Abstract
In cell cultures, HSV-1 replication is initiated by recruitment by virion protein 16 of transcriptional factors and histone-modifying enzymes to immediate early (α) gene promoters. HSV establishes latent infections characterized by suppression of viral gene expression except for latency-associated transcripts (LATs) and miRNAs. The latent virus reactivates in stressed neurons. A fundamental question is how reactivation initiates in the absence of virion protein 16. We report the following findings in the ganglion explant model. (i) Anti-nerve growth factor antibody accelerated the reactivation of latent virus. Viral mRNAs were detected as early as 9 h after explantation. (ii) After explantation the amounts of viral mRNAs increased whereas amounts of miRNAs and LATs decreased. The decrease in miRNAs and LATs required ongoing protein synthesis, raising the possibility that LAT and miRNAs were degraded by a viral gene product. (iii) The expression of viral genes in explanted ganglia was disordered rather than sequentially ordered as in infected cells in culture. These findings suggest that in reactivating ganglia gene expression is totally derepressed and challenge the current models in that establishment of or exit from latency could not be dependent on the suppression or activation of single or small clusters of viral genes. Finally, miRNAs and LATs reached peak levels 9–11 d after corneal inoculation, thus approximating the pattern of virus replication in these ganglia. These findings suggest that the patterns of accumulation of LATs and miRNAs reflect many different stages in the infection of neurons.
To initiate infection in cell culture and presumably also at the portal of entry into the body, the HSV-1 capsid delivers the DNA to the nucleus. Concurrently, virion protein 16 (VP16), a key viral protein packaged in the virion and delivered to the nucleus, recruits the cellular factors octamer-binding protein 1 (Oct1), host cell factor 1 (HCF1), lysine-specific demethylase 1 (LSD1), circadian locomotor output cycles kaput (CLOCK) histone acetyl transferase, and other transcriptional factors to initiate a cascade of viral gene expression that begins with immediate early (α) genes followed by early (β) and late (γ) genes (1–5). VP16 is encoded by a γ gene expressed late in infection (1). In all, the transcriptional program yields almost 100 different transcripts. In contrast, in latently infected neurons viral gene expression is limited to the latency-associated transcript (LAT) and approximately six or more miRNAs (6, 7). Given the requirement for VP16 to initiate viral replication in productively infected cells, the question arises as to how virus replication initiates in stressed neurons harboring latent virus. Among the many hypotheses, two stand out: That VP16 is expressed first or that in neurons α gene expression can ensue in the absence of VP16.
Resolution of this problem requires analyses of the reactivation in ganglia under conditions that are physiologically similar to those that occur in vivo. In the studies reported here we modified the ganglion explantation model used for many years to study reactivation. In brief, latent virus reactivates and is readily demonstrable in trigeminal ganglia excised from mice and explanted in appropriate medium at least 28 d after virus inoculation by the corneal route. The relevance of this protocol for the study of reactivation dates back to the pioneering studies by Cushing (8) and others (9–12) showing that in patients suffering from trigeminal neuralgia the severance of the connection between the ganglion and the brain results in virus reactivation and atrophy of the ganglion. Extraction of ganglia failed to yield infectious virus (9). In effect, excised ganglia bathed in nutrient medium resemble in substance the severed human ganglion left to atrophy and reactivate virus. The modification we introduced in this model is based on the observation reported by Wilcox et al. (13, 14) and Camarena et al. (15) that nerve growth factor (NGF) blocks virus replication in explanted neurons in vitro. Because a key requirement of our model is to accelerate reactivation and compress the interval of maintenance of explanted ganglia, we tested the effects of both NGF and anti-NGF antibody on viral gene expression. We report that NGF retarded whereas anti-NGF antibody accelerated the accumulation of viral mRNAs.
The key finding reported here is that the expression of viral genes following excision leading to a decrease in the amounts of LATs and viral miRNAs requires protein synthesis and concurs with the expression of genes encoding viral proteins and that the transcription of infected cell protein 0 (ICP0), a key α regulatory gene, thymidine kinase (TK), a β gene, and VP16 and UL41 [virion host shutoff protein (VHS)], both γ genes, takes place concurrently and in the absence of prior protein synthesis.
Results
Incubation of Excised Ganglia in Medium Containing Anti-NGF Antibody Accelerates the Accumulation of Viral mRNA.
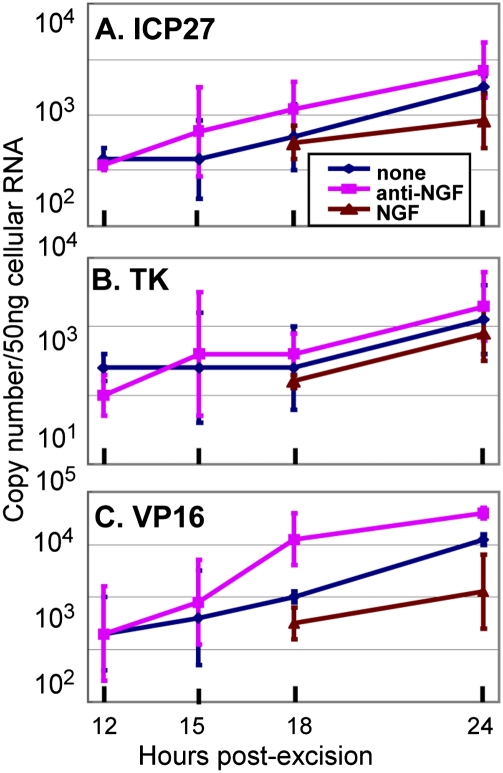
In this series of experiments trigeminal ganglia removed from mice maintained for 30 d after corneal inoculation were incubated in medium 199V alone (blue line in Fig. 1) or supplemented with NGF (brown line in Fig. 1) or anti-NGF antibody (purple line in Fig. 1). At 12, 15, 18, or 24 h after excision the ganglia from three mice were extracted individually, and the amounts of mRNAs encoding infected cell protein 27 (ICP27), TK, and VP16 were quantified as described in Materials and Methods. The results (Fig. 1) were as follows: mRNAs encoding ICP27 and VP16 were readily detected and quantifiable by 12 h after excision and incubation of the ganglia. The TK mRNAs were detected at 18 h after explantation of the ganglia. As expected, the detected mRNA levels were lowest in medium containing NGF. The mRNA levels were next highest in unsupplemented medium and were at least two- to threefold higher in medium containing anti-NGF antibody. To enable unambiguous quantification of viral transcripts at the earliest time after excision, the excised ganglia were incubated in medium containing anti-NGF antibody in all experiments described below.
Fig. 1.
NGF accelerates the accumulation of viral mRNAs from explanted trigeminal ganglia. Trigeminal ganglia excised from mice 30 d after corneal inoculation were incubated in 199V medium alone or in 199V medium supplemented with NGF or anti-NGF antibody. The ganglia were processed individually at times shown, as described in Materials and Methods. The results shown are the geometric mean amounts of viral mRNAs encoding ICP27 (A), TK (B), and VP16 (C) per groups of six ganglia selected at random.
Concurrent Accumulation of Viral mRNAs and LAT or Viral microRNAs Appears to Be Incompatible.
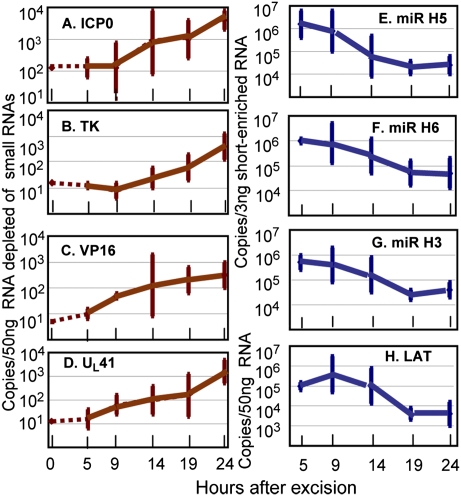
In these experiments ganglia of mice maintained for 30 d after corneal inoculation were incubated in medium containing anti-NGF antibody. At the time of the excision (time 0) and at 5, 9, 14, 19, or 24 h after excision the ganglia from three mice were extracted, and the amounts of viral mRNAs encoding ICP0, TK, VP16, UL41, LAT, microRNA (miR)-H3, miR-H5, or miR-H6 were quantified as described in Materials and Methods. The results (Fig. 2) were as follows: All viral mRNAs quantified in this study increased in amount between 5 and 9 h after excision. In the time span between 9 and 24 h the amounts of viral mRNAs increased 30- to 100-fold. We estimate that the limits of detection of viral mRNAs were ∼100 copies per 50 ng of RNA depleted of small mRNAs. In the same time span, between 5 and 9 h after excision, LAT, miR-H3, miR-H5, and miR-H6 decreased 30- to 100-fold in amount. The results suggest that the accumulation of mRNAs encoding vial proteins is incompatible with the accumulation of LAT and the three selected miRNAs.
Fig. 2.
In explanted ganglia the accumulation of viral mRNAs takes place concurrently with a decrease in the accumulation of LATs and miRNAs. Ganglia were excised from mice 30 d after corneal inoculation and incubated in 199V medium containing anti-NGF antibody. At times shown individual ganglia were processed as described in Materials and Methods. The figure shows the geometric mean amounts of viral mRNAs (A–D) and miRNAs and LAT (E–H) per group of six ganglia.
Decrease in the Accumulation of LAT and of miRNAs Is Blocked by Inhibitors of RNA or Protein Synthesis.
The objective of this series of experiments was to determine whether the decrease in the amounts of LAT or miRNAs tested in these studies was related to the synthesis of proteins initiated after explantation of the ganglia. We report two series of experiments.
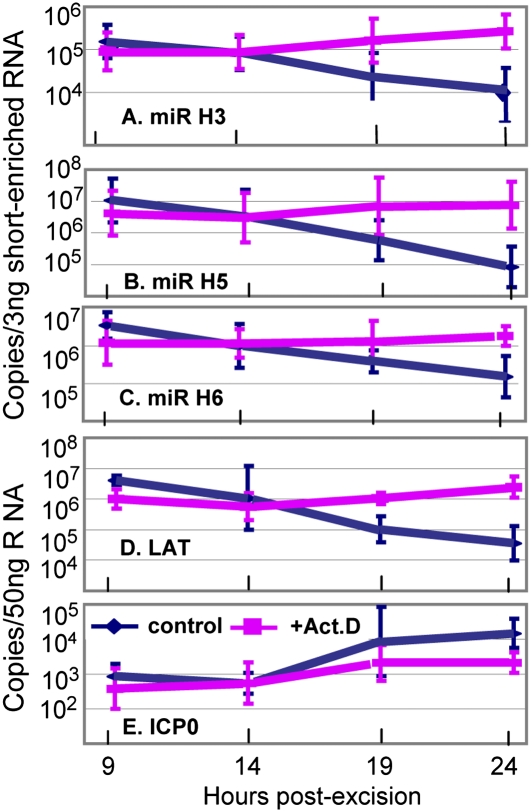
In the first series, ganglia excised from mice maintained for 30 d after corneal inoculation were incubated in medium containing only anti-NGF antibody or supplemented with 10 μg/mL of actinomycin D. At 9, 14, 19, and 24 h after excision, sets of six randomly selected ganglia, either untreated or treated with actinomycin D, were processed individually as described in Materials and Methods, and the miRNA were isolated and quantified. The results (Fig. 3) were as follows: In the presence of actinomycin D the amounts of miRNA detected in these studies were unchanged [e.g., miR-H5 (Fig. 3B) and miR-H6 (Fig. 3C)] or increased slightly [e.g., miR-H3 (Fig. 3A) or LAT (Fig. 3D)]. In contrast, with the exception of LAT (Fig. 3D), all miRNAs decreased 20- to100-fold in amount in ganglia incubated in medium without actinomycin D.
Fig. 3.
Actinomycin D blocks the decrease in miRNAs and LATs in ganglia of mice explanted 30 d after corneal inoculation. The ganglia were incubated in 199V medium containing anti-NGF antibody alone or supplemented with 10 μg/mL of actinomycin D. At times shown individual ganglia were processed as described in Materials and Methods. The results shown are the geometric mean amounts of miRNAs (A–C), LAT (D), and ICP0 (E) per group of six ganglia selected at random.
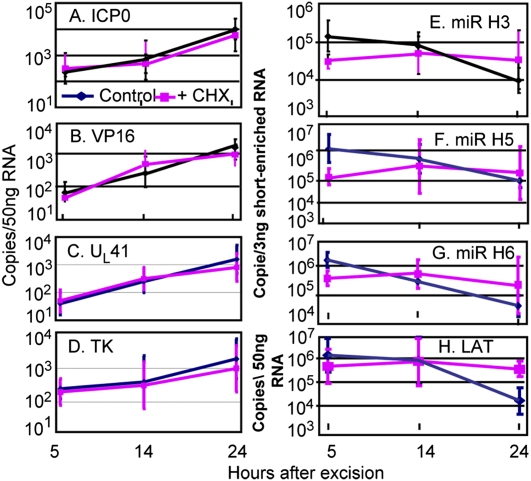
In the second series of experiments, ganglia from mice maintained for 30 d after corneal inoculation were excised and incubated in medium containing only anti-NGF antibody or supplemented with 100 μg of cycloheximide per mL of medium. At 5, 14, or 24 h after excision, sets of four randomly selected ganglia either untreated or treated with cycloheximide were processed individually as described in Materials and Methods. In these studies we quantified mRNAs encoding ICP0, VP16, UL41, TK, miR-H3, miR-H5, miR-H6, and LAT. The results (Fig. 4) were as follows: The mRNAs encoding ICP0, VP16, UL41, or TK (Fig. 4 A–D) increased at approximately equal rates in the presence or absence of cycloheximide. The levels of miRNAs or of LAT (Fig. 4 E–H) remained unchanged in medium containing cycloheximide but decreased in medium lacking the protein synthesis inhibitor.
Fig. 4.
The effect of cycloheximide on the accumulation of mRNAs encoding ICP0 (A), VP16 (B), UL41 (C), and TK (D), miRNAs (E–G), and LATs (H). The explanted ganglia were incubated in 199V medium containing anti-NGF antibody alone or supplemented with 100 μg/mL of cycloheximide (CHX). At times shown individual ganglia were processed as described in Materials and Methods. The results shown are geometric means of four randomly selected ganglia processed individually.
We conclude the following: (i) Protein synthesis is not required after excision to activate the transcription of viral genes. (ii) The results do not show evidence that protein synthesis is required for the transcription of ICP0, TK, VP16, or UL41 genes. The mRNAs of all of the genes tested increased in amount in the presence of cycloheximide. (iii) The decrease in the accumulation of miRNAs and of LAT requires both transcription and protein synthesis.
Pattern of Accumulation of LAT and HSV miRNAs in Trigeminal Ganglia After Corneal Inoculation.
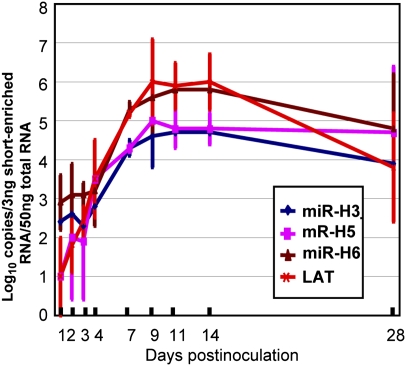
The purpose of this series of experiments was to measure the accumulation of LAT and miRNAs in trigeminal ganglia of mice inoculated by the corneal route with 105 pfu in each eye. On the days after corneal inoculation indicated in Fig. 5, trigeminal ganglia were excised and processed as described in Materials and Methods. Fig. 5 shows amounts of miR-H3, miR-H5, and miR-H6 per 3 ng RNA enriched by small RNAs and the copy number of LAT per 50 ng of cellular RNA. The results were as follows: The copy numbers of both miRNAs and LAT increase with time and reach peak levels 9–14 d after inoculation. The amounts detected at 28 d after infection were ∼10-fold lower than those quantified on days 9–14.
Fig. 5.
Accumulation of LAT and miRNAs in ganglia removed from mice inoculated by the corneal route with 105 pfu of HSV-1(F) virus in each eye. The ganglia were harvested on days shown and processed as described in Materials and Methods. The results shown are geometric mean per groups of six ganglia.
Discussion
The objective of the studies reported here was to determine the pattern of expression of selected viral genes and the fate of miRNAs and LAT on reactivation of virus from latent state. We used the ganglion explantation model. To accelerate the reactivation of virus, we included anti-NGF antibody in the medium bathing the explanted murine trigeminal ganglia. The key findings reported here are as follows: (i) Inclusion of anti-NGF antibody accelerated the reactivation of virus from explanted ganglia. We could detect the appearance of viral mRNAs as early as 9 h after explantation. (ii) Viral mRNAs increased in amount after explantation, but the amounts of miRNAs and LATs decreased. The accumulation of viral mRNAs and persistence of miRNAs and of LATs appeared to be incompatible. The decrease in the amounts of miRNAs and of LATs could not be attributed to a short half-life because they were stable in ganglia maintained in medium containing actinomycin D or cycloheximide. The observed results are consistent with the hypothesis that miRNAs and LATs were degraded by a viral gene product. (iii) A key finding was that accumulation of ICP0, VP16, UL41, or TK mRNAs does not require de novo protein synthesis. The results also indicate that in excised ganglia the expression of viral genes is disordered; that is, it does not resemble the sequentially ordered expression observed in cultured cells. Finally, we report that miRNAs and LATs reached peak levels 9–11 d after corneal inoculation, thus approximating the pattern of virus replication in these ganglia.
Relevant to this report are the following:
i) A key finding of this study is that accumulation of viral transcripts and maintenance of miRNAs and of LATs are mutually exclusive. Evidence that expression of LATs and expression of ICP0 gene may be incompatible was reported by Amelio et al. (16) and Maillet et al. (17). The studies with actinomycin D and cycloheximide suggest that these two events are linked. The prime viral gene product that could be responsible for the decrease in LATs or miRNAs is the viral endoribonuclease encoded by the UL41 gene. In cell culture, modified forms of LATs accumulate late in infection (18), after the endoribonuclease is neutralized by two viral proteins, VP16 and virion protein 22 (19). Whether the degradation of LATs or miRNAs is caused by endoribonuclease encoded by the UL41 gene remains to be determined.
ii) Studies designed to determine which viral genes are key to initiation of reactivation produced discordant results. Studies carried on in infected cells in culture indicated that the expression of viral genes is ordered sequentially in a cascade fashion: α genes exemplified in these studies by ICP0 are expressed first, followed by expression of β (e.g., TK) and ultimately γ1 (e.g., VP16), and γ2 (e.g., UL41) genes (20). The expression of α genes primarily requires the recruitment of transcriptional factors and histone-modifying enzymes, whereas the expression of β and γ genes requires prior synthesis of viral α proteins and, in the case of γ2 genes (e.g., UL41), the initiation of synthesis of viral DNA. In this report, we show that mRNAs of ICP0, TK, VP16, and UL41 accumulate in the presence of cycloheximide added to the medium at the time of explantation. More recent studies indicate that viral genes are derepressed sequentially in a cascade fashion (21). The studies presented here suggest that in the explanted ganglia viral gene expressions is disordered—potentially as the consequence of total derepression of the stressed, excised ganglion—rather than sequentially ordered. A fundamental conclusion of this report is that synthesis of viral proteins and assembly of virions in ganglia subjected to a level of stress sufficiently high to reactivate latent virus would not require an initiator protein such as VP16.
The fundamental question raised by this report is whether disordered gene expression is characteristic of neurons deprived of regulatory NGF harboring latent virus, a general characteristic of neurons triggered to reactivate virus, or a general characteristic of neurons infected with HSV-1. Reports of aberrant HSV gene expression in neurons based on studies in vivo and in vitro abound (22–25). If this aberrant gene expression is a general characteristic of neurons, the model in which retention of VP16 in the cytoplasm results in silencing of α genes, and hence the expression of all viral genes, becomes untenable. Disordered gene expression as a characteristic solely of neurons reactivating virus suggests that neurons triggered to reactivate virus undergo extensive changes in cell gene regulation, resulting in total derepression of neuronal genes. The significance of the questions arising from these studies cannot be overstated; resolving these questions will require extensive analyses of the infected neurons at both the entry and exit from latent state.
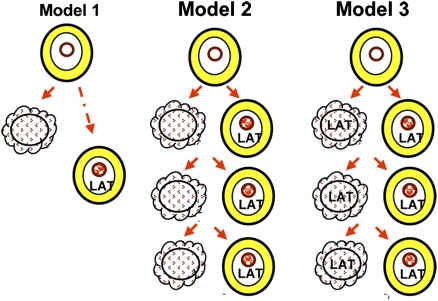
iii) The analyses of the accumulation of LAT and of miRNAs after corneal inoculation were undertaken with the expectation that, if LAT and miRNAs are incompatible with productive infection, the patterns of accumulation of LATs, miRNAs, and infectious virus would differ. In our studies reported elsewhere (26), infectious virus reached peak titers ∼7 d after corneal inoculation and then declined. The patterns of accumulation of virus, LATs, and miRNAs are similar, at least in part, between days 1 and 7. This phenomenon raises the question of when neurons acquire a latent infection. Model 1 (Fig. 6) predicts that on entry into the ganglion some neurons become infected and ultimately express LATs, whereas others succumb to productive infection. This model would be valid only if LATs were slower to accumulate in neurons on their way to establishing a latent infection than in neurons undergoing lytic infection, as has been suggested (27). The alternative model is that the virus spreads from neuron to neuron and in the course of this spread enters both permissive and nonpermissive cells. If the decision to establish latency were instantaneous after entry, and the synthesis of miRNAs and LATs followed, the data would suggest that in the vast majority of neurons LATs would be synthesized several days after the neurons committed themselves to maintain a latent infection (model 1). If the bulk of neurons committed themselves after serial virus propagation, 4–11 d after corneal infection rather than on day 1, the appropriate model would be model 2. Last, there is the additional question of whether LATs are present only in latently infected neurons or also are present in productively infected neurons. The studies presented here do not exclude the possibility that LATs and miRNAs also accumulate in productively infected neurons (model 3).
Fig. 6.
Working models of accumulation of LATs and miRNAs in ganglia after virus inoculation. In model 1 neurons destined to harbor latent virus are infected immediately on entry of virus into the ganglion. LATs may appear in latently infected neurons after some delay as described in ref. 27. Model 2 predicts that virus spreads in neurons; some cells become productively infected and are destroyed, whereas others are committed to harbor latent virus. Model 3 is similar to model 2 except that both productively infected and latently infected neurons exhibit LAT and miRNAs. Model 3 suggests that the majority of LAT-expressing cells are productively infected.
Materials and Methods
Virus Strain and Murine Model of Ocular Infection.
The properties of HSV-1(F), the prototype strain used in this laboratory were reported elsewhere (28). Four-week-old inbred female CBA/J mice (Jackson Laboratories) receiving unrestricted access to food and water were infected by the corneal route as reported previously (29). All animal studies were performed according to protocols approved by the University Animal Care Committee.
Virus Reactivation from Latently Infected Trigeminal Ganglia.
Trigeminal ganglia were removed 30 d after infection and were incubated at 37 °C, plus 5% CO2 in 199V medium consisting of mixture 199 supplemented with 1% serum (medium 199V) alone or with NGF (2.5S, 1 μg/mL; Invitrogen) or rabbit Ab against NGF (10 μg/mL; Abcam). In some experiments the medium included cycloheximide (100 μg/mL) or actinomycin D (10 μg/mL).
RNA Isolation and Assays.
RNAs depleted and enriched by small RNAs (<200 nt) were extracted from trigeminal ganglia with mirVana miRNA isolation kit (Ambion) according to the manufacturer's instructions. After DNase I treatment, RNA depleted of small RNAs was reverse transcribed with Oligo d(T) by SuperScript III RT kit (Invitrogen) according to the manufacturer's instructions. LAT was transcribed with LAT-specific primer (5′CGCTGTGACCGCTGCGGCTAC3′). Quantitative real-time PCR analysis for mRNA expression was performed by custom TaqMan assays. Viral genes and the neuronal-specific expression gene MAP2 as reference gene were quantified with primers and probes sequences as described in Table 1. HSV-1(F) miRNAs (miR-H3, miR-H5, miR-H6) were reverse transcribed and quantified (6). The assays were performed using a StepOnePlus system (Applied Biosystems) and were analyzed with software provided by the supplier. Viral genes and LATs copies were normalized to 50 ng RNAs depleted of small RNAs, and miRNAs were normalized to 3 ng RNAs enriched by small RNAs which were measured by let-7a.
Table 1.
Sequences of primers and probes used in Taqman assays
| Gene | Forward primer | Reverse primer | Probe |
| ICP0 | 5′GGCCCCCTTGTCAACAGA3′ | 5′GGGAGTCGCTGATCACTATGG3′ | 5′CCCCTTGCAAACAACA3′ |
| ICP27 | 5′TCATGCACGACCCCTTTGG3′ | 5′CTTGGCCCGCCAACAC3′ | 5′TTCCCGCCGCGAATAG3′ |
| TK | 5′AAGGTCGGCGGGATGAG3′ | 5′CGGCCGCGCGATAC3′ | 5′CTTATGGGCAGCATGACCCC3′ |
| UL41 | 5′GGACATCCGCGACGAAAAC3′ | 5′AGAAACCTGTCGGCGATATCAG3′ | 5′CTGGCGCGATCTATC3′ |
| VP16 | 5′CCGGGTCCGGGATTTACC3′ | 5′CTCGAAGTCGGCCATATCCA3′ | 5′CCCCACGACTCCGCC3′ |
| MAP2 | 5′GCAAGAGCCCAGAGAAACGT3′ | 5′GATACACCCCTGCGAGGAG3′ | 5′CCTCCCAAGACCTTCC3′ |
| LAT | 5′GCATAGAGAGCCAGGCACAAAA3′ | 5′ACGTACTCCAAGAAGGCATGTG3′ | 5′TCCCACCCCGCCTGTG3′ |
Acknowledgments
These studies were supported by National Cancer Institute Grant 5R37CA078766-12.
Footnotes
The authors declare no conflict of interest.
References
- 1.Roizman B, Knipe DM, Whitley RJ. The replication of Herpes simplex viruses. In: Knipe DM, et al., editors. Fields’ Virology. 5th Ed. New York: Lippincott-Williams and Wilkins; 2007. pp. 2501–2601. [Google Scholar]
- 2.Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat Med. 2009;15:1312–1317. doi: 10.1038/nm.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalamvoki M, Roizman B. The histone acetyltransferase CLOCK is an essential component of the herpes simplex virus 1 transcriptome that includes TFIID, ICP4, ICP27, and ICP22. J Virol. 2011;85:9472–9477. doi: 10.1128/JVI.00876-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stevens JG, Wagner EK, Devi-Rao GB, Cook ML, Feldman LT. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science. 1987;235:1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- 5.Deatly AM, Spivack JG, Lavi E, Fraser NW, Fraser NW. RNA from an immediate early region of the type 1 herpes simplex virus genome is present in the trigeminal ganglia of latently infected mice. Proc Natl Acad Sci USA. 1987;84:3204–3208. doi: 10.1073/pnas.84.10.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Umbach JL, et al. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Umbach JL, Nagel MA, Cohrs RJ, Gilden DH, Cullen BR. Analysis of human alphaherpesvirus microRNA expression in latently infected human trigeminal ganglia. J Virol. 2009;83:10677–10683. doi: 10.1128/JVI.01185-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cushing H. The surgical aspects of major neuralgia of the trigeminal nerve. JAMA. 1905;44 773–779, 860–865, 920–929, 1002–1008, 1088–1093. [Google Scholar]
- 9.Carton CA, Kilbourne ED. Activation of latent herpes simplex by trigeminal sensory-root section. N Engl J Med. 1952;246(5):172–176. doi: 10.1056/NEJM195201312460503. [DOI] [PubMed] [Google Scholar]
- 10.Carton CA. Effect of previous sensory loss on the appearance of herpes simplex following trigeminal sensory root section. J Neurosurg. 1953;10:463–468. doi: 10.3171/jns.1953.10.5.0463. [DOI] [PubMed] [Google Scholar]
- 11.Behrman S, Knight G. Herpes simplex associated with trigeminal neuralgia. Neurology. 1954;4:525–530. doi: 10.1212/wnl.4.7.525. [DOI] [PubMed] [Google Scholar]
- 12.Ellison SA, Carton CA, Rose HM. Studies of recurrent herpes simplex infections following section of the trigeminal nerve. J Infect Dis. 1959;105(2):161–167. doi: 10.1093/infdis/105.2.161. [DOI] [PubMed] [Google Scholar]
- 13.Wilcox CL, Johnson EM., Jr Nerve growth factor deprivation results in the reactivation of latent herpes simplex virus in vitro. J Virol. 1987;61:2311–2315. doi: 10.1128/jvi.61.7.2311-2315.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilcox CL, Smith RL, Freed CR, Johnson EM., Jr Nerve growth factor-dependence of herpes simplex virus latency in peripheral sympathetic and sensory neurons in vitro. J Neurosci. 1990;10:1268–1275. doi: 10.1523/JNEUROSCI.10-04-01268.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camarena V, et al. Nature and duration of growth factor signaling through receptor tyrosine kinases regulates HSV-1 latency in neurons. Cell Host Microbe. 2010;8:320–330. doi: 10.1016/j.chom.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amelio AL, Giordani NV, Kubat NJ, O'neil JE, Bloom DC. Deacetylation of the herpes simplex virus type 1 latency-associated transcript (LAT) enhancer and a decrease in LAT abundance precede an increase in ICP0 transcriptional permissiveness at early times postexplant. J Virol. 2006;80:2063–2068. doi: 10.1128/JVI.80.4.2063-2068.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maillet S, et al. Herpes simplex virus type 1 latently infected neurons differentially express latency-associated and ICP0 transcripts. J Virol. 2006;80:9310–9321. doi: 10.1128/JVI.02615-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang W, et al. Characterization of a spliced exon product of herpes simplex type-1 latency-associated transcript in productively infected cells. Virology. 2006;356(1–2):106–114. doi: 10.1016/j.virol.2006.07.033. [DOI] [PubMed] [Google Scholar]
- 19.Taddeo B, Sciortino MT, Zhang W, Roizman B. Interaction of herpes simplex virus RNase with VP16 and VP22 is required for the accumulation of the protein but not for accumulation of mRNA. Proc Natl Acad Sci USA. 2007;104:12163–12168. doi: 10.1073/pnas.0705245104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honess RW, Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roizman B. The checkpoints of viral gene expression in productive and latent infection: The role of the HDAC/CoREST/LSD1/REST repressor complex. J Virol. 2011;85:7474–7482. doi: 10.1128/JVI.00180-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nichol PF, Chang JY, Johnson EM, Jr, Olivo PD. Herpes simplex virus gene expression in neurons: Viral DNA synthesis is a critical regulatory event in the branch point between the lytic and latent pathways. J Virol. 1996;70:5476–5486. doi: 10.1128/jvi.70.8.5476-5486.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tal-Singer R, et al. Gene expression during reactivation of herpes simplex virus type 1 from latency in the peripheral nervous system is different from that during lytic infection of tissue cultures. J Virol. 1997;71:5268–5276. doi: 10.1128/jvi.71.7.5268-5276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson RL, Shieh MT, Sawtell NM. Analysis of herpes simplex virus ICP0 promoter function in sensory neurons during acute infection, establishment of latency, and reactivation in vivo. J Virol. 2003;77:12319–12330. doi: 10.1128/JVI.77.22.12319-12330.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson RL, Preston CM, Sawtell NM. De novo synthesis of VP16 coordinates the exit from HSV latency in vivo. PLoS Pathog. 2009;5:e1000352. doi: 10.1371/journal.ppat.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du T, Zhou G, Khan S, Gu H, Roizman B. Disruption of HDAC/CoREST/REST repressor by dnREST reduces genome silencing and increases virulence of herpes simplex virus. Proc Natl Acad Sci USA. 2010;107:15904–15909. doi: 10.1073/pnas.1010741107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith RL, Escudero JM, Wilcox CL. Regulation of the herpes simplex virus latency-associated transcripts during establishment of latency in sensory neurons in vitro. Virology. 1994;202(1):49–60. doi: 10.1006/viro.1994.1321. [DOI] [PubMed] [Google Scholar]
- 28.Ejercito PM, Kieff ED, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 29.Lagunoff M, Randall G, Roizman B. Phenotypic properties of herpes simplex virus 1 containing a derepressed open reading frame P gene. J Virol. 1996;70:1810–1817. doi: 10.1128/jvi.70.3.1810-1817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]