Abstract
Sickle cell disease (SCD) is a hematologic disorder caused by a missense mutation in the adult β-globin gene. Higher fetal hemoglobin (HbF) levels in red blood cells of SCD patients have been shown to improve morbidity and mortality. We previously found that nuclear receptors TR2 and TR4 repress expression of the human embryonic ε-globin and fetal γ-globin genes in definitive erythroid cells. Because forced expression of TR2/TR4 in murine adult erythroid cells paradoxically enhanced fetal γ-globin gene expression in transgenic mice, we wished to determine if forced TR2/TR4 expression in a SCD model mouse would result in elevated HbF synthesis and thereby alleviate the disease phenotype. In a “humanized” sickle cell model mouse, forced TR2/TR4 expression increased HbF abundance from 7.6% of total hemoglobin to 18.6%, accompanied by increased hematocrit from 23% to 34% and reticulocyte reduction from 61% to 18%, indicating a significant reduction in hemolysis. Moreover, forced TR2/TR4 expression reduced hepatosplenomegaly and liver parenchymal necrosis and inflammation in SCD mice, indicating alleviation of usual pathophysiological characteristics. This article shows that genetic manipulation of nonglobin proteins, or transcription factors regulating globin gene expression, can ameliorate the disease phenotype in a SCD model animal. This proof-of-concept study demonstrates that modulating TR2/TR4 activity in SCD patients may be a promising therapeutic approach to induce persistent HbF accumulation and for treatment of the disease.
Keywords: repression, NR2C1, NR2C2, β-thalassemia, therapeutic target
The β-globinopathies [β-thalassemia and sickle cell disease (SCD)] together comprise the most common inherited disorders in man (1, 2). The β-thalassemias can be caused by mutations in (or near), or complete deletion of, the adult β-globin gene, and the disorders differ in severity depending on the precise nature of the mutation, whereas SCD is caused by inheritance of two alleles bearing a single nucleotide change in the adult β-globin coding sequence, thereby generating sickle hemoglobin (HbS, α2βS2) (3). SCD can also be heterogeneous in terms of clinical consequences: the vast majority of SCD patients present early in childhood when adult hemoglobin normally replaces fetal hemoglobin (HbF, α2γ2), but the severity of the disease can differ markedly, correlating most strongly with the level of HbF present in red cells (4, 5). Indeed, HbF was one of the sole predictors of survival in the Cooperative Study of Sickle Cell Disease cohort study of 3,764 patients (6). Biochemically, HbF is known to competitively inhibit the formation of HbS polymers (7), thought to be responsible for the unusual, characteristic shape of the red cells, which leads to their rigidity, fragility, and premature destruction (2). Given these observations, it has been an expectation of the biomedical community for decades that if we could find a safe, effective drug that would induce high levels of fetal γ-globin synthesis in both SCD and β-thalassemia patients, in the absence of significant side effects, that these diseases could be medically managed.
The human β-globin locus consists of ε- (embryonic), Gγ- and Aγ- (fetal), and δ- and β-globin (adult) genes, which are spatially arranged from 5′ to 3′ and developmentally expressed in the same order (8). A little more than a decade ago, we serendipitously identified the repressor for the ε- and γ-globin genes, which was named a direct repeat erythroid definitive (DRED) protein (9). In analysis of mechanisms governing ε- and γ-globin gene silencing in definitive erythroid cells using transgenic mice bearing a yeast artificial chromosome (YAC) containing the entire human β-type globin locus, direct repeat (DR) elements, consensus binding sites for nonsteroidal nuclear receptors, in their proximal promoters were identified as essential silencer elements for both genes (9, 10). Consistent with this notion, hereditary persistence of fetal hemoglobin (HPFH), a human genetic condition in which the fetal γ-globin gene is abundantly transcribed in adulthood with elevated synthesis of HbF (≤50% of total hemoglobin) (8), is often associated with mutations located within the γ-globin DR elements (six of 16 documented nondeletional HPFH cases) (11). Further biochemical analyses indicated that the same DRED protein could bind to both the ε- and γ-globin promoter DR elements (9, 12). The initial hypothesis we entertained was that if one could biochemically identify the putative repressor and then devise ways to inactivate it, that condition would lead to induced γ-globin gene expression. Once the precise nature of the repressor was known, one could then screen for drugs that would inactivate its activity.
Using the ε-globin promoter binding site as an affinity reagent, we purified a candidate repressor from an adult erythroid cell line; fortuitously, the DNA binding core of the repressor turned out to be a pair of nuclear receptor “orphans” (meaning that at that time they had no known ligands) called TR2 and TR4 (in updated nomenclature, NR2C1 and NR2C2, respectively) (12). The possibility that these two nuclear receptors generated a heterodimeric γ-globin repressor (13) led to exciting expectations, because nuclear receptors have been exploited for decades as therapeutic targets for numerous diseases through mimetic ligand binding, whereas transcription factors themselves have never been effectively targeted. Our recent finding that the TR4 ligand binding domain can perfectly accommodate specific retinoid ligands (14), and the demonstration that both retinol and all-trans retinoic acid are capable of activating TR4 in a dose-dependent manner, suggests that at least TR4 is a bona fide receptor, and thus could be an effective target for defining a new ligand with which we might be able to effectively control embryonic or HbF production.
We demonstrated that germ-line compound null mutations of both TR2 and TR4 genes led to enhanced expression of both the human ε- and γ-globin genes, phenocopying the DR mutation phenotypes (9, 10), in definitive erythroid cells of the β-type globin YAC transgenic mice, which clearly indicated central roles for TR2/TR4 in the silencing of both the ε- and γ-globin genes (13). However, forced transgenic expression of TR2/TR4 using a Gata1 transcriptional control region in erythroid cells unexpectedly led to induction of the fetal γ-globin gene in adult erythroid cells in YAC transgenic mice (13). We hypothesized that the elevated abundance of the two receptors in these mice led to “squelching” (15) of the high molecular-weight repressor complexes (12, 16) by sequestering limiting components of those complexes, thereby effectively inactivating repressor function. Alternatively, TR2 and TR4 may bear an inherent context-dependent transcriptional activator function that is specific to the γ-globin gene in definitive erythroid cells, in keeping with the dual functionality of most nuclear receptors.
Here we report the consequences of forced TR2/TR4 expression on hematological and pathophysiological characteristics of a “humanized” SCD model mouse in which the murine adult α- and β-globin genes were replaced in the germ line by fragments of the human α- and sickle βS-globin loci (17). In this model, the human fetal Aγ-globin gene was placed in a position that would mimic its location and orientation between the murine embryonic and human adult βS-globin genes. We found that elevated levels of the TR2 and TR4 enhanced fetal γ-globin gene expression and HbF synthesis in adult erythroid cells of the humanized SCD mice, and led in turn to effective reversal of the disease phenotype that describes the usual cellular pathophysiology associated with SCD: alleviation of anemia, diminished hemolysis, and reduction of hepatosplenomegaly and liver necrosis and inflammation. These data unambiguously support the hypothesis that altering the activity of the TR2 and TR4 nuclear receptors could be an effective therapeutic strategy for the treatment of SCD and β-thalassemia.
Results and Discussion
Transgenic Expression of TR2 and TR4 in SCD Mice.
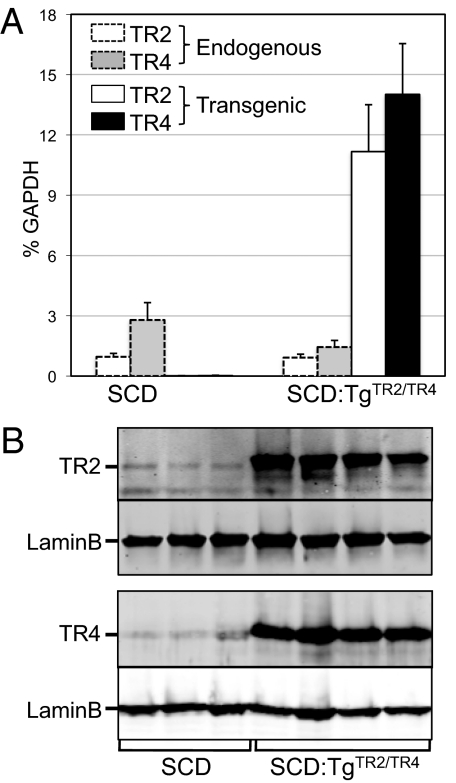
To assess the effects of forced TR2/TR4 expression in erythroid cells on hematological and pathological characteristics of SCD, we used the model mouse generated by Wu et al. (17), in which the murine adult α-globin genes were replaced with the human α-globin gene (genotype: Hba hα/hα), and the murine adult β-type globin genes were replaced with human sickle β (βS)- and fetal Aγ-globin gene fragments linked together (genotype: Hbb hγβS/hγβS); hereafter, we refer to this model as the SCD mouse. We bred the SCD mouse to a transgenic line in which TR2 and TR4 were forcibly expressed exclusively in the erythroid lineage by a Gata1 transcriptional regulatory region (13, 18) to generate compound mutant mice, represented here as SCD:TgTR2/TR4 mice, where the TR2/TR4 transgene is hemizygous. TR2 and TR4 mRNA levels in both the SCD mice and the SCD:TgTR2/TR4 mice were quantified. Reverse transcription and real-time quantitative PCR (RT-qPCR) analysis, which distinguishes endogenous and transgene-derived mRNAs, of spleen RNA of these mice showed that the transgenes were indeed expressed at significantly greater abundance than the endogenous genes (Fig. 1A). Transgenic TR2 mRNA abundance normalized to GAPDH mRNA in SCD:TgTR2/TR4 mice was about 12-fold more abundant than the endogenous TR2 mRNA levels in SCD mice, whereas TR4 mRNA in SCD:TgTR2/TR4 mice was approximately fivefold higher than endogenous TR4 in SCD animals. Curiously, we noted that in SCD:TgTR2/TR4 mice, the endogenous TR4 mRNA level was reduced to ∼50% of that in SCD mice, suggesting that the endogenous TR4 gene may be regulated in a negative feedback loop by TR2 or TR4.
Fig. 1.
Expression of TR2 and TR4 in SCD and SCD:TgTR2/TR4 mice. (A) RT-qPCR quantification of mRNA abundance of endogenous or transgenic TR2 and TR4 in the spleens of SCD and SCD:TgTR2/TR4 mice, normalized to an internal control (GAPDH). Four mice of each genotype (3–7 mo old) were analyzed. Error bars represent the SEM. (B) Quantification of TR2 and TR4 proteins in the spleens of three SCD mice and four SCD:TgTR2/TR4 compound mutant mice (8–12 wk old) by Western blotting.
The levels of TR2 and TR4 protein expression in the two different genotypes were also quantified by Western blot analysis (Fig. 1B), where expression was normalized to the ubiquitously expressed protein, lamin B. The spleens of the SCD:TgTR2/TR4 mice had 12-fold higher levels of TR2 compared with SCD (control) spleens; TR4 was also expressed at 17-fold–higher levels in the compound mutant mouse spleens than in SCD control spleens. These analyses confirmed that the TR2 and TR4 nuclear receptors were expressed at elevated levels in the definitive erythroid cells of compound mutant SCD mice. We speculate that the significantly greater enhancement of TR4 protein expression than of mRNA expression in SCD:TgTR2/TR4 mice, compared with endogenous levels in SCD mice, may be due to sequence differences in the untranslated regions of the transgenic and endogenous mRNA transcripts (e.g., the presence of a perfect Kozak consensus sequence only in the transgenic mRNAs) or in spliced (endogenous) vs. cDNA (transgenic)-expressed copies.
Expression of γ-Globin in SCD and SCD:TgTR2/TR4 Mice.
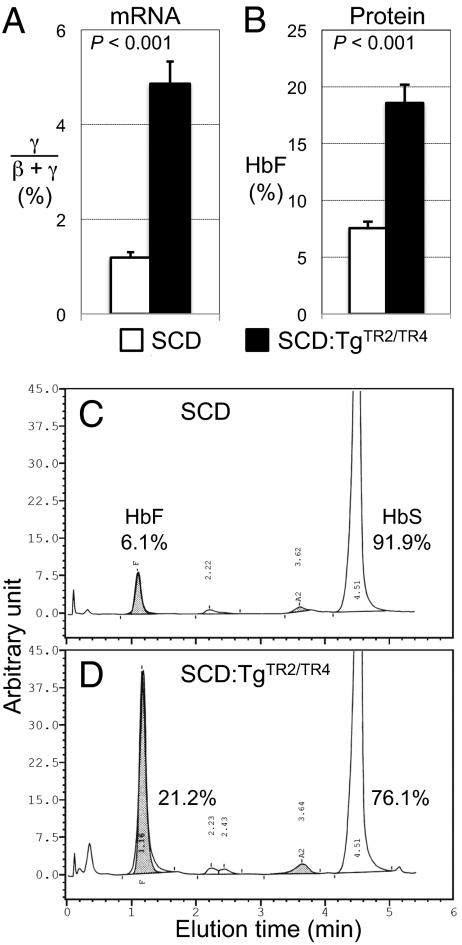
Mononuclear cells were isolated from bone marrow and spleens of SCD and SCD:TgTR2/TR4 mice, and analyzed for γ- and β-globin mRNA expression by RT-qPCR (normalized to 18S rRNA). Erythroid cells defined by cell surface Ter119 expression accounted for ∼95% of the spleen cells of both SCD and SCD:TgTR2/TR4 mice, and ∼75% and 85% of the bone marrow cells of SCD and SCD:TgTR2/TR4 mice, respectively (Fig. S1). The abundance of γ-globin mRNA as a fractional percentage of the total of γ- and β-globin mRNA abundance in SCD:TgTR2/TR4 mouse bone marrow cells averaged about fourfold higher than in SCD mice (Fig. 2A). We also examined peripheral blood for hemoglobin content. HbS and HbF abundance in whole blood from mice of the different genotypes was measured by HPLC. HbF (Fig. 2B and Table 1) was ∼2.5-fold higher in the SCD:TgTR2/TR4 mice (18.6% of total hemoglobin on average) than in SCD mice (7.6% on average). In fact, the most abundant HbF level reached 24% in one of the SCD:TgTR2/TR4 mice, whereas the highest HbF level in the SCD mice was 9.8%. Individual representative HPLC profiles displaying the hemoglobin constitution of SCD and SCD:TgTR2/TR4 mice are shown (Fig. 2 C and D). The SCD:TgTR2/TR4 mouse (Fig. 2D) exhibited significantly elevated HbF (to 21% of total hemoglobin) with concomitantly diminished HbS (76%) compared with the SCD mouse (Fig. 2C). In another sibling pair of 2-mo-old mice, the HbF of an SCD mouse bearing the TR2/TR4 transgenes was once again significantly higher (17.5%) than its SCD littermate (7.2%).
Fig. 2.
Expression of fetal γ-globin in SCD and SCD:TgTR2/TR4 mice. (A) RT-qPCR quantification of γ-globin mRNA abundance as a fraction of the total of γ- and β-globin mRNAs in SCD mice (open box) or SCD:TgTR2/TR4 compound mutant mice (black box). Four mice of each genotype (3–7 mo old) were analyzed. (B) Quantification of HbF (by HPLC) as a fraction of total hemoglobin in SCD mice (open box) or SCD:TgTR2/TR4 compound mutant mice (black box). Seven SCD mice and six SCD:TgTR2/TR4 mice 8–12 wk old were analyzed. In A and B, error bars represent the SEM. P values were calculated by Student t test. (C and D) Representative HPLC chromatograms to quantify hemoglobin tetramers in whole-blood lysates of SCD and SCD:TgTR2/TR4 mice. HbF (shaded areas) in the SCD mouse (C) and the SCD:TgTR2/TR4 mouse (D) represented 6.1% and 21%, respectively, of total hemoglobin.
Table 1.
Hematological parameters of SCD and SCD:TgTR2/TR4 mice
| Genotype |
|||||||||||
| Mouse | Hba* | Hbb† | TgTR2/TR4 | No.‡ | Age (mo) | HbF (%) | Hct (%) | MCV (fL) | MCHC (g/dL) | Platelet (×103 cells/μL) | WBC (×103 cells/μL) |
| Wild type (C57BL/6) | +/+ | +/+ | – | 7 | 2–3 | 0.1 ± 0.1§ | 48 ± 2.3 | 63.3 ± 1.8 | 20.8 ± 0.9 | 730 ± 146 | 4.8 ± 1.3 |
| Heterozygous SCD | hα/hα | hγβS/+ | – | 10 | 2–3 | 2.1 ± 0.7 | 41 ± 5.7 | 55.0 ± 2.4 | 21.2 ± 1.9 | 830 ± 324 | 8.8 ± 5.8 |
| SCD | hα/hα | hγβS/hγβS | – | 8 | 2–3 | 7.6 ± 1.6 | 23 ± 6.3 | 74.1 ± 8.7 | 14.7 ± 2.7 | 394 ± 186 | 19.3 ± 12.8 |
| SCD:TgTR2/TR4 | hα/hα | hγβS/hγβS | + | 6 | 2–3 | 18.6 ± 3.9 | 34 ± 8.9 | 59.9 ± 3.3 | 16.8 ± 1.7 | 421 ± 125 | 15.7 ± 4.8 |
| P value¶ | <0.001 | <0.01 | <0.01 | 0.068 | 0.38 | 0.27 | |||||
*For the genotype of the Hba locus, +/+ represents wild type (bearing the endogenous murine α-type globin genes), whereas hα/hα represents mutants homozygous for the human α-globin gene knockin (17).
†For the Hbb locus, +/+ represents wild type (the endogenous murine β-type globin genes), whereas Hbb hγβS/+ or hγβS/hγβS represents heterozygous or homozygous mutants bearing the human fetal Aγ- and βS-globin genes (17).
‡Number of mice analyzed for complete blood cell counts. For HbF analysis, five wild-type, nine heterozygous SCD, seven SCD, and six SCD:TgTR2/TR4 mice were examined.
§Numbers following ± refer to SDs of the stated numbers of mice.
¶P values refer to the comparison between the two final listed genotypes (SCD vs. SCD:TgTR2/TR4) by Student t test.
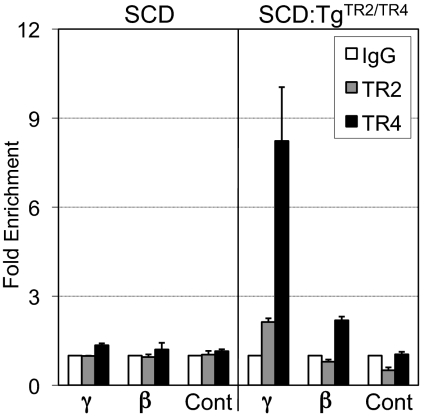
To address the underlying mechanism of the γ-globin gene induction in the SCD:TgTR2/TR4 mice, we performed a ChIP assay to determine whether TR2 and TR4 are bound in vivo to the γ-globin gene proximal promoter region that contains the DR element (Fig. 3), to which TR2 and TR4 can bind in vitro (13). In spleen cells of the SCD mice, association of TR2 or TR4 with the fetal γ-globin promoter was not detectable. In the SCD:TgTR2/TR4 mice, highly significant enrichment of TR4 in the γ-globin promoter (about eightfold over control IgG) was observed, whereas TR2 was also enriched on the same region to a lower extent (about twofold). Interestingly, also in the adult β-globin promoter, TR4, but not TR2, was also enriched to a lower extent (about twofold), although a specific binding sequence for TR4 in the β-globin promoter has not been identified. These data suggest that the overexpressed TR2 and TR4 directly bind to the fetal γ-globin promoter and thus activate its transcription in the SCD:TgTR2/TR4 mice, but a possible contribution of the “squelching” (15) or dominant negative effects by overexpressed receptors is not formally ruled out.
Fig. 3.
TR2 and TR4 bind to the fetal γ-globin gene promoter in spleen cells of SCD:TgTR2/TR4 mice. Binding of TR2 and TR4 to the human fetal γ-globin gene proximal promoter region containing the DR sequence, as well as the adult β-globin gene promoter in spleen cells of SCD or SCD:TgTR2/TR4 mice, was analyzed by ChIP assay using anti-TR2 or -TR4 antibody, or control IgG. For a negative control (Cont), an intronic region of the murine CHMP4B gene was also analyzed. Two mice of each genotype were used for the analysis. Error bars represent SD.
Reduction of Anemia and Reticulocytosis in SCD:TgTR2/TR4 Mice.
We next investigated the effect of forced TR2/TR4 expression on the hematologic parameters of the SCD mice. An automated hematology analyzer was used to perform complete blood cell counts. The WBC count, hematocrit (Hct), MCV, and platelet count were analyzed using peripheral blood from wild-type, heterozygous SCD, SCD, or SCD:TgTR2/TR4 mice (Table 1). Here, wild-type refers to C57BL/6 mice, whereas heterozygous SCD refers to mice homozygous for the human α-globin gene (Hba hα/hα), and heterozygous for the hγβS mutant allele and the wild-type murine β-type globin locus (genotype: Hbb hγβS/+). The SCD mice exhibited moderately severe anemia (Hct 23%), accurately reproducing the disease phenotype of human patients, as originally reported (17). In comparison with the SCD mice, the SCD:TgTR2/TR4 mice exhibited significantly higher Hct and lower MCV values, both of which were closer to wild-type or heterozygous SCD levels, indicating alleviation of the hematological deficiencies observed in SCD mice. WBC and platelet counts of SCD:TgTR2/TR4 mice were not significantly different from SCD mice, suggesting that the effects of forced TR2/TR4 expression under Gata1 transcriptional control (18) was limited to the erythroid lineage, as anticipated.
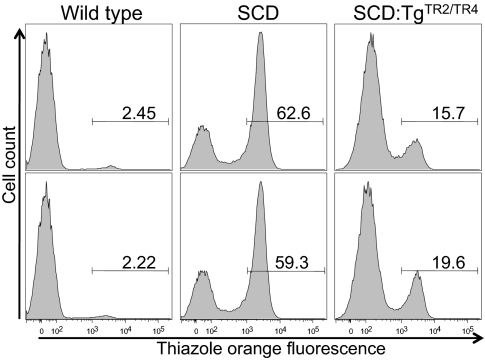
In hemolytic anemia, reticulocyte count represents the rate of red blood cell production and reflects the extent of prior red cell destruction, and therefore SCD is usually associated with increased reticulocyte counts (reticulocytosis). Reticulocyte counts in wild-type, SCD, and SCD:TgTR2/TR4 mice were measured by flow cytometry (Fig. 4) using thiazole orange for reticulocyte staining of whole blood. Wild-type mice exhibited the lowest reticulocyte count (2.3% in average), as expected. High reticulocyte counts in SCD mice (61%) were dramatically reduced in SCD:TgTR2/TR4 mice to 18% (P < 0.01 by Student t test). Because reticulocytosis is a robust indicator of increased blood cell turnover, these results support the hypothesis that forcibly expressed TR2 and TR4 reduce hemolysis and thereby alleviate hematological deficiencies in the SCD mice. We also examined differentiation profiles of erythroid precursor cells in the bone marrow and spleen of wild-type, SCD, and SCD:TgTR2/TR4 mice by flow cytometric analysis of cell surface expression of Ter119 and the transferrin receptor (CD71) (19) (Fig. S1). The data showed a significant increase of early erythroid progenitors (Ter119+CD71+) accompanied by a decrease of late erythroblasts (Ter119+CD71−) in SCD mice compared with wild-type mice in both the spleen and bone marrow, probably reflecting a compensatory erythropoietic response to hemolysis. In SCD:TgTR2/TR4 mice, compared with SCD mice, early erythroid progenitors were decreased, whereas late erythroblasts were increased, consistent with the notion that forced expression of TR2 and TR4 alleviated hematological deficiencies of SCD mice by reduction of hemolysis.
Fig. 4.
Reticulocytes in SCD and SCD:TgTR2/TR4 mice. The relative abundance of reticulocytes was measured by flow cytometry after thiazole orange staining of the whole blood of wild-type C57BL/6 (Left), SCD (Center), or SCD:TgTR2/TR4 (Right) mice. The number shown above the horizontal bar in each box represents the fractional percentage of reticulocytes in total red blood cells in each sample.
Pathophysiological and Histological Characteristics of SCD and SCD:TgTR2/TR4 Mice.
Weights of the livers, spleens, and kidneys of wild-type, heterozygous SCD, SCD, and SCD:TgTR2/TR4 mice were determined and expressed as a percentage of body weight (Table 2). These data revealed a decrease of hematopoietic organ weight in SCD:TgTR2/TR4 mice in comparison with SCD controls. The SCD mice exhibited enlargement of the spleen, liver, and kidneys, which are 13-, 1.8-, and 1.4-fold heavier, respectively, on average than those of wild-type mice when normalized to body weight, recapitulating the pathophysiology of human SCD patients (20). In comparison with SCD mice, SCD:TgTR2/TR4 mice had significantly smaller spleens and livers, weighing 23% and 14% less (P < 0.05), respectively, than those of the SCD mice, indicating amelioration of pathophysiological conditions of SCD.
Table 2.
Organ weight of SCD and SCD:TgTR2/TR4 mice
| Genotype |
||||||||
| Mouse | Hba | Hbb | TgTR2/TR4 | No. | Age (mo) | Spleen (% body weight) | Liver (% body weight) | Kidneys (% body weight) |
| Wild-type (C57BL/6) | +/+ | +/+ | – | 6 | 2.5 | 0.36 ± 0.16 | 5.0 ± 0.79 | 1.6 ± 0.23 |
| Heterozygous SCD | hα/hα | hγβS/+ | – | 10 | 2–3 | 1.2 ± 0.44 | 7.0 ± 1.31 | 2.1 ± 0.82 |
| SCD | hα/hα | hγβS/hγβS | – | 8 | 2–3 | 4.8 ± 1.34 | 8.8 ± 1.18 | 2.2 ± 0.33 |
| SCD:TgTR2/TR4 | hα/hα | hγβS/hγβS | + | 6 | 2–3 | 3.7 ± 0.46 | 7.6 ± 0.69 | 2.2 ± 0.35 |
| P value | <0.05 | <0.05 | 0.42 | |||||
Numbers following ± in the Spleen, Liver, and Kidney columns refer to SDs among the stated numbers of mice. P values refer to the comparison between the two final listed genotypes (SCD vs. SCD:TgTR2/TR4) by Student t test.
Finally, histological examination of the liver, spleen, and kidneys of wild-type, SCD, and SCD:TgTR2/TR4 mice was performed. Significant degrees of parenchymal necrosis and inflammation were observed in the liver of SCD mice, as reported (17) (Fig. S2), and those lesions were reduced in SCD:TgTR2/TR4 mice. The SCD mice exhibited 13.0 necrotic foci per 10 high-power fields (HPF; average of six mice), whereas none of four wild-type mice exhibited liver necrosis. In comparison with the SCD mice, the number of necrotic foci in the SCD:TgTR2/TR4 mice was reduced to 5.6 foci/10 HPF (average of five mice; P < 0.05, Mann–Whitney U test). As yet another indicator for alleviation of the SCD-induced pathophysiology, the SCD mice exhibited 5.3 inflammation foci/10 HPF, whereas wild-type mice showed 3.0 foci/10 HPF. Compared with the SCD mice, the number of inflammation foci in the SCD:TgTR2/TR4 mice was reduced to 1.6 foci/HPF (P < 0.02, Mann–Whitney U test). In the kidneys of SCD mice, medullary congestion was observed in five of six animals examined, as previously reported (17), whereas none of five SCD:TgTR2/TR4 mice or any wild-type mice exhibited congestion (P < 0.01, by Mann–Whitney U test). As previously reported (17), the spleen of the SCD mice exhibited congestion and extensive effacement of the normal splenic architecture (predominantly loss of the red pulp), which were similarly observed in the SCD:TgTR2/TR4 mice.
In conclusion, the accompanying data show, in one of the closest sickle cell animal models that is currently available, that forced transgenic TR2 and TR4 expression leads to elevated γ-globin mRNA accumulation, HbF synthesis, and alleviation of many hematological and pathological indications of SCD without causing any apparent adverse effects. This article demonstrates that genetic manipulation of nonglobin proteins or transcription factors can significantly ameliorate hematological defects in an SCD model mouse. This proof-of-concept study provides firm experimental evidence to support the hypothesis that therapeutic manipulation of TR2 and/or TR4 activities in SCD patients could lead to persistently elevated HbF synthesis and thus significantly lessen both the underlying cause and the secondary pathologies ascribed to the disease without adverse effects.
Materials and Methods
Transgenic Mice.
All the animal experiments were approved by the University Committee on Use and Care of Animals at the University of Michigan. The transgenic mouse line expressing murine TR2 and TR4 transgenes under the control of the Gata1 gene promoter and enhancers (TgTR2/TR, line 2) was previously characterized (13). The humanized sickle cell model mouse was also characterized previously (17). Genotyping was performed by PCR on tail biopsies. Primer sequences used to detect the murine wild-type adult α-globin gene, the murine adult βmajor-globin gene, the human α-globin gene, the human Aγ-globin gene, the TR2 transgene, and the TR4 transgene are shown in Table S1. Genotyping for the globin genes was independently confirmed by HPLC analysis (Hercules) of whole-blood lysates for the presence of mouse, human, and hybrid hemoglobin tetramers.
Real-Time RT-qPCR Analysis for Quantifying TR2, TR4, and Human Globin mRNAs.
Total RNA was extracted from the spleen or bone marrow and then subjected to first-strand cDNA synthesis with SuperScript II using 1 μg total RNA in a 20-μL reaction. Real-time PCR analysis was performed with 0.1 μL cDNA in a 25-μL reaction using SYBR Green PCR Master Mix (Applied Biosystems) on the ABI Prism 7000 Sequence Detection System. Sequences of the primers for endogenous and transgenic TR2 and TR4 cDNAs were previously described (13). Primer sequences used to quantitatively amplify GAPDH, 18S rRNA, and human α-, γ-, and β-globin cDNAs are shown in Table S1. All of the primer sets, except for the 18S rRNA amplicon, were designed to span introns. The abundance of each cDNA was determined based on cycle threshold values and the experimentally determined amplification efficiency for each primer set, and then normalized to the abundance of GAPDH, 18S, or α-globin cDNAs as the internal control.
Western Blotting.
Splenocytes were lysed in 1× Laemmli sample buffer and subjected to SDS/PAGE. Proteins in the gel were then transferred to a nitrocellulose membrane (Li-Cor) and probed with rabbit polyclonal antibodies against TR2 and TR4 (13) and fluorescence-conjugated secondary antibodies (Li-Cor). Proteins were visualized and quantified on the Odyssey Infrared Imaging System (Li-Cor).
Hemoglobin Analysis by HPLC.
Blood was collected by retro-orbital bleeding after anesthesia with ketamine. About 20 μL of whole blood was used for analysis on a Bio-Rad Variant II Hemoglobin Testing System using an ion-exchange HPLC column (Hercules). Separated hemoglobin fractions were detected by absorbance at 415 nm. The testing system was calibrated each day of use based on the retention time of human HbA2 (α2δ2) and the known percentages of HbA2 and HbF of a BioRad HbA2/F Calibrator consisting of pooled human red blood cells. Each calibration was checked for acceptability with two levels of BioRad Lyphochek HbA2 control material containing normal or elevated levels of HbA2, HbF, and HbS. EDTA-treated mouse whole blood (20 μL) was added to 1 mL of BioRad Variant II Wash/Diluent solution, vortexed for 30 s, and then rocked for 10 min to hemolyze red blood cells. The hemolysate was centrifuged at 13,000 × g for 5 min to remove debris, diluted further with the wash/diluent solution until the final concentration was close to that of the HbA2/F calibrator, and then subjected to HPLC analysis.
ChIP assay.
ChIP assay was performed essentially as described previously (16). A single-cell suspension of spleen cells was treated with 2 mM ethylene glycol bis(succinimidyl succinate) (EGS; Pierce) before fixation with 1% formaldehyde (Polysciences). Rabbit polyclonal antibodies against TR2 and TR4 (13) were used for immunoprecipitation. Primers for real-time qPCR to quantify human genomic DNA sequences in the fetal γ- and adult β-globin gene proximal promoters as well as in the third intron of the CHMP4B gene are shown in Table S1.
Complete Blood Cell Count and Reticulocyte Counting.
EDTA-treated whole blood (20 μL) was diluted 10-fold with 180 μL of 5% BSA in PBS and subjected to analysis with an Advia120 Multispecies Hematology Analyzer (Bayer Diagnostics). Reticulocyte counting was performed by flow cytometry as previously reported (21). Briefly, 5 μL of EDTA-treated whole blood was added to 0.5 mL PBS containing 0.2 μg/mL thiazole orange (Sigma-Aldrich) or PBS only for an unstained control. After 90 min of incubation at room temperature, samples were analyzed using a FACSCanto II Flow Cytometer (BD Biosciences).
Flow Cytometric Analysis of Bone Marrow and Spleen Cells.
Bone marrow or spleen cells were stained with phycoerythrin-conjugated anti-mouse CD71 (eBioscience) and allophycocyanin-conjugated anti-mouse Ter119 (eBioscience) on ice for 20–30 min in the dark. Cells were washed twice and resuspended in PBS with 0.5% BSA. Analytical FACS was performed using a FACSCanto II instrument (BD Biosciences).
Pathological Analysis.
The spleen, liver, and kidneys were removed immediately postmortem and then weighed. Tissue preparations from the organs were fixed in 70% alcoholic formalin, embedded in paraffin, sectioned, and stained with H&E. The pathology scoring of the livers, spleens, and kidneys was based on several histopathologic findings by light microscopy. The liver was scored based on necrosis, parenchymal inflammation, hemosiderin phagocytosis, extramedullary hematopoiesis, dilated veins (portal and central), and cytomegaly. The spleen was scored based on congestion, architectural distortion, hemosiderin phagocytosis, and extramedullary hematopoiesis. The kidney was scored based on medullary congestion, cortical cysts, inflammation, and increased glomerular cellularity. Degrees of inflammation and necrosis were determined based on numbers of foci per 10 HPF by light microscopy.
Supplementary Material
Acknowledgments
We thank David Ginsburg for the use of his Advia120 Multispecies Hematology Analyzer; Kelly Labarge (Helena Laboratories, Beaumont, TX); Tamika Cross, David Shepard, Monica Yalamanchilli, Carmen Yu, and Michael Sierant for their assistance with aspects of this work; and our laboratory colleagues for the many insightful comments and helpful suggestions on this work and its presentation. Support for this work was provided by the American Heart Association (L. Shi), a Cooley's Anemia Foundation postdoctoral fellowship (to K.T. and O.T.), and National Institutes of Health Grants DK086956 (to O.T. and J.D.E.) and HL24415 (to J.D.E.). Career development support was provided by National Heart, Lung and Blood Institute/National Institutes of Health Grant K01 HL03011 and The Robert Wood Johnson Foundation Grant RWJ-AMFDP 051891 (to A.D.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104964108/-/DCSupplemental.
References
- 1.Weatherall DJ. In: The Molecular Basis of Blood Diseases. Stamatoyannopoulos G, Majerus PW, Perlmutter RM, Varmus H, editors. Philadelphia: Saunders; 2001. pp. 183–226. [Google Scholar]
- 2.Bunn HF. In: The Molecular Basis of Blood Diseases. Stamatoyannopoulos G, Majerus PW, Perlmutter RM, Varmus H, editors. Philadelphia: Saunders; 2001. pp. 227–273. [Google Scholar]
- 3.Ingram VM. Gene mutations in human haemoglobin: The chemical difference between normal and sickle cell haemoglobin. Nature. 1957;180:326–328. doi: 10.1038/180326a0. [DOI] [PubMed] [Google Scholar]
- 4.Powars DR, Weiss JN, Chan LS, Schroeder WA. Is there a threshold level of fetal hemoglobin that ameliorates morbidity in sickle cell anemia? Blood. 1984;63:921–926. [PubMed] [Google Scholar]
- 5.Miller BA, et al. Molecular analysis of the high-hemoglobin-F phenotype in Saudi Arabian sickle cell anemia. N Engl J Med. 1987;316:244–250. doi: 10.1056/NEJM198701293160504. [DOI] [PubMed] [Google Scholar]
- 6.Platt OS, et al. Mortality in sickle cell disease. Life expectancy and risk factors for early death. N Engl J Med. 1994;330:1639–1644. doi: 10.1056/NEJM199406093302303. [DOI] [PubMed] [Google Scholar]
- 7.Sunshine HR, Hofrichter J, Eaton WA. Requirement for therapeutic inhibition of sickle haemoglobin gelation. Nature. 1978;275:238–240. doi: 10.1038/275238a0. [DOI] [PubMed] [Google Scholar]
- 8.Stamatoyannopoulos G, Grosveld F. In: The Molecular Basis of Blood Diseases. Stamatoyannopoulos G, Majerus PW, Perlmutter RM, Varmus H, editors. Philadelphia: Saunders; 2001. pp. 135–182. [Google Scholar]
- 9.Tanimoto K, Liu Q, Grosveld F, Bungert J, Engel JD. Context-dependent EKLF responsiveness defines the developmental specificity of the human ε-globin gene in erythroid cells of YAC transgenic mice. Genes Dev. 2000;14:2778–2794. doi: 10.1101/gad.822500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Omori A, Tanabe O, Engel JD, Fukamizu A, Tanimoto K. Adult stage γ-globin silencing is mediated by a promoter direct repeat element. Mol Cell Biol. 2005;25:3443–3451. doi: 10.1128/MCB.25.9.3443-3451.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huisman THJ, Carver MFH, Baysal E. A Syllabus of Thalassemia Mutations. Augusta, GA: Sickle Cell Anemia Foundation; 1997. [Google Scholar]
- 12.Tanabe O, et al. An embryonic/fetal β-type globin gene repressor contains a nuclear receptor TR2/TR4 heterodimer. EMBO J. 2002;21:3434–3442. doi: 10.1093/emboj/cdf340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanabe O, et al. Embryonic and fetal β-globin gene repression by the orphan nuclear receptors, TR2 and TR4. EMBO J. 2007;26:2295–2306. doi: 10.1038/sj.emboj.7601676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou XE, et al. The orphan nuclear receptor TR4 is a vitamin A-activated nuclear receptor. J Biol Chem. 2011;286:2877–2885. doi: 10.1074/jbc.M110.168740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gill G, Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- 16.Cui S, et al. Nuclear receptors TR2 and TR4 recruit multiple epigenetic transcriptional corepressors that associate specifically with the embryonic β-type globin promoters in differentiated adult erythroid cells. Mol Cell Biol. 2011;31:3298–3311. doi: 10.1128/MCB.05310-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu LC, et al. Correction of sickle cell disease by homologous recombination in embryonic stem cells. Blood. 2006;108:1183–1188. doi: 10.1182/blood-2006-02-004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Onodera K, et al. GATA-1 transcription is controlled by distinct regulatory mechanisms during primitive and definitive erythropoiesis. Proc Natl Acad Sci USA. 1997;94:4487–4492. doi: 10.1073/pnas.94.9.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Socolovsky M, et al. Ineffective erythropoiesis in Stat5a−/−5b−/− mice due to decreased survival of early erythroblasts. Blood. 2001;98:3261–3273. doi: 10.1182/blood.v98.12.3261. [DOI] [PubMed] [Google Scholar]
- 20.Serjeant GR, Serjeant BE. Sickle Cell Disease. 3rd Ed. Oxford: Oxford Univ Press; 2001. [Google Scholar]
- 21.Nobes PR, Carter AB. Reticulocyte counting using flow cytometry. J Clin Pathol. 1990;43:675–678. doi: 10.1136/jcp.43.8.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.