (n = 66) that showed specific binding to prohibitin was subjected to individual database searches for similarity to human proteins, leading to identification of a smaller peptide subset (n = 14) similar to the prohibitin-binding sequence CKGGRAKDC that matched a region of human ANXA2 (Fig. S3C). Reciprocal analysis revealed a wide range of sequences among all human WAT-selected peptides that mimicked prohibitin and that were clustered within the prohibitin N-terminal segment (Fig. S3C), a region involved in lipid raft location and interaction with other proteins such as ANXA2 (24). To map the ANXA2 domains involved in the binding to prohibitin, we performed automated alignment of human WAT-homing peptides against ANXA2 and identified two similarity hotspots in the N-terminal domain of the protein (Fig. S3D). Strikingly, these segments correspond to the connector loops between ANXA2 repeats 1/2 and 2/3, which interact with membrane-bound proteins (24).
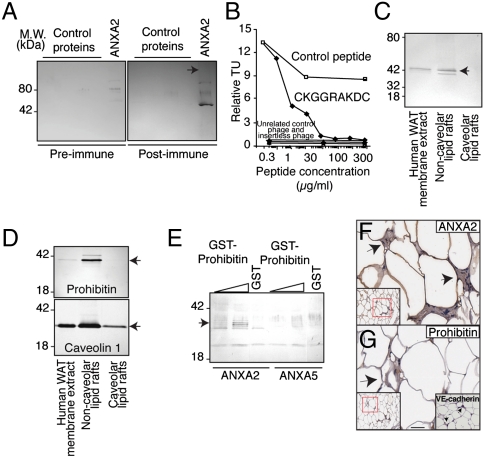
To confirm that CKGGRAKDC mimics the candidate ligand ANXA2 and binds to prohibitin, we showed that antibodies against KLH-conjugated CKGGRAKDC recognize recombinant ANXA2 (Fig. 4A). Phage-CKGGRAKDC also bound to prohibitin in vitro, an interaction specifically inhibited by the synthetic peptide (Fig. 4B). Next, membrane fractions extracted from WAT (Fig. 4C) showed that ANXA2 and prohibitin are located in noncaveolar lipid rafts (Fig. 4D). Lastly, we used recombinant glutathione S-transferase (GST)-conjugated protein to show that prohibitin binds to ANXA2, but not to the control ANXA5 (Fig. 4E).
Fig. 4.
Discovery of ANXA2/prohibitin as a tissue-specific ligand-receptor targeting normal human tissue. (A) Immunoblotting of His6-ANXA2 or control proteins with antiserum against CKGGRAKDC or control preimmune serum, as indicated. Arrow: His6-ANXA2. (B) Binding of CKGGRAKDC-displaying phage is specifically inhibited by the synthetic peptide. Binding of unrelated control phage, insertless phage, binding to BSA and inhibition with an unrelated peptide served as controls. (C and D) Association of prohibitin and ANXA2 with membrane lipid rafts. Membrane proteins extracted from human WAT were subjected to immunoblotting or to fractionation enriching for noncaveolar or caveolar lipid rafts. Proteins recognized by anti-ANXA2 (C), antiprohibitin (D, upper box), and anticaveolin 1 antibodies (D, lower box) are indicated by arrows. (E) Binding of prohibitin and ANXA2 in vitro. Increasing concentrations of GST-prohibitin or GST control were captured with His6-ANXA2 or control His6-ANXA5. Specific binding was assessed with anti-GST antibodies. Arrow indicates GST-prohibitin (migrating as several bands). (F and G) Vascular expression of ANXA2 in human WAT. Immunohistochemistry with anti-ANXA2 and antiprohibitin antibodies on human WAT demonstrated colocalization of ANXA2 and prohibitin in the vasculature. Blood vessels identity was confirmed by staining with anti-VE-cadherin antibody (G, inset). Arrows point to blood vessels. Red insets show lower magnification of the corresponding area. (Scale bar, 100 μm).
Expression of ANXA2 on the surface of endothelial cells has been reported (25), but without organ comparisons. The expression pattern of prohibitin and ANXA2 in human tissue samples appeared coincident in human WAT but not in non-WAT (Fig. 4 F and G, Fig. S3E). We conclude that the vascular coexpression and interaction between ANXA2 and prohibitin are likely restricted to WAT.
Discovery of a Specific Ligand-Receptor in Tumor-Containing Human Bone Marrow.
Human bone marrow is often affected by primary hematologic tumors or metastatic solid tumors. In all three patients selected in this study, bone marrows were replete with tumor cells. Within the bone marrow microenvironment, we assumed that the molecular crosstalk between nonmalignant cells of the vascular endothelium and cancer cells might have at least some common elements, independent of the tumor type.
Statistical analysis revealed tripeptides enriched in bone marrow after three rounds of selection (Table S1). In particular, selected peptides containing the motif Gly-Gly-Gly-Pro were identified within RAGE, the receptor for advanced glycation end-products (Table S2). A computer-assisted modeling of RAGE and three 7-mer peptides containing an embedded Gly-Gly-Gly-Pro motif (CWELGGGPC, CHVLGGGPC, and CVQGGGGPC) showed high similarity to an exposed surface of the ligand-binding extracellular domain of the protein (Fig. S4A).
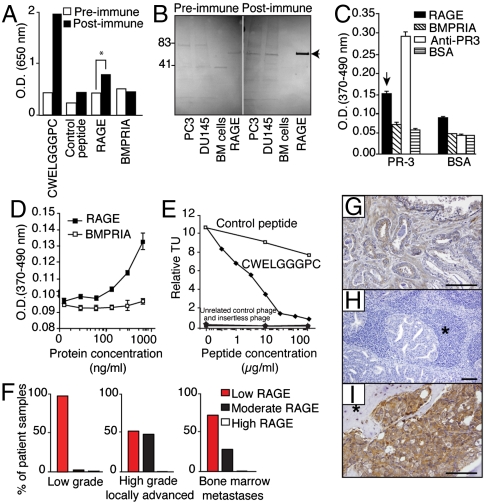
To query whether CWELGGGPC behaves as a molecular mimic of RAGE, we developed a polyclonal antibody against KLH-conjugated CWELGGGPC and used ELISA to evaluate binding to immobilized RAGE. We showed that the anti-CWELGGGPC antibody recognizes the segment of RAGE containing CWKLGGGPC (Fig. 5A), whereas preimmune serum produces only a background signal. Immunoblotting with the anti-CWKLGGGPC antibody confirmed reactivity with the native protein extracted from human prostate cancer cells (Fig. 5B, arrow).
Fig. 5.
Discovery of RAGE/PR-3 as a ligand-binding targeting human bone marrow containing cancer cells. (A) Anti-CWELGGGPC antibodies recognize human recombinant RAGE. ELISA with post- and preimmune polyclonal antibodies against CWELGGGPC was performed on immobilized CWELGGGPC, a control peptide, recombinant Fc-tagged proteins, and a control protein. (B) Anti-CWELGGGPC antibodies recognize native human RAGE. Protein extracts from human prostate cancer cell lines PC3 and DU145, or from human bone marrow mononuclear control cells, along with recombinant RAGE protein, were immunoblotted with post- and preimmune polyclonal antibodies against CWELGGGPC. Arrow points to RAGE. (C) Validation of RAGE binding to PR-3 in vitro. Either immobilized PR-3 or control protein (BSA) was subjected to RAGE, BMPRIA, BSA, and anti-PR-3 antibody. Bars represent mean ± SEM (D) RAGE binding to active PR-3 is concentration-dependent. (E) Binding of CWELGGGPC-phage is specifically inhibited by the synthetic peptide. Binding of unrelated control phage, insertless phage, binding to BSA and inhibition with an unrelated peptide served as controls. (F) Relative quantification of RAGE expression on prostate cancer patient samples. Expression of RAGE is represented as low, moderate and high expression according to a standardized pathology score. (G–I) Immunohistochemistry with RAGE-specific antibodies performed on human tissue sections derived from a panel of prostate cancer patients. (G) Organ-confined prostate cancer; (H), lymph node metastasis; and (I), bone marrow metastasis. Asterisks represent lymphoid (H) and bone (I) tissues. (Scale bar, 100 μm).
Next, we identified the human leukocyte proteinase-3 (PR-3) by peptide affinity chromatography and mass spectrometry as a candidate receptor for CWKLGGGPC (Fig. S4B). This result was confirmed through a second affinity purification with human bone marrow cell membrane extracts as the protein source (Fig. S4C). While PR-3 is a serine protease abundant within the bone marrow of patients with chronic myelogenous leukemia (26), it has not yet been implicated in bone metastases. In support of the hypothesis that PR-3 interacts with RAGE, protein analysis demonstrated that PR-3 does share epitopes with other established RAGE ligands (27), such as the advanced glycosylation end-products (AGE), high mobility group protein B1 (HMGB1), and S100 calcium-binding protein A12 (EN-RAGE) (Fig. S4D). Indeed, 7 of 15 HMGB1 residues (47%) and 13 of 21 EN-RAGE residues (60%) critical for RAGE binding (28) were identical or conserved within PR-3. Moreover, the C-terminal α-helix of PR-3 is highly similar to the corresponding part of EN-RAGE (Fig. S4D), revealing a previously unrecognized structural relationship between PR-3 and RAGE partners.
For functional characterization of this ligand-receptor, we performed binding experiments in vitro between the extracellular portion of human RAGE and endogenous PR-3. RAGE—but not control proteins—bound to immobilized PR-3 (Fig. 5C).
We showed that binding between PR-3 and RAGE is concentration-dependent (Fig. 5D) and that competition assays with targeted phage and the synthetic peptide (Fig. 5E) support the specificity of this interaction. These results indicate that the selected human bone marrow-targeting motif mimics a functional site within the extracellular domain of RAGE and that RAGE binds to PR-3 through its WKLGGGP-spanning region. Notably, elevation of RAGE mRNA transcripts has been reported in human prostate cancer (29), which can metastasize to the bone marrow, and the expression of RAGE in prostate cancer patients has been documented at the protein level (www.proteinatlas.org). We assessed RAGE expression in a large annotated panel of human tumor samples (n = 164) from prostate cancer patients (Fig. 5F), including low-grade (n = 76) and high-grade locally advanced primary tumors (n = 76), and prostate cancer-infiltrated bone marrow biopsy samples (n = 12). We applied a linear regression model to assess biomarker expression and distribution among the groups. Significant differences in RAGE expression were observed between low-grade (Fig. 5F, left box) and high-grade (Fig. 5F, middle box) tumors (t-test, P < 0.0001). Moreover, expression of RAGE was significantly higher in bone marrow-infiltrated metastases compared to low-grade tumors (Fig. 5F, right and left boxes; t-test, P = 0.0002 for the black bars). We did not observe statistically significant differences between high-grade primary tumors and bone marrow metastases (t-test, P = 0.61). Finally, we detailed RAGE expression immunohistochemically in human prostate cancer, in a representative patient sample set (n = 12) including primary tumors (Fig. 5G), lymph node metastases (Fig. 5H), and bone marrow metastases (Fig. 5I). While RAGE expression was barely detectable in normal prostate glands (Fig. 5G), it was strongly expressed and widespread in tumor cells within the marrow cavity of all cases of prostate cancer patients with bone metastatic disease (Fig. 5I), but not within lymph node metastases (Fig. 5H).
These results are consistent with the hypothesis that RAGE-expressing tumor cells can arise focally in primary tumors and might provide at least part of an advantageous setting for bone marrow metastases.
Discussion
We have initiated the creation of a ligand/receptor-based molecular map of human blood vessels towards a targeted vascular pharmacology. Our first patient screening (12, 13) served as a foundation for an ongoing clinical trial of a ligand-directed drug, and we have improved quantitative and qualitative methodology (16, 17) in an ethics framework that includes cancer center-specific guidelines (14) and national recommendations (15) to harmonize this line of patient-oriented research with current practices of transplantation medicine. Our efforts may lead to an improved understanding of vascular proteomics with clinical implications.
Four ligand-receptors were validated functionally in shared or tissue-specific settings. Two shared human vascular addresses were found by our selection. First, ANXA4, a membrane protein identified in Ca+2-dependent membrane trafficking (30), was characterized as a marker of the human vasculature that interacts with α4 integrins. Among the α4 integrin family, α4β1 (31) and/or α4β7 (32) are membrane constituents of leukocytes that mediate cell adhesion to the vascular endothelium and affect inflammation, immune response, and tumor dissemination (31). Vascular cell adhesion molecule-1 and fibronectin are among the molecules that mediate the binding of α4β1-expressing lymphocytes to endothelial and dendritic cells (31). Unlike α4β1, the integrin α4β7 is active mainly in intestinal immune response, mediated through interaction with mucosal cell adhesion molecule-1 (32). Whether the association between ANXA4 in human vascular endothelium and heterodimers of α4βx integrin on the circulating cells occurs in widespread (α4β1) or localized (α4β7) patterns remains to be determined.
Cathepsin B and ApoE3 were also validated as an unrecognized ligand-receptor shared in several tissues. ApoE is a secreted protein that acts through the low-density lipoprotein receptor to mediate lipoprotein binding and catabolism (33). Although ApoE has not been generally considered a conventional cell surface receptor, our studies revealed a presence of ApoE3 on the luminal surface of blood vessels in normal human tissues. As documented for other secreted proteins, binding of ApoE3 to its conventional receptors or to the extracellular matrix surrounding the vascular endothelium plausibly explains the isolation of ApoE3 as a receptor for CMGGHGWGC.
We also uncovered two tissue-specific vascular targeting systems. ANXA2 and prohibitin were found as a ligand-receptor in human WAT vasculature. Studies with yeast two-hybrid methodology confirmed ANXA2 and prohibitin as interacting components in lipid rafts (24, 34). Given the weight loss in obese rodents by the targeting of prohibitin in the vasculature with an apoptotic agent (8, 35), we predict that the selective mapping of this protein in human WAT will have translational value.
Lastly, we report RAGE and PR-3 as molecular partners in human tumor-containing bone marrow resulting from tumorigenesis or metastasis. RAGE and PR-3 appeared unexpectedly as a molecular complex, mediating the homing of human metastatic prostate cancer cells to the bone marrow. These proteins have, until now, been considered to be active in unrelated pathways and therefore, functionally distinct. PR-3 is secreted by activated bone marrow-derived polymorphonuclear leukocytes (26, 27) and deposited on the surface of endothelial cells in inflammation (36). Given our additional data, PR-3 appears functionally relevant to bone marrow-specific tumorigenesis and metastasis. Finally, one should note that established vascular markers such as aminopeptidase N and cadherin-5 have also been affinity-purified (Table S2) further supporting this screening strategy.
In summary, it is clear that a large-scale analysis of protein interactions in blood vessels of healthy and diseased organs can uncover many as yet unidentified or unique molecular networks in the human vasculature. As such, generation and annotation of a comprehensive molecular map based on accessible ligand-receptors in blood vessels may be used as a starting point for functional discovery and elucidation of protein networks in the human vasculature.
Materials and Methods
The series of primary and secondary antibody reagents are detailed in the SI Text. Selection of the three patients and their study adhered strictly to institutional guidelines specified in the SI Text, where administration of the phage display library, sample collection, postbiopsy processing of human tissue samples, statistical methods, chemical analytical methods, and immunostaining methods are also fully described.
Supplementary Material
Acknowledgments.
We thank Drs. Ricardo Brentani, Webster Cavenee, Roy Lobb, and Helene Sage for manuscript reading and David Bier, Pauline Dieringer, Cherie Perez, and Dallas Williams for infrastructure. This work was supported by grants from the National Institutes of Health, National Cancer Institute, Department of Defense, and by awards from AngelWorks, the Gillson-Longenbaugh Foundation, and the Marcus Foundation.
Footnotes
Conflict of interest statement: The University of Texas M.D. Anderson Cancer Center and some of its researchers (W.A. and R.P.) have equity positions in and are paid consultants for Alvos Therapeutics and Ablaris Therapeutics, which are subjected to certain restrictions under university policy; the university manages and monitors the terms of these arrangements in accordance with its conflict-of-interest policy.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114503108/-/DCSupplemental.
References
- 1.Barrios-Rodiles M, et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- 2.Stelzl U, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 3.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 4.Breitkreutz A, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zetter BR. The cellular basis of site-specific tumor metastasis. N Engl J Med. 1990;322:605–612. doi: 10.1056/NEJM199003013220907. [DOI] [PubMed] [Google Scholar]
- 6.Risau W, Flamme I. Vasculogenesis. Annu Rev Cell Dev Biol. 1995;11:73–91. doi: 10.1146/annurev.cb.11.110195.000445. [DOI] [PubMed] [Google Scholar]
- 7.Folkman J. Angiogenesis: an organizing principle for drug discovery? Nat Rev Drug Discov. 2007;6:273–286. doi: 10.1038/nrd2115. [DOI] [PubMed] [Google Scholar]
- 8.Kolonin MG, Saha PK, Chan L, Pasqualini R, Arap W. Reversal of obesity by targeted ablation of adipose tissue. Nat Med. 2004;10:625–632. doi: 10.1038/nm1048. [DOI] [PubMed] [Google Scholar]
- 9.Staquicini FI, Pasqualini R, Arap W. Ligand-directed profiling: applications to target drug discovery in cancer. Expert Opin Drug Dis. 2009;4:51–59. doi: 10.1517/17460440802628152. [DOI] [PubMed] [Google Scholar]
- 10.Pasqualini R, Ruoslahti E. Organ targeting in vivo using phage display peptide libraries. Nature. 1996;380:364–366. doi: 10.1038/380364a0. [DOI] [PubMed] [Google Scholar]
- 11.Smith GP, Scott JK. Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 1993;217:228–257. doi: 10.1016/0076-6879(93)17065-d. [DOI] [PubMed] [Google Scholar]
- 12.Arap W, et al. Steps toward mapping the human vasculature by phage display. Nat Med. 2002;8:121–127. doi: 10.1038/nm0202-121. [DOI] [PubMed] [Google Scholar]
- 13.Zurita AJ, et al. Combinatorial screenings in patients: the interleukin-11 receptor alpha as a candidate target in the progression of human prostate cancer. Cancer Res. 2004;64:435–439. doi: 10.1158/0008-5472.can-03-2675. [DOI] [PubMed] [Google Scholar]
- 14.Pentz RD, Flamm AL, Pasqualini R, Logothetis CJ, Arap W. Revisiting technical guidelines for research with terminal wean and brain-dead patients. Hastings Cent Rep. 2003;33:20–26. [PubMed] [Google Scholar]
- 15.Pentz RD, et al. Ethics guidelines for research with the recently dead. Nat Med. 2005;11:1145–1149. doi: 10.1038/nm1105-1145. [DOI] [PubMed] [Google Scholar]
- 16.Kolonin MG, et al. Synchronous selection of homing peptides for multiple tissues by in vivo phage display. FASEB J. 2006;20:979–981. doi: 10.1096/fj.05-5186fje. [DOI] [PubMed] [Google Scholar]
- 17.Dias-Neto E, et al. Next-generation phage display: integrating and comparing available molecular tools to enable cost-effective high-throughput analysis. PLoS One. 2009;4:1–11. doi: 10.1371/journal.pone.0008338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vendruscolo M, Paci E, Dobson CM, Karplus M. Three key residues form a critical contact network in a protein folding transition state. Nature. 2001;409:641–645. doi: 10.1038/35054591. [DOI] [PubMed] [Google Scholar]
- 19.Swairjo MA, Seaton BA. Annexin structure and membrane interactions: a molecular perspective. Annu Rev Biophys Biomol Struct. 1994;23:193–213. doi: 10.1146/annurev.bb.23.060194.001205. [DOI] [PubMed] [Google Scholar]
- 20.Burgoyne RD, Geisow MJ. The annexin family of calcium-binding proteins. Review article. Cell Calcium. 1989;10:1–10. doi: 10.1016/0143-4160(89)90038-9. [DOI] [PubMed] [Google Scholar]
- 21.Rose DM, Alon R, Ginsberg MH. Integrin modulation and signaling in leukocyte adhesion and migration. Immunol Rev. 2007;218:126–134. doi: 10.1111/j.1600-065X.2007.00536.x. [DOI] [PubMed] [Google Scholar]
- 22.van Vlijmen BJ, et al. Diet-induced hyperlipoproteinemia and atherosclerosis in apolipoprotein E3-Leiden transgenic mice. J Clin Invest. 1994;93:1403–1410. doi: 10.1172/JCI117117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lutgens SPM, Cleutjens KB, Daemen MJ, Heeneman S. Cathepsin cysteine proteases in cardiovascular disease. FASEB J. 2007;21:3029–3041. doi: 10.1096/fj.06-7924com. [DOI] [PubMed] [Google Scholar]
- 24.Liu J, Deyoung SM, Zhang M, Dold LH, Saltiel AR. The stomatin/prohibitin/flotillin/HflK/C domain of flotillin-1 contains distinct sequences that direct plasma membrane localization and protein interactions in 3T3-L1 adipocytes. J Biol Chem. 2005;280:16125–16134. doi: 10.1074/jbc.M500940200. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, McCrae KR. Annexin A2 mediates endothelial cell activation by antiphospholipid/anti-beta2 glycoprotein I antibodies. Blood. 2005;105:1964–1969. doi: 10.1182/blood-2004-05-1708. [DOI] [PubMed] [Google Scholar]
- 26.Molldrem JJ, et al. Evidence that specific T lymphocytes may participate in the elimination of chronic myelogenous leukemia. Nat Med. 2000;6:1018–1023. doi: 10.1038/79526. [DOI] [PubMed] [Google Scholar]
- 27.Campanelli D, et al. Cloning of cDNA for proteinase 3: a serine protease, antibiotic, and autoantigen from human neutrophils. J Exp Med. 1990;172:1709–1715. doi: 10.1084/jem.172.6.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huttunen HJ, Fages C, Kuja-Panula J, Ridley AJ, Rauvala H. Receptor for advanced glycation end products-binding COOH-terminal motif of amphoterin inhibits invasive migration and metastasis. Cancer Res. 2002;62:4805–4811. [PubMed] [Google Scholar]
- 29.Ishiguro H, et al. Receptor for advanced glycation end products (RAGE) and its ligand, amphoterin are overexpressed and associated with prostate cancer development. Prostate. 2005;64:92–100. doi: 10.1002/pros.20219. [DOI] [PubMed] [Google Scholar]
- 30.Jeon YJ, et al. Annexin A4 interacts with NF-kappaB p50 subunit and modulates NF-kappaB transcriptional activity in a Ca+-dependent manner. Cell Mol Life Sci. 2010;67:2271–2281. doi: 10.1007/s00018-010-0331-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lobb RR, Hemler ME. The pathophysiologic role of α4 integrins in vivo. J Clin Invest. 1994;94:1722–1728. doi: 10.1172/JCI117519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorfu G, Rivera-Nieves J, Ley K. Role of β7 integrins in intestinal lymphocyte homing and retention. Curr Mol Med. 2009;9:836–850. doi: 10.2174/156652409789105525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hatters DM, Peters-Libeu CA, Weisgraber KH. Apolipoprotein E structure: insights into function. Trends Biochem Sci. 2006;31:445–454. doi: 10.1016/j.tibs.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 34.Bacher S, Achatz G, Schmitz ML, Lamers MC. Prohibitin and prohibitone are contained in high-molecular weight complexes and interact with alpha-actinin and annexin A2. Biochimie. 2002;84:1207–1220. doi: 10.1016/s0300-9084(02)00027-5. [DOI] [PubMed] [Google Scholar]
- 35.Kim DH, Woods SC, Seeley RJ. Peptide designed to elicit apoptosis in adipose tissue endothelium reduces food intake and body weight. Diabetes. 2010;59:907–915. doi: 10.2337/db09-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uehara A, Sugawara Y, Sasano T, Takada H, Sugawara S. Proinflammatory cytokines induce proteinase 3 as membrane-bound and secretory forms in human oral epithelial cells and antibodies to proteinase 3 activate the cells through protease-activated receptor-2. J Immunol. 2004;173:4179–4189. doi: 10.4049/jimmunol.173.6.4179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.