Abstract
Arrestins are regulatory molecules for G-protein coupled receptor function. In visual rhodopsin, selective binding of arrestin to the cytoplasmic side of light-activated, phosphorylated rhodopsin (P-Rh*) terminates signaling via the G-protein transducin. While the “phosphate-sensor” of arrestin for the recognition of receptor-attached phosphates is identified, the molecular mechanism of arrestin binding and the involvement of receptor conformations in this process are still largely hypothetic. Here we used fluorescence pump-probe and time-resolved fluorescence depolarization measurements to investigate the kinetics of arrestin conformational changes and the corresponding nanosecond dynamical changes at the receptor surface. We show that at least two sequential conformational changes of arrestin occur upon interaction with P-Rh*, thus providing a kinetic proof for the suggested multistep nature of arrestin binding. At the cytoplasmic surface of P-Rh*, the structural dynamics of the amphipathic helix 8 (H8), connecting transmembrane helix 7 and the phosphorylated C-terminal tail, depends on the arrestin interaction state. We find that a high mobility of H8 is required in the low-affinity (prebinding) but not in the high-affinity binding state. High-affinity arrestin binding is inhibited when a bulky, inflexible group is bound to H8, indicating close interaction. We further show that this close steric interaction of H8 with arrestin is mandatory for the transition from prebinding to high-affinity binding; i.e., for arrestin activation. This finding implies a regulatory role for H8 in activation of visual arrestin, which shows high selectivity to P-Rh* in contrast to the broad receptor specificity displayed by the two nonvisual arrestins.
Keywords: membrane receptor, protein conformational change, binding kinetics
G-protein coupled receptors (GPCRs) are central for the function of biological systems as they transmit various external stimuli, including hormones, neurotransmitters, and photons, to intracellular signaling cascades (1). Termination of GPCR signaling is mediated by arrestin molecules. Two arrestin forms (arrestin-2 and arrestin-3) with broad receptor specificity were found, while visual arrestin (arrestin-1), which binds to the photoreceptor rhodopsin in rod outer segments, displays high selectivity to its active phosphorylated receptor (2). Nonvisual arrestins serve in their receptor-bound form as interaction platforms for further molecules involved in GPCR internalization and G-protein-independent signaling (3, 4). Visual arrestin (Fig. 1A) binds to the cytosolic face of phosphorylated active metarhodopsin-II (Meta-II), thereby hindering G-protein binding and in this way quenching rhodopsin activity (5–7). Phosphorylation of serine and/or threonine residues at rhodopsin’s C-terminal tail (Fig. 1 A–C) by rhodopsin kinase (8) is a prerequisite for arrestin binding (6, 9).
Fig. 1.
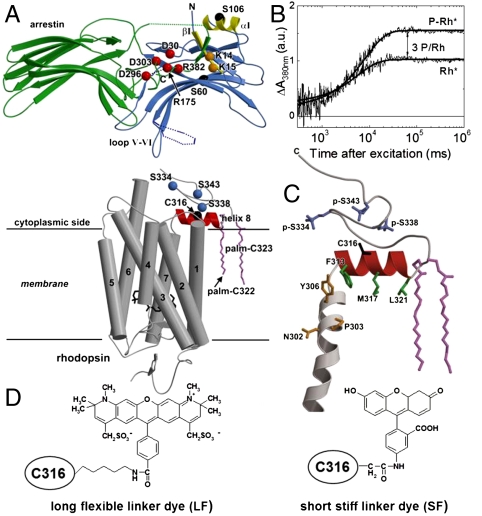
Structures of arrestin and rhodopsin. (A) Crystal structural models of rhodopsin (gray, PDB entry 1U19) and arrestin (blue: N-domain, green: C-domain, PDB entry 1CF1), generated with MOLSCRIPT (49). Rhodopsin: blue- three possible phosphorylation sites S334, S338, and S343, red- H8, violet- palmitoylated cysteines (palm-C322, palm-C323). Arrestin: dark red—interacting charged residues in the polar core of arrestin, yellow- helix αI and β-strand βI of the three-element interaction, dark yellow—external phosphate-sensors K14 and K15. Loop V-VI of arrestin is presented in its extended (solid line) and “closed” (dashed line) conformation (coordinates are taken from different molecules in the asymmetric unit of the arrestin crystal). The labeling positions C316 (rhodopsin), S106C and S60C (arrestin) are indicated in black. (B) Time traces of Meta-II formation at 380 nm (3 °C, pH 7.5). The increase in Meta-II concentration of P-Rh* membranes corresponds to an average of three phosphates per rhodopsin. (C) Close-up of the C-terminal region of rhodopsin, including part of transmembrane helix 7 (gray), helix 8  (red), and three phosphorylated serines, p-S334, p-S338, and p-S343 (blue). Dark yellow- residues belonging to the NPxxY motif. Green- side chains of the hydrophobic residues F313, M317, and L321. (D) Chemical structure of 5-IAF (SF) and Alexa594 (LF) bound to C316.
(red), and three phosphorylated serines, p-S334, p-S338, and p-S343 (blue). Dark yellow- residues belonging to the NPxxY motif. Green- side chains of the hydrophobic residues F313, M317, and L321. (D) Chemical structure of 5-IAF (SF) and Alexa594 (LF) bound to C316.
It has been suggested that multisite binding of visual arrestin for sensing the activation and phosphorylation state of the receptor is necessary for the multistep transition to the active receptor-bound conformation of arrestin (10–15). This proposed transition is thought to involve the disruption of three intramolecular interactions in arrestin (11, 13): (i) the hydrophobic interface between the two dome-shaped domains (green/blue), (ii) the polar core, consisting of five shielded interacting charged residues (red), and (iii) the three-element interaction between β-strand I and α-helix I (yellow) in the N-terminal domain (N-domain, blue), and the C-terminal tail (Fig. 1A). The multistep binding model includes the recognition of receptor-bound phosphates by arrestin’s “external” phosphate binding sites, Lys14 and Lys15, (referred to as binding step 1) and by the main phosphate-sensor Arg175 in the polar core of visual arrestin (referred to as binding step 2) (12, 15). The external phosphate binding sites are conserved among the different arrestins, which act as antennas collecting the receptor-bound phosphates at the entrance of the N-domain cupola (15). Disruption of the salt bridge Arg175-Asp296 by phosphate binding in the polar core is suggested to result in conformational changes; i.e., arrestin activation, leading to the high-affinity arrestin binding state (9–14). A direct kinetic proof of multistep arrestin binding, however, is not provided so far.
The arrestin binding interface was shown to involve loop V-VI of arrestin and further residues lining mostly the inner surfaces of the two concave domains (Fig. 1A), based on X-ray crystal structural models of visual arrestin (11, 13), biochemical (9, 16–18), and biophysical (19–24) data. Information about specific interaction structures of the receptor with arrestin is limited (24, 25), although the major activation-dependent conformational changes of rhodopsin are known from biophysical [e.g., (26–30)] and crystallographic data (31, 32). In particular, rhodopsin’s amphipathic helix 8 (H8) is a prime candidate for the development of dynamic interaction structures with arrestin. H8 starts after the conserved NPxxY motif in transmembrane helix 7 and connects the membrane domain via the two palmitoylated cysteines 322 and 323 with the phosphorylated C-terminal tail (Fig. 1 A and C). Because the first X-ray crystal structure of rhodopsin (33) resolved this helical stretch as lying parallel to the cytoplasmic membrane surface (Fig. 1 A and C), it was anticipated that H8 plays an important role in the GPCR signaling process due to its amphipathic properties. Indeed, it was shown that H8 may act as a membrane dependent conformational switch (34) and is coupled by its conformational dynamics to the ligand-binding site (29, 30, 35) via the conserved NPxxY motif (Fig. 1C).
As the receptor-bound conformation of arrestin and its interplay with receptor conformations determine arrestin function, we answer in this study the question whether there are sequential arrestin conformational changes upon binding to rhodopsin and whether H8 is involved in termination of receptor signaling via arrestin binding. Using a combination of site-directed mutagenesis, fluorescence labeling and time-correlated single-photon counting we determined the kinetics of arrestin conformational changes after light activation of phosphorylated rhodopsin (P-Rh). The structural dynamics of H8 was investigated by time-resolved fluorescence depolarization in the inactive and active receptor state, before and after receptor phosphorylation, as well as with and without arrestin. We show that the dynamics and conformational changes of H8 are intimately involved in the binding process of visual arrestin. The direct role of H8 in the arrestin binding process is supported by our finding that chemical modifications of H8 can inhibit arrestin binding. Kinetic measurements of rhodopsin-induced conformational changes of arrestin provide direct proof for the multistep nature of arrestin binding to the activated phosphorylated receptor (P-Rh*), thereby allowing a correlation between the individual steps of arrestin binding and structural changes of rhodopsins binding interface.
Results
Direct Observation of Multistep Arrestin Binding.
We set out to test the anticipated multistep binding of arrestin and to understand the molecular nature of the process by following the kinetics of arrestin conformational changes upon interaction with P-Rh*. First, we used fluorescently labeled visual arrestin in combination with multidimensional time-correlated single-photon counting for fluorescence pump-probe measurements (36). In these experiments the pump pulse activates rhodopsin and the fluorescence excitation pulse probes the resulting conformational changes of arrestin via fluorescence changes of the reporter group. As a sensitive site to detect arrestin conformational changes we chose position 106 within arrestin α-helix I (Fig. 1A). Ser106 is supposedly not involved in the arrestin-rhodopsin binding interface (11, 24), but is located directly above the three-element interaction domain (Fig. 1A). Interaction of the receptor-bound phosphates with the external phosphate binding sites at β-strand I is thought to destabilize the three-element interaction and to result in a guiding of the negatively charged phosphates to the polar core (15, 37, 38). Site-directed fluorescence labeling was achieved via the single-cysteine arrestin mutant ArrCA-S106C (22) to which the fluorescent dye Lucifer Yellow (LY) iodoacetamide was covalently bound (ArrCA-S106C-LY, Fig. S1A). ArrCA-S106C-LY shows the same binding affinity to P-Rh* as unlabeled arrestin (Fig. S1B). All experiments were performed under standardized conditions using prephosphorylated rhodopsin disk membranes with a phosphorylation level of ∼3 phosphates/rhodopsin (Fig. 1B). This number of receptor-attached phosphates was shown to be sufficient to trigger high-affinity arrestin binding (6, 9).
Using LY as the reporter group, we observed changes in the fluorescence lifetime curve upon addition of P-Rh and P-Rh* to ArrCA-S106C-LY, including a pronounced fast picosecond decay component and specific changes in intensity as compared to ArrCA-S106C-LY alone (Fig. 2A). Thus, LY bound to position 106 is indeed sensitive to arrestin interactions with the phosphorylated receptor in both its inactive and active state. Furthermore, this result supports recent models (10), which suggest that initial phosphate recognition by the external phosphate binding sites already occurs in a low-affinity interaction state, as probed here using P-Rh.
Fig. 2.
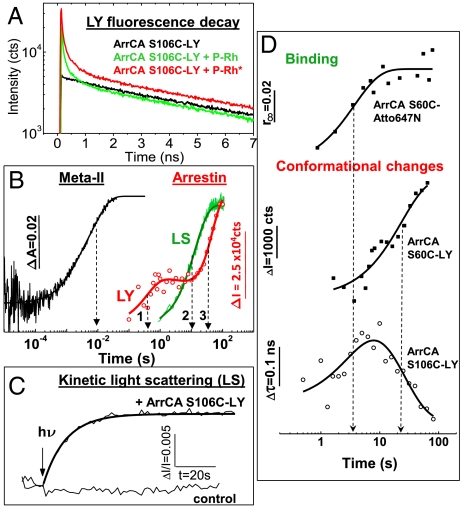
Kinetics of arrestin binding and conformational changes after activation of rhodopsin by a flash of light. (A) Fluorescence lifetime curves of LY covalently bound to S106C of arrestin in the absence and presence of P-Rh and P-Rh*, respectively. (B) Time trace of Meta-II formation (black). Time trace of arrestin binding to P-Rh* as measured by kinetic light scattering (LS, green). Time trace of arrestin conformational changes as monitored by the integral fluorescence changes of bound LY to position 106 of arrestin (LY, red). The solid lines represent multiexponential fits. Arrows and numbers indicate the transitions explained in the text. Conditions: 1 μM rhodopsin, 10 μM arrestin, 150 mM NaCl, 50 mM potassium phosphate buffer pH 7.5, 20 °C. (C) Time trace of arrestin binding as shown in (B) together with the control (light scattering without arrestin) on a linear time scale. (D) Time trace of arrestin binding to P-Rh* as measured by the increase in final anisotropy r∞ of the covalently bound dye Atto647N to position 60 of arrestin (top; τ = 4 s). Conformational changes of arrestin at position 60 and 106 as revealed by the changes in integral fluorescence intensity and lifetime of LY, respectively (bottom; ArrCA-S106C-LY: τ1 = 4 s, τ2 = 24 s, ArrCA-S60C-LY: τ = 29 s). The fluorescence decay times of ArrCA-S106C-LY are 0.1 ns, 1.6 ns, and 7.8 ns and the changes of the slowest decay time (lifetime) upon binding to P-Rh* are shown. Conditions: 1 μM arrestin, 3–5 μM rhodopsin, 150 mM NaCl, 50 mM potassium phosphate buffer pH 7.5, 20 °C. Fluorescence excitation was at 428 nm (LY) or 617 nm (Atto647N) and the emission was detected after passing through a cut-off filter OG495 and RG665, respectively.
To follow the kinetics of receptor-induced conformational changes in arrestin, we measured the LY-fluorescence as a function of time after light activation of rhodopsin with a short flash of light (Fig. 2B, red curve). As initial interaction between arrestin and P-Rh was observed, prebound arrestins may directly proceed to binding step 2 after light activation of P-Rh, undergoing the large conformational change proposed for arrestin activation. Soluble arrestin molecules, which are recruited to the rhodopsin surface after light activation, are anticipated to interact first with P-Rh* via their external phosphate recognition sites (binding step 1) before recognition of the phosphorylated rhodopsin C terminus in the polar core and arrestin activation occurs (binding step 2). Consequently, a superposition of the corresponding kinetics is expected. After light activation of P-Rh (Fig. 2B, black curve, τ ∼ 15 ms) we indeed observed three kinetic phases of arrestin conformational changes (Fig. 2B, red curve), in agreement with the above described multistep binding scenario. Assuming a sequential reaction scheme, the time constants for the conformational transitions to the high-affinity binding state of arrestin were obtained from a three-exponential fit yielding τ1 = 0.4 ± 0.1 s, τ2 = 11 ± 1.6 s, and τ3 = 36 ± 2.3 s (Fig. 2B). To discriminate between conformational changes connected to the binding of soluble arrestins and those connected to arrestin activation, we performed kinetic light scattering experiments to follow the binding of arrestin (22). The binding curve is based on the increase of light scattering upon arrestin binding to rhodopsin disk membranes (Fig. 2B, green curve, Fig. 2C). Its timing correlates with the second transition (τ2 = 11 s) in the LY-fluorescence change (Fig. 2B). This result supports a tentative assignment of transition 1 to arrestin conformational changes connected to arrestin activation of prebound arrestin molecules, transition 2 to conformational changes occurring upon light-induced binding of soluble arrestins and transition 3 to conformational changes connected to arrestin activation of the latter.
To further corroborate the multistep nature of the arrestin binding process, we performed additional experiments with different concentrations and excess P-Rh (1 μM arrestin, 3–5 μM P-Rh). Under these conditions the binding of arrestin to P-Rh can be observed using fluorescence anisotropy. Binding of fluorescently labeled arrestin with a rotational correlation time of about 40 ns (Fig. S2) to rhodopsin disk membranes with a rotational correlation time in the milliseconds time range results in an anisotropy that virtually decays to a constant end value r∞ on the fluorescence observation time scale. For the anisotropy experiment we used arrestin labeled in position 60 at the inside of the N-domain dome (Fig. 1A). Binding of a dye to a single cysteine in this position (ArrCA-S60C) does not interfere with arrestin binding (Fig. S3). From the polarized fluorescence pump-probe experiment we extracted r∞, whose increase is indicative of arrestin binding to rhodopsin membranes (Fig. 2D, top). The corresponding changes of LY fluorescence and lifetime were measured for ArrCA-S60C-LY and ArrCA-S106C-LY, respectively (Fig. 2D, bottom). Two conformational transitions of arrestin are clearly visible in the time range between 1 and 100 s (Fig. 2D, bottom). The conformational transition in the 1–10 s range (ArrCA-S106C-LY) correlates with binding kinetics from anisotropy. The later transitions in the 20–30 s range (ArrCA-S106C-LY and ArrCA-S60C-LY) are indicative of conformational changes connected with the transition of arrestin to its high-affinity binding conformation.
Phosphorylation and Arrestin Interaction Affect H8 Dynamics.
To further understand the molecular mechanism behind the individual kinetic steps of arrestin binding, we investigated the dynamic conformations of rhodopsin involved in arrestin-rhodopsin interaction, in particular of rhodopsin’s amphipathic H8. In earlier studies, we already showed a correlation of H8 dynamics with different receptor states (28, 30, 39). To obtain information about the nanosecond dynamics of H8, we measured the time-resolved fluorescence anisotropy of the reporter group iodoacetamidofluorescein covalently bound to Cys316 in H8 (Fig. 1 C and D). As the dye is bound via a short acetamido linker, we termed the construct Rh-H8-SF [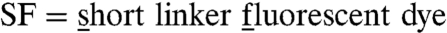 (Fig. 1D; Fig. S4A)]. Because the probed dynamics of the dye is affected by the motion of the protein segment to which it is covalently attached, the anisotropy decay curve yields information on global and local protein dynamics as well as on the protein structure and conformational changes. The analysis of H8-SF anisotropy decay curves (Fig. 3) was performed as described previously (35, 40). The main decay components were assigned to the dynamics of (i) the dye itself with a correlation time in the 100–300 ps range, (ii) the H8 segment the dye is bound to, and (iii) a constant end value of the anisotropy (r∞), which represents a measure of steric hindrance of H8 motion (Fig. S5, Table S1).
(Fig. 1D; Fig. S4A)]. Because the probed dynamics of the dye is affected by the motion of the protein segment to which it is covalently attached, the anisotropy decay curve yields information on global and local protein dynamics as well as on the protein structure and conformational changes. The analysis of H8-SF anisotropy decay curves (Fig. 3) was performed as described previously (35, 40). The main decay components were assigned to the dynamics of (i) the dye itself with a correlation time in the 100–300 ps range, (ii) the H8 segment the dye is bound to, and (iii) a constant end value of the anisotropy (r∞), which represents a measure of steric hindrance of H8 motion (Fig. S5, Table S1).
Fig. 3.
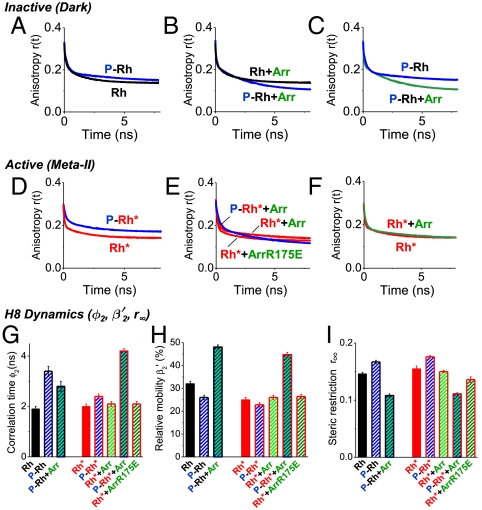
H8 dynamics in response to rhodopsin phosphorylation and arrestin interaction. Time-resolved anisotropy decay curves of SF covalently bound to rhodopsin in position Cys316 for dark (A–C) and light-activated rhodopsin membranes (D–F): (A), (D) phosphorylated and unphosphorylated rhodopsin, (B), (E) phosphorylated and unphosphorylated rhodopsin in the presence of arrestin or the arrestin mutant R175E, (C) phosphorylated rhodopsin in the absence and presence of arrestin, (F) rhodopsin in the absence and presence of arrestin. The corresponding fluorescence anisotropy decay parameters are presented in (G–I). (G) rotational correlation time of H8 (ϕ2), (H) conformational space of H8 expressed as relative mobility 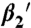 in percentage (
in percentage (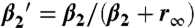 ), (I) steric restriction r∞ of H8. Black—dark (inactive) rhodopsin, red—light-activated rhodopsin, blue—phosphorylated rhodopsin, green—presence of arrestin. Conditions: 5 μM rhodopsin, 50 μM arrestin, 150 mM NaCl, 50 mM potassium phosphate buffer pH 7.5, 20 °C. The fluorescence excitation was at 470 nm and the emission was detected after passing through a cut-off filter OG515.
), (I) steric restriction r∞ of H8. Black—dark (inactive) rhodopsin, red—light-activated rhodopsin, blue—phosphorylated rhodopsin, green—presence of arrestin. Conditions: 5 μM rhodopsin, 50 μM arrestin, 150 mM NaCl, 50 mM potassium phosphate buffer pH 7.5, 20 °C. The fluorescence excitation was at 470 nm and the emission was detected after passing through a cut-off filter OG515.
Phosphorylation of the receptor results in a slower and more restricted motion of H8 in the inactive receptor state (Fig. 3 A, G–I). H8 correlation time (ϕ2) increases from 1.9 ± 0.1 ns in unphosphorylated rhodopsin (Rh) membranes to 3.4 ± 0.2 ns in P-Rh (Fig. 3G). Using the corresponding amplitude (β2) as a measure (Fig. 3H), we observed that the conformational space of H8 movement decreases slightly upon phosphorylation. Qualitatively similar phosphorylation-induced changes were found for P-Rh* (Fig. 3 D, G–I).
To probe the conformational changes of H8 upon arrestin interaction, we first added arrestin to inactive Rh and P-Rh (Fig. 3B). The difference between the anisotropy curves shown in Fig. 3B suggests that arrestin interacts differentially with Rh and P-Rh and indicates an involvement of H8 in the low-affinity phosphate recognition state of arrestin with P-Rh (compare Fig. 2A). Arrestin interaction with P-Rh leads to faster H8 motion (ϕ2,P-Rh = 3.4 ± 0.2 ns, ϕ2,P-Rh+Arr = 2.8 ± 0.2 ns) and an increase in conformational space of H8 motion (Fig. 3 C, G–I).
Next, we investigated the involvement of H8 in arrestin binding to light-activated nonphosphorylated rhodopsin (Rh*). Virtually no dynamics changes of H8 were detected (Fig. 3 F, G–I). When adding arrestin to P-Rh*, however, we observed a specific interaction of H8 with arrestin as indicated by the difference in the anisotropy curves of P-Rh*+Arr and Rh*+Arr (Fig. 3D). Arrestin interaction with P-Rh* leads to an approximately twofold slower motion of H8 as well as a larger conformational space sampled by H8, primarily induced by a reduction in steric restriction (r∞) from the surrounding constituents (Fig. 3 G–I).
H8 Is Directly Involved in Arrestin Binding.
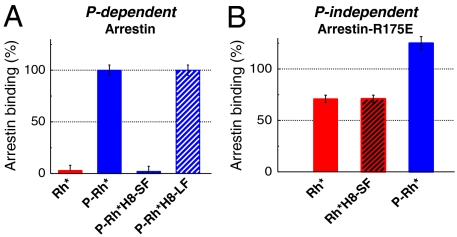
Functional arrestin binding to P-Rh* can be measured via its stabilizing effect on the active receptor state Meta-II (24, 41) (Fig. S6). Although arrestin interaction with P-Rh* has been shown by distinct changes in H8 dynamics (Fig. 3E), we observed no Meta-II stabilization for P-Rh*H8-SF (Fig. 4A). To solve this puzzle, we used another bulky dye, Alexa594-maleimide (Fig. 1D, Fig. S4B), which contains a long and flexible C5-linker (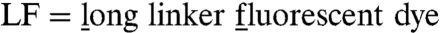 ) In contrast to P-Rh*H8-SF, rhodopsin labeled with Alexa594 (P-Rh*H8-LF) displays normal arrestin binding affinity (Fig. 4A). Thus, we concluded that the inflexible, short linker dye SF hinders functional arrestin binding by steric means, while the long flexible linker dye LF presumably is pushed aside and therefore permits the H8-arrestin interaction.
) In contrast to P-Rh*H8-SF, rhodopsin labeled with Alexa594 (P-Rh*H8-LF) displays normal arrestin binding affinity (Fig. 4A). Thus, we concluded that the inflexible, short linker dye SF hinders functional arrestin binding by steric means, while the long flexible linker dye LF presumably is pushed aside and therefore permits the H8-arrestin interaction.
Fig. 4.
Arrestin binding based on Meta-II stabilization. Binding affinity of (A) arrestin WT, and (B) Arrestin-R175E. The arrestin binding affinity to unlabeled P-Rh* was set to 100%. Red—unphosphorylated rhodopsin, blue—phosphorylated rhodopsin, black hatched—SF-labeled rhodopsin, white hatched—LF-labeled rhodopsin.
Close Interaction of H8 with Arrestin Is Required for Arrestin Activation.
To test whether this close interaction of H8 with arrestin occurs in the high-affinity binding conformation, we investigated the binding of SF-labeled rhodopsin to the “preactivated” arrestin mutant R175E. In this mutant protein the crucial salt bridge Arg175-Asp296 is already disrupted (12). As shown in Fig. 4B, Rh*H8-SF exhibits the same binding affinity to preactivated arrestin as unlabeled rhodopsin, ruling out the hypothesis that SF inhibits H8-arrestin interaction in the high-affinity binding conformation. Therefore, a close interaction of H8 with arrestin is assumed for the binding process of the phosphorylated receptor C terminus to the phosphate-sensor Arg175, resulting in arrestin activation.
H8 Dynamics in the Prebinding and High-Affinity Binding State.
Meta-II stabilization and experiments with preactivated arrestin R175E indicated that the binding process between P-Rh*H8-SF and arrestin is trapped before arrestin activation occurs. Thus, the anisotropy experiment of P-Rh*H8-SF in the presence of arrestin (Fig. 3E) describes H8 dynamics in a light-activated prebinding state of arrestin. The dynamics of H8 in the high-affinity binding state between light-activated rhodopsin and arrestin, however, were measured using the preactivated arrestin mutant R175E (Fig. 3 E, G–I). Summarizing H8 dynamics, we observed that the interaction of arrestin with P-Rh* in the prebinding state leads to a larger conformational space and a slower motion of H8. Modest conformational space and fast motion of H8 were observed in the high-affinity arrestin binding state (Fig. 3 G and H).
Discussion
Mechanistic Model of Arrestin Binding.
The current multistep model of arrestin binding (10, 12, 15) suggests that two major binding steps facilitate arrestin binding. Our kinetic experiments to elucidate arrestin conformational changes upon receptor binding showed that two distinct transitions occur after light activation of rhodopsin, supporting a sequential multistep binding mechanism (Fig. 2). As illustrated in Fig. 5, initial binding (step 1) of soluble arrestin by light-activated rhodopsin coincides with a conformational change observed with a fluorescent reporter group attached to α-helix I of arrestin (Fig. 2 B–D, 1–11 s transition). This observation is consistent with a destabilization of the three-element interaction between β-strand I, α-helix I, and the arrestin C-terminal tail, induced by prebinding of the receptor-bound phosphates to the external binding sites Lys14 and Lys15 (15) in binding step 1. A separate conformational transition of arrestin emerges at later times (∼30 s transition), as clearly evident from the biphasic fluorescence trace in Fig. 2D (bottom). This transition involves the region of the three-element interaction (probed at position 106) as well as the inside of the N-domain dome (probed at position 60) and is consistent with conformational changes connected to binding of the receptor-bound phosphates to Arg175 in the polar core resulting in arrestin activation (binding step 2, Fig. 5). Our observations generally agree with recent findings from EPR spectroscopy, which show that the local structure of arrestin around α-helix I changes upon binding to rhodopsin (42). While in the natural situation phosphorylation of rhodopsin by rhodopsin kinase occurs after light activation, we performed our experiments with prephosphorylated rhodopsin. Here, we observed low-affinity interaction between arrestin and P-Rh. Our data suggest that light-induced binding of soluble arrestins, which occurs with significantly slower kinetics than the conformational transition of already prebound arrestin molecules to the high-affinity arrestin binding state (Fig. 2B), seems to be a rate-limiting step in the arrestin binding process. The timing of arrestin conformational changes, as discussed above, provides clear kinetic evidence for a sequential multistep process of arrestin binding.
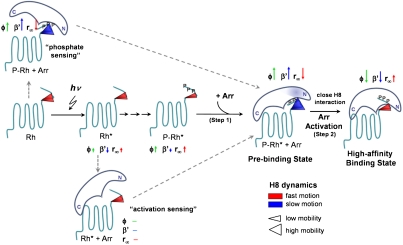
Fig. 5.
Model for the activation of arrestin (Arr). After phosphorylation of Rh* by rhodopsin kinase (not shown), the prebinding and high-affinity binding state are successively formed. See Discussion for details. The conformational changes around α-helix I in the prebinding state of arrestin are indicated by a transparent blue coloring. H8 dynamics is visualized by the size and color of the cone representing H8 conformational space (mobility) and motion (ϕ < 2.5 ns: red, ϕ > 2.5 ns: blue), respectively. The dynamics changes compared to the preceding state are indicated by the corresponding anisotropy parameters φ (correlation time), β′ (conformational space/relative mobility), and r∞ (steric restriction) presented as arrows. The arrow length represents the magnitude of the change and the direction indicates the increase or decrease of the respective value.
Combining the kinetic data of the arrestin binding process with surface dynamics changes of rhodopsin, we propose a mechanistic model of arrestin binding, which highlights rhodopsin’s H8 as a crucial element in arrestin activation (illustrated in Fig. 5). Only recently, this short amphipathic H8, located at the start of the C-terminal tail of class A GPCRs, is gaining recognition for its importance in GPCR function (43), as it might serve as a transmitter of signaling states or is itself involved in regulating C-terminal structures of rhodopsin. Our data support that notion and show that H8 is intimately involved in the multistep binding process of arrestin by providing specific conformational and structural constraints for initial phosphate recognition and activation of the phosphate-sensor Arg175 in the polar core. We showed that binding of a bulky inflexible group (SF) to H8 traps the arrestin—P-Rh* complex in a prebinding state (Fig. 4). We further concluded that a close steric interaction of H8 with arrestin is mandatory for the transition from pre- to high-affinity binding. The reaction model in Fig. 5 shows how the different arrestin interacting forms of H8 develop. The model starts in the Rh state and depicts the dynamical changes of H8 in the Rh* and P-Rh* states preceding arrestin interaction. Upon initial binding of arrestin to P-Rh* (binding step 1) a prebinding state forms, that is characterized by slower motion and an increase in conformational space (high mobility) of H8. Arrestin itself exhibits subtle conformational changes around α-helix I. Binding step 2 requires a close interaction with H8 to permit arrestin activation; i.e., disruption of the Arg175-Asp296 salt bridge in the polar core and the subsequent large-scale conformational change. H8 adopts a modest mobility in the resulting high-affinity binding state, consistent with a tight binding between rhodopsin and arrestin. Because multisite binding of arrestin for sensing the activation and phosphorylation state of the receptor was suggested (9, 10), the respective dynamic interactions were revealed by investigating the effect of arrestin on P-Rh and Rh*. Low-affinity interaction of arrestin with P-Rh results in a similar increase of H8 conformational space as observed for arrestin prebinding to P-Rh*. Thus, our data suggest that changes of H8 dynamics are involved in the “phosphate sensing” process. In contrast, “activation sensing” determined via Rh* interaction was not observed to affect H8 dynamics, provided that the bound reporter group does not interfere. To distinguish these data in the reaction scheme in Fig. 5, dashed gray arrows were used.
Role of H8 in Visual Arrestin Binding Specificity.
The question which remains to be answered is how a large conformational space of H8 motion as well as a specific interaction between H8 and arrestin could regulate the delivery of the phosphorylated C-terminal tail of the receptor to the polar core of arrestin. Visual arrestin shows the highest preference for the active phosphorylated receptor form over the inactive phosphoreceptor in comparison to the two known nonvisual arrestins, which display broad receptor specificity (2). It has been suggested that the higher rigidity of rod arrestin and an additional positively charged residue (Arg18) in the loop following β-strand I, which contains Lys14 and Lys15 (Fig. 1A), is responsible for the better discrimination between the functional forms of rhodopsin (10, 44). Our results indicate that the rigidity of visual arrestin is counterbalanced by an increased mobility (conformational space) of H8 after phosphorylation and arrestin interaction (Fig. 3). In contrast to nonvisual arrestins, whose flexibility seems to allow them to fit the active receptor face right away, the higher rigidity of visual arrestin seems to require the active rhodopsin to fit its H8, and presumably its phosphorylated C terminus, into a conformation facilitating “phosphate-sensor” activation and inducing a higher flexibility of the N-domain allowing arrestin to adopt its active structure. This picture is supported by the finding of different crystallographic visual arrestin conformers, where a correlation between the flexibility in the N-domain and the conformation of loop V-VI, spanning the highly conserved residues 68–78, was observed (11). A higher flexibility of the N-domain correlates with an extended conformation of loop V-VI (11), the latter was assumed to develop upon arrestin binding to P-Rh* (23). A lower flexibility of the N-domain correlates with a conformation in which this loop is folded back towards the N-cupola (“closed” conformation, Fig. 1A), preventing an interaction between the receptor-attached phosphates and the phosphate-sensor due to steric hindrance (11, 23)—unless a higher mobility of H8 may circumvent this obstacle.
In summary, our results show that H8 is a crucial contributor in the phosphate sensing process of arrestin. We suggest that the function of H8 is to provide a receptor-activation-mediated feedback, allowing the guiding process of the phosphates to the polar core and thus arrestin activation. The combination of fluorescence pump-probe and depolarization experiments used here is a versatile method to obtain dynamic information of binding partners during the different stages of their engagement. This information is not possible to extract from static X-ray structures and provides a dynamic framework for understanding the molecular mechanism underlying protein-protein interaction.
Material and Methods
The details of arrestin expression, purification, rhodopsin phosphorylation, site-directed fluorescence labeling and the spectroscopic techniques are described in SI Text.
Sample Preparation.
11-cis-retinal was prepared as described (45). Rod outer segment disk membranes were prepared from bovine retinae (W.L. Lawson Corp) as described (35, 46). Multi-phosphorylated rhodopsin was prepared according to ref. 8. Determination of the phosphorylation stoichiometry was performed in a spectroscopic assay based on the stabilization of Meta-II by phosphorylation. The concentration of Meta-II formed at low temperature and high pH increases by a factor of ∼2 for a phosphorylation stoichiometry of six phosphates/rhodopsin (47). Large-scale expression of arrestin was done as described (22). Labeling of rhodopsin’s Cys316 with the fluorescent dyes 5-iodoacteamidofluorescein (5-IAF, SF) and Alexa594-C5-maleimide (LF) (Molecular Probes/Invitrogen) and labeling of the single-cysteine arrestin mutants CA-S106C and CA-S60C (22) with Lucifer yellow (LY) iodoacetamide (Molecular Probes/Invitrogen) and Atto647N-maleimide (Atto-Tec) was carried out essentially as described (35).
Spectroscopy.
Flash spectroscopy to measure Meta-II kinetics and data analysis were performed as described (35). Fluorescence lifetime and anisotropy were measured employing a tunable Ti:Sapphire laser/microchannel plate based single-photon counting apparatus equipped with a TCSPC Module (SPC-830, B&H GmbH) (35, 36). A forward kinetic light scattering instrument ( ) was build up according to (48). Reactions were triggered by a flash of light (478 nm).
) was build up according to (48). Reactions were triggered by a flash of light (478 nm).
Supplementary Material
Acknowledgments.
We thank Dr. R. Batra Safferling (ICS) and C. Seidler (FU Berlin) for help with sample and measurements, and S. Lehmann for building up the light-scattering instrument. The work was supported by the Deutsche Forschungsgemeinschaft, Sfb 449, (to U.A.) and ONEXIM (to G.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015461108/-/DCSupplemental.
References
- 1.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 2.Gurevich VV, et al. Arrestin interactions with G protein-coupled receptors. Direct binding studies of wild type and mutant arrestins with rhodopsin, beta 2-adrenergic, and m2 muscarinic cholinergic receptors. J Biol Chem. 1995;270:720–731. doi: 10.1074/jbc.270.2.720. [DOI] [PubMed] [Google Scholar]
- 3.Goodman OB, Jr, et al. Beta-arrestin acts as a clathrin adaptor in endocytosis of the beta2-adrenergic receptor. Nature. 1996;383:447–450. doi: 10.1038/383447a0. [DOI] [PubMed] [Google Scholar]
- 4.Lefkowitz RJ, Shenoy SK. Transduction of receptor signals by beta-arrestins. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- 5.Sakmar TP. Structure of rhodopsin and the superfamily of seven-helical receptors: the same and not the same. Curr Opin Cell Biol. 2002;14:189–195. doi: 10.1016/s0955-0674(02)00306-x. [DOI] [PubMed] [Google Scholar]
- 6.Mendez A, et al. Rapid and reproducible deactivation of rhodopsin requires multiple phosphorylation sites. Neuron. 2000;28:153–164. doi: 10.1016/s0896-6273(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 7.Kuhn H, Hall SW, Wilden U. Light-induced binding of 48-kDa protein to photoreceptor membranes is highly enhanced by phosphorylation of rhodopsin. FEBS Lett. 1984;176:473–478. doi: 10.1016/0014-5793(84)81221-1. [DOI] [PubMed] [Google Scholar]
- 8.Wilden U, Kuhn H. Light-dependent phosphorylation of rhodopsin: number of phosphorylation sites. Biochemistry. 1982;21:3014–3022. doi: 10.1021/bi00541a032. [DOI] [PubMed] [Google Scholar]
- 9.Gurevich VV, Benovic JL. Visual arrestin interaction with rhodopsin. Sequential multisite binding ensures strict selectivity toward light-activated phosphorylated rhodopsin. J Biol Chem. 1993;268:11628–11638. [PubMed] [Google Scholar]
- 10.Gurevich VV, Gurevich EV. The structural basis of arrestin-mediated regulation of G-protein-coupled receptors. Pharmacol Ther. 2006;110:465–502. doi: 10.1016/j.pharmthera.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Granzin J, et al. X-ray crystal structure of arrestin from bovine rod outer segments. Nature. 1998;391:918–921. doi: 10.1038/36147. [DOI] [PubMed] [Google Scholar]
- 12.Gurevich VV, Benovic JL. Visual arrestin binding to rhodopsin. Diverse functional roles of positively charged residues within the phosphorylation-recognition region of arrestin. J Biol Chem. 1995;270:6010–6016. doi: 10.1074/jbc.270.11.6010. [DOI] [PubMed] [Google Scholar]
- 13.Hirsch JA, Schubert C, Gurevich VV, Sigler PB. The 2.8 A crystal structure of visual arrestin: a model for arrestin’s regulation. Cell. 1999;97:257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- 14.Schleicher A, Kuhn H, Hofmann KP. Kinetics, binding constant, and activation energy of the 48-kDa protein-rhodopsin complex by extra-metarhodopsin II. Biochemistry. 1989;28:1770–1775. doi: 10.1021/bi00430a052. [DOI] [PubMed] [Google Scholar]
- 15.Vishnivetskiy SA, et al. An additional phosphate-binding element in arrestin molecule. Implications for the mechanism of arrestin activation. J Biol Chem. 2000;275:41049–41057. doi: 10.1074/jbc.M007159200. [DOI] [PubMed] [Google Scholar]
- 16.Ascano MT, Smith WC, Gregurick SK, Robinson PR. Arrestin residues involved in the functional binding of arrestin to phosphorylated, photolyzed rhodopsin. Mol Vis. 2006;12:1516–1525. [PubMed] [Google Scholar]
- 17.Smith WC, et al. Identification of regions of arrestin that bind to rhodopsin. Biochemistry. 1999;38:2752–2761. doi: 10.1021/bi982643l. [DOI] [PubMed] [Google Scholar]
- 18.Vishnivetskiy SA, Hosey MM, Benovic JL, Gurevich VV. Mapping the arrestin-receptor interface. Structural elements responsible for receptor specificity of arrestin proteins. J Biol Chem. 2004;279:1262–1268. doi: 10.1074/jbc.M308834200. [DOI] [PubMed] [Google Scholar]
- 19.Hanson SM, et al. Differential interaction of spin-labeled arrestin with inactive and active phosphorhodopsin. Proc Natl Acad Sci USA. 2006;103:4900–4905. doi: 10.1073/pnas.0600733103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotake S, Hey P, Mirmira RG, Copeland RA. Physicochemical characterization of bovine retinal arrestin. Arch Biochem Biophys. 1991;285:126–133. doi: 10.1016/0003-9861(91)90338-j. [DOI] [PubMed] [Google Scholar]
- 21.Mokarzel-Falcon L, Padron-Garcia JA, Carrasco-Velar R, Berry C, Montero-Cabrera LA. In silico study of the human rhodopsin and meta rhodopsin II/S-arrestin complexes: impact of single point mutations related to retina degenerative diseases. Proteins. 2008;70:1133–1141. doi: 10.1002/prot.21873. [DOI] [PubMed] [Google Scholar]
- 22.Skegro D, et al. N-terminal and C-terminal domains of arrestin both contribute in binding to rhodopsin. Photochem Photobiol. 2007;83:385–392. doi: 10.1562/2006-08-25-RA-1014. [DOI] [PubMed] [Google Scholar]
- 23.Sommer ME, Farrens DL, McDowell JH, Weber LA, Smith WC. Dynamics of arrestin-rhodopsin interactions: loop movement is involved in arrestin activation and receptor binding. J Biol Chem. 2007;282:25560–25568. doi: 10.1074/jbc.M702155200. [DOI] [PubMed] [Google Scholar]
- 24.Pulvermuller A, Schroder K, Fischer T, Hofmann KP. Interactions of metarhodopsin II. Arrestin peptides compete with arrestin and transducin. J Biol Chem. 2000;275:37679–37685. doi: 10.1074/jbc.M006776200. [DOI] [PubMed] [Google Scholar]
- 25.Raman D, Osawa S, Gurevich VV, Weiss ER. The interaction with the cytoplasmic loops of rhodopsin plays a crucial role in arrestin activation and binding. J Neurochem. 2003;84:1040–1050. doi: 10.1046/j.1471-4159.2003.01598.x. [DOI] [PubMed] [Google Scholar]
- 26.Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 27.Hubbell WL, Altenbach C, Hubbell CM, Khorana HG. Rhodopsin structure, dynamics, and activation: a perspective from crystallography, site-directed spin labeling, sulfhydryl reactivity, and disulfide cross-linking. Adv Protein Chem. 2003;63:243–290. doi: 10.1016/s0065-3233(03)63010-x. [DOI] [PubMed] [Google Scholar]
- 28.Kim TY, Moeller M, Winkler K, Kirchberg K, Alexiev U. Dissection of environmental changes at the cytoplasmic surface of light-activated bacteriorhodopsin and visual rhodopsin: sequence of spectrally silent steps. Photochem Photobiol. 2009;85:570–577. doi: 10.1111/j.1751-1097.2008.00525.x. [DOI] [PubMed] [Google Scholar]
- 29.Lehmann N, Alexiev U, Fahmy K. Linkage between the intramembrane H-bond network around aspartic acid 83 and the cytosolic environment of helix 8 in photoactivated rhodopsin. J Mol Biol. 2007;366:1129–1141. doi: 10.1016/j.jmb.2006.11.098. [DOI] [PubMed] [Google Scholar]
- 30.Kirchberg K, Kim TY, Haase S, Alexiev U. Functional interaction structures of the photochromic retinal protein rhodopsin. Photochem Photobiol Sci. 2010;9:226–233. doi: 10.1039/b9pp00134d. [DOI] [PubMed] [Google Scholar]
- 31.Scheerer P, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 32.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 33.Palczewski K, et al. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 34.Krishna AG, Menon ST, Terry TJ, Sakmar TP. Evidence that helix 8 of rhodopsin acts as a membrane-dependent conformational switch. Biochemistry. 2002;41:8298–8309. doi: 10.1021/bi025534m. [DOI] [PubMed] [Google Scholar]
- 35.Alexiev U, Rimke I, Pohlmann T. Elucidation of the nature of the conformational changes of the EF-interhelical loop in bacteriorhodopsin and of the helix VIII on the cytoplasmic surface of bovine rhodopsin: a time-resolved fluorescence depolarization study. J Mol Biol. 2003;328:705–719. doi: 10.1016/s0022-2836(03)00326-7. [DOI] [PubMed] [Google Scholar]
- 36.Kim TY, Winkler K, Alexiev U. Picosecond multidimensional fluorescence spectroscopy: a tool to measure real-time protein dynamics during function. Photochem Photobiol. 2007;83:378–384. doi: 10.1562/2006-06-21-RA-943. [DOI] [PubMed] [Google Scholar]
- 37.Gurevich VV, Gurevich EV. The molecular acrobatics of arrestin activation. Trends Pharmacol Sci. 2004;25:105–111. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 38.Palczewski K, Buczylko J, Imami NR, McDowell JH, Hargrave PA. Role of the carboxyl-terminal region of arrestin in binding to phosphorylated rhodopsin. J Biol Chem. 1991;266:15334–15339. [PubMed] [Google Scholar]
- 39.Kim TY, Schlieter T, Haase S, Alexiev U. Activation and molecular recognition of the GPCR rhodopsin—Insights from time-resolved fluorescence depolarisation and single molecule experiments. Eur J Cell Biol. 2011 doi: 10.1016/j.ejcb.2011.03.009. doi: 10.1016/j.ejcb.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 40.Schroder GF, Alexiev U, Grubmuller H. Simulation of fluorescence anisotropy experiments: probing protein dynamics. Biophys J. 2005;89:3757–3770. doi: 10.1529/biophysj.105.069500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hofmann KP. Effect of GTP on the rhodopsin-G-protein complex by transient formation of extra metarhodopsin II. Biochim Biophys Acta. 1985;810:278–281. doi: 10.1016/0005-2728(85)90143-4. [DOI] [PubMed] [Google Scholar]
- 42.Vishnivetskiy SA, et al. The role of arrestin alpha-helix I in receptor binding. J Mol Biol. 2010;395:42–54. doi: 10.1016/j.jmb.2009.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huynh J, Thomas WG, Aguilar MI, Pattenden LK. Role of helix 8 in G protein-coupled receptors based on structure-function studies on the type 1 angiotensin receptor. Mol Cell Endocrinol. 2009;302:118–127. doi: 10.1016/j.mce.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 44.Sutton RB, et al. Crystal structure of cone arrestin at 2.3 A: evolution of receptor specificity. J Mol Biol. 2005;354:1069–1080. doi: 10.1016/j.jmb.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 45.Knowles A, Priestley A. The preparation of 11-cis-retinal. Vision Res. 1978;18:115–116. doi: 10.1016/0042-6989(78)90086-x. [DOI] [PubMed] [Google Scholar]
- 46.McDowell JH, Kuhn H. Light-induced phosphorylation of rhodopsin in cattle photoreceptor membranes: substrate activation and inactivation. Biochemistry. 1977;16:4054–4060. doi: 10.1021/bi00637a018. [DOI] [PubMed] [Google Scholar]
- 47.Gibson SK, Parkes JH, Liebman PA. Phosphorylation stabilizes the active conformation of rhodopsin. Biochemistry. 1998;37:11393–11398. doi: 10.1021/bi980933w. [DOI] [PubMed] [Google Scholar]
- 48.Heck M, Pulvermuller A, Hofmann KP. Light scattering methods to monitor interactions between rhodopsin-containing membranes and soluble proteins. Methods Enzymol. 2000;315:329–347. doi: 10.1016/s0076-6879(00)15852-5. [DOI] [PubMed] [Google Scholar]
- 49.Kraulis P. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.